A Radiomics Approach Based on Follow-Up CT for Pathological Subtypes Classification of Pulmonary Ground Glass Nodules
Abstract
:1. Introduction
2. Materials and Methods
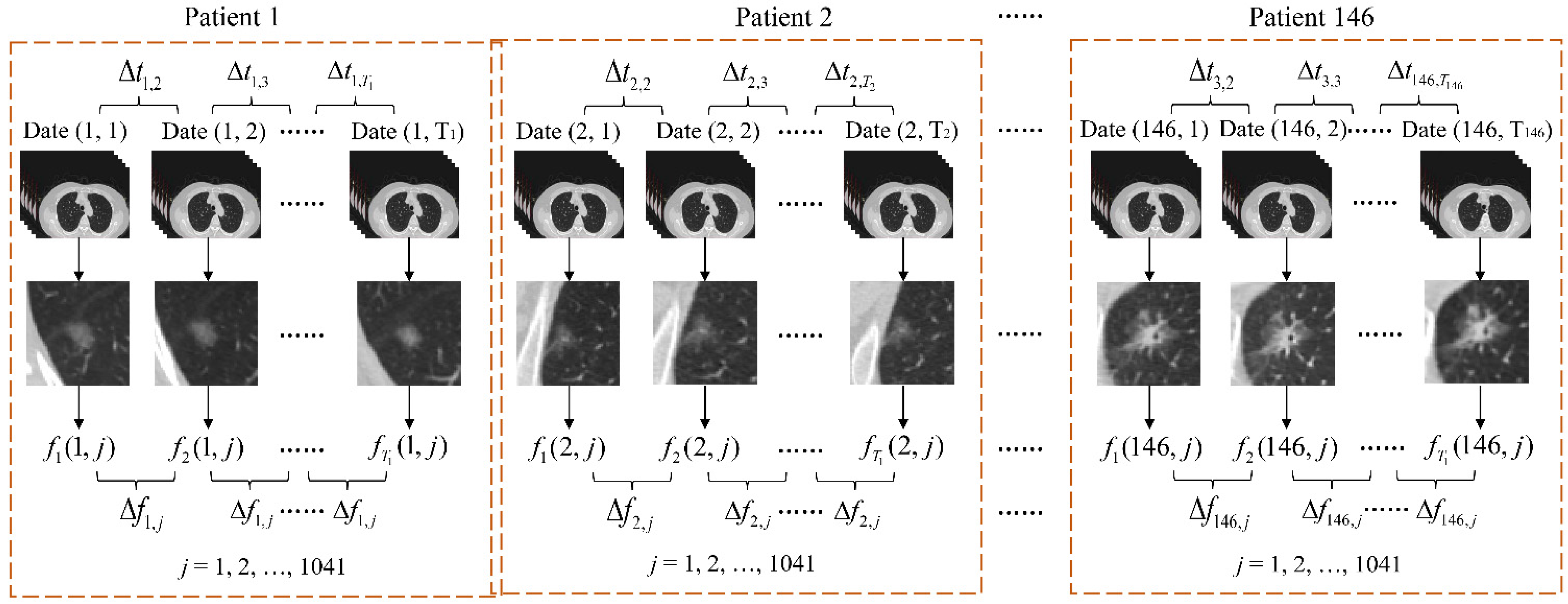
2.1. Sample Acquisition and Labeling
- (1)
- Import a set of CTIs for each patient into 3D Slicer and locate the GGNs.
- (2)
- Select CTIs that contain GGNs and then find the CTI with the largest area among these selected CTIs.
- (3)
- Segment the GGN with the largest area and save it as sequential classification.
- (4)
- Label the GGNs subtype with pathology reports.
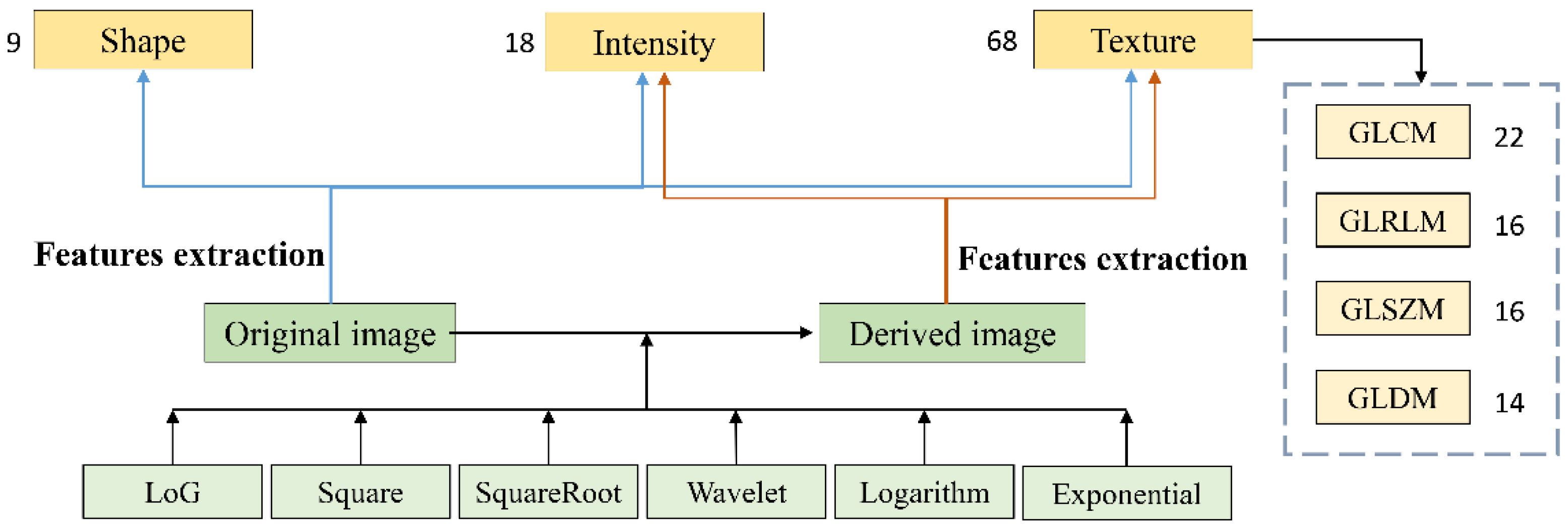
2.2. Radiomics Feature Extraction
2.3. Feature Selection and Data Augmentation
- (1)
- The number of samples is much less than that of the features, and some features are unnecessary.
- (2)
- The sample distribution is imbalanced; Table 1 shows that the number of samples in the majority class is 96, but there are only eight in the minority class.
- (1)
- For each sample a in the minority class, five nearest neighbors are found.
- (2)
- For each randomly selected nearest neighbor b, a new sample c is constructed with the original sample a according to the following equation:
- (1)
- The new sample set is thus obtained by the original and generated samples.
2.4. Performance Assessment
3. Results and Discussion
3.1. Classification Comparison
3.2. Different Subtypes Development Based on the Follow-Up Radiomics Features
- (1)
- ‘wavelet-L_glcm_MaximumProbability’ reflects the probability of the highest frequency of adjacent gray pairs in ROI. The smaller the probability, the more complex the texture pattern. The texture complexity of GGNs manifested as IA and benign became uncomplicated over time, and benign changed faster than IA. In contrast, the texture complexity of GGNs gradually increased in MIA and AIS stage.
- (2)
- ‘log-sigma-5-0-mm-3D_glszm_GrayLevelVariance’ reflects the discreteness of each pixel gray, relative to the average gray. The greater the value, the greater the image contrast. Among the four pathological results, only the contrast of GGNs in the MIA stage was gradually increased, and IA changed the fastest in other gradually decreasing stages.
- (3)
- ‘exponential_glszm_SmallAreaLowGrayLevelEmphasis’ measures the distribution of low gray values in small regions of ROI. The larger the value, the more emphasis is placed on the range of low gray values in small regions. In addition to the gradual increase in benign eigenvalues, the values of the other three pathological stages gradually decreased, and the IA stage changed the slowest.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2016. CAA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kodama, K.; Higashiyama, M.; Yokouchi, H.; Takami, K.; Kuriyama, K.; Kusunoki, Y.; Nakayama, T.; Imamura, F. Natural history of pure ground-glass opacity after long-term follow-up of more than 2 years. Ann. Thorac. Surg. 2002, 73, 386–392. [Google Scholar] [CrossRef]
- MacMahon, H.; Naidich, D.P.; Goo, J.M.; Lee, K.S.; Leung, A.N.C.; Mayo, J.R.; Mehta, A.C.; Ohno, Y.; Powell, C.A.; Prokop, M.; et al. Guidelines for management of incidental Pulmonary Nodules Detected on CT Images: From the Fleischner society. Radiology 2017, 284, 228–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-H.; Chang, C.-K.; Tu, C.-Y.; Liao, W.-C.; Wu, B.-R.; Chou, K.-T.; Chiou, Y.-R.; Yang, S.-N.; Zhang, G.; Huang, T.-C. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS ONE 2018, 13, e0192002. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Goo, J.M.; Lee, C.H.; Park, C.M.; Kim, K.G.; Park, E.-A. Predictive CT findings of malignancy in ground-glass nodules on thin-section chest CT: The effects on radiologist performance. Eur. Radiol. 2009, 19, 552–560. [Google Scholar] [CrossRef]
- Qing, C.W.; Shou, Z.R.; Peter, D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to the 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.Q.; Yue, S.H. Ground Glass Nodule Segmentation Based on Regional Adaptive MRF Model. In Proceedings of the 39th Chinese Control Conference, Xi’an, China, 27–29 July 2020. [Google Scholar]
- Dong, T.; Wei, L.; Ye, X.; Chen, Y.; Hou, X.; Nie, S. Segmentation of ground glass pulmonary nodules using full convolution residual network based on atrous spatial pyramid pooling structure and attention mechanism. J. Biomed. Eng. 2022, 39, 441–451. [Google Scholar]
- Statnikov, A.; Wang, L.; Aliferis, C.F. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinform. 2008, 9, 319. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using ad-vanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Schabath, M.; Balagurunathan, Y.; Dmitry, G.; Lawrence, H.; Samuel, H.; Stringfield, O.; Li, Q.; Liu, Y.; Gillies, R. Radiomics of lung cancer. J. Thorac. Oncol. 2016, 11, S5–S6. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, S.; Dong, D.; Wei, J.; Fang, C.; Zhou, X.; Sun, K.; Li, L.; Li, B.; Wang, M.; et al. The application of radiomics in precision diagnosis and treatment of oncology: Opportunities and challenges. Theranostics 2021, 9, 1303–1322. [Google Scholar] [CrossRef]
- Thawani, R.; McLane, M.; Beig, N.; Ghose, S.; Prasanna, P.; Velcheti, V.; Madabhushi, A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 155, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Fornacon-Wood, I.; Faivre-Finn, C.; O’Connor, J.P.; Price, G. Radiomics as a personalized medicine tool in lung cancer: Separating the hope from the hype. Lung Cancer 2020, 146, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.F.; Gong, J.; Zhou, W. Computer-aided diagnosis of ground glass pulmonary nodule by fusing deep learning and radiomics features. Phys. Med. Biol. 2021, 66, 065015. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, Q.C.; Cai, J.L. Predicting the invasiveness of lung adenocarcinomas appearing as ground-glass nodule on CT scan using multi-task learning and deep radiomics. Transl. Lung Cancer Res. 2020, 9, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Yang, Y.; Zheng, D.; Xie, Z.; Huang, H.; Wang, W. The invasiveness classification of ground-glass nodules using 3D attention network and HRCT. J. Digit. Imaging 2020, 33, 1144–1154. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, K.; Wang, H.; Lu, Y.; Wang, Z.; Xiao, R.; Lu, T. TMSF-Net: Multi-series fusion network with treeconnect for colorectal tumor segmentation. Comput. Methods Programs Biomed. 2022, 215, 106613. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, T.; Li, M.; Bueno, R.; Jayender, J. 3D deep learning based classification of pulmonary ground glass opacity nodules with automatic segmentation. Comput. Med. Imaging Graph. 2021, 88, 101814. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, Q.; Gao, Y.; He, K.; Wang, S.; Tang, X.; Tang, J.; Shen, D. Weakly Supervised Segmentation of COVID-19 Infection with Scribble Annotation on CT Images. Pattern Recognit. 2022, 122, 108341. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.Z.; Estepar, R.S.J.; Folch, E.; Onieva, J.; Gangadharan, S.; Majid, A. Three-dimensional printing and 3D slicer powerful tools in understanding and treating structural lung disease. Chest 2016, 149, 1136–1142. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Ye, W.; Li, Z.; Chen, C.; Cheng, S.; Lv, X.; Weng, W.; Li, J.; Weng, Q.; Pang, P.; et al. Non-invasive evaluation for benign and malignant subcentimeter pulmonary ground-glass nodules (≤1 cm) based on CT texture analysis. Br. J. Radiol. 2020, 93, 20190762. [Google Scholar] [CrossRef]
- Cao, P.; Liu, X.; Yang, J.; Zhao, D.; Li, W.; Huang, M.; Zaiane, O. A multi-kernel based framework for heterogeneous feature selection and over-sampling for computer-aided detection of pulmonary nodules. Pattern Recognit. 2017, 64, 327–346. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Ma, W.; Sun, Y.; Gao, P.; Huang, X.; Lu, J.; Chen, W.; Wu, Y.; Jin, L.; Tang, L.; et al. Prediction of the Growth Rate of Early-Stage Lung Adenocarcinoma by Radiomics. Front. Oncol. 2021, 11, 1141. [Google Scholar] [CrossRef]
- Monica, L.; Gaddis, P.D. Statistical Methodology: IV. Analysis of Variance, Analysis of Covariance, and Multivariate Analysis of Variance. Acad. Emerg. Med. 1998, 5, 258–265. [Google Scholar]
- Abu Bakar, Z.; Ispawi, D.I.; Ibrahim, N.F.; Tahir, N.M. Classification of Parkinson’s disease based on Multilayer Perceptrons (MLPs) Neural Network and ANOVA as a feature extraction. In Proceedings of the 2012 IEEE 8th International Colloquium on Signal Processing and Its Applications, Malacca, Malaysia, 23–25 March 2012. [Google Scholar]
- Gong, J.; Liu, J.-Y.; Hao, W.; Nie, S.-D.; Wang, S.; Peng, W. Computer-aided diagnosis of ground-glass opacity pulmonary nodules using radiomic features analysis. Phys. Med. Biol. 2019, 64, 135015. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-J.; Chen, Y.-S.; Lu, H.-Y. Imbalances data classification using random subspace method and SMOTE. In Proceedings of the 13th International Symposium on Advanced Intelligence Systems, Kobe, Japan, 20–24 November 2012. [Google Scholar]
- Sánchez-Crisostomo, J.P.; Alejo, R.; López-González, E.; Valdovinos, R.M.; Pacheco-Sánchez, J.H. Empirical analysis of assessments metrics for multi-class imbalance learning on the back-propagation contest. In Advances in Swarm Intelligence; Springer: Cham, Switzerland, 2014. [Google Scholar]
- García, S.; Zhang, Z.-L.; Altalhi, A.; Alshomrani, S.; Herrera, F. Dynamic ensemble selection for multi-class imbalanced datasets. Inf. Sci. 2018, 445, 22–37. [Google Scholar] [CrossRef]
- Zhang, Z.-L.; Luo, X.-G.; González, S.; García, S.; Herrera, F. DRCW-ASEG: One-versus-One distance-based relative competence weighting with adaptive synthetic example generation for multi-class imbalanced datasets. Neurocomputing 2018, 285, 176–187. [Google Scholar] [CrossRef]
- Pavan, M.R.; Jayagopal, P. A preprocessing method combined with an ensemble framework for the multiclass imbalanced data classification. Int. J. Comput. Appl. 2019, 64, 1–8. [Google Scholar]
- Swarna, K.; Vinayagam, A.; Ananth, M.B.J.; Kumar, P.V.; Veerasamy, V.; Radhakrishnan, P. A KNN based random subspace ensemble classifier for detection and discrimination of high impedance fault in PV integrated power network. Measurement 2022, 187, 110333. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, J.; Sun, Y.; Li, C.; Wu, W.; Jin, L.; Yang, Z.; Ni, B.; Gao, P.; Wang, P.; et al. 3D deep learning from CT scans predicts tumor invasiveness of subcentimeter pulmonary adenocarcinomas. Cancer Res. 2018, 78, 6881–6889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Wang, N.; Huang, N.; Liu, X.; Zheng, Y.; Fu, Y.; Li, X.; Wu, H.; Xu, J.; Cheng, J. Determining the invasiveness of ground-glass nodules using a 3D multi-task network. Eur. Radiol. 2021, 31, 7162–7171. [Google Scholar] [CrossRef] [PubMed]




| Pathological Subtypes | Number of Patients | Number of Follow-Ups | |
|---|---|---|---|
| Malignant | IA | 96 | 249 |
| MIA | 21 | 50 | |
| AIS | 8 | 37 | |
| Benign | AAH | 21 | 47 |
| Experiments | IA | MIA | AIS | Benign | ||
|---|---|---|---|---|---|---|
| FFDC | Before augmentation | Training set | 143 | 19 | 19 | 16 |
| Test set | 10 | 10 | 10 | 10 | ||
| After augmentation | Training set | 143 | 95 | 95 | 80 | |
| Test set | 10 | 10 | 10 | 10 | ||
| OFDC | Before augmentation | Training set | 100 | 14 | 14 | 18 |
| Test set | 10 | 10 | 10 | 10 | ||
| After augmentation | Training set | 100 | 70 | 70 | 90 | |
| Test set | 10 | 10 | 10 | 10 | ||
| Confusion Matrix | Predicted Class | ||
|---|---|---|---|
| Positive | Negative | ||
| True class | Positive | True positive (TP) | False negative (FN) |
| Negative | False positive (FP) | True negative (TN) | |
| Metrics | Equation |
|---|---|
| True positive rate (TPR) or recall | |
| True negative rate (TNR) | |
| False positive rate (FPR) | |
| False negative rate (FNR) | |
| precision |
| Year | Number of Classes | Method | Diagnostic Performance | |||
|---|---|---|---|---|---|---|
| Pre-Invasive | MIA | IA | Accuracy | AUC | ||
| 2018 [37] | 205 | 316 | 130 | OFDC + DenseSharp | 64.1% | — |
| 2021 [21] | 225 | 335 | 180 | OFDC + joint deep learning model | 58.67% | 0.81 |
| 2021 [38] | 302 | 349 | 258 | OFDC + 3D multi-task deep learning network | 64.9% | 0.82 |
| 2022 [ours] | 52 | 24 | 110 | OFDC + traditional classifier | 70% | 0.89 |
| 55 | 29 | 153 | FFDC + traditional classifier | 80% | 0.88 | |
| Features | Pathology | p Value | |||
|---|---|---|---|---|---|
| IA | MIA | AIS | Benign | ||
| wavelet-L_glcm_MaximumProbability | 4.15 × 10−5 | −1.23 × 10−4 | −5.32 × 10−6 | 2.38 × 10−4 | 0.00003 |
| log-sigma-5-0-mm-3D_glszm_GrayLevelVariance | −8.84 × 10−3 | 3.95 × 10−2 | −1.55 × 10−3 | −5.06 × 10−3 | 0.00008 |
| exponential_glszm_SmallAreaLowGrayLevelEmphasis | −6.26 × 10−5 | −2.03 × 10−4 | −1.09 × 10−4 | 3.21 × 10−4 | 0.00010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Yue, S.; Sun, C. A Radiomics Approach Based on Follow-Up CT for Pathological Subtypes Classification of Pulmonary Ground Glass Nodules. Appl. Sci. 2022, 12, 10587. https://doi.org/10.3390/app122010587
Ma C, Yue S, Sun C. A Radiomics Approach Based on Follow-Up CT for Pathological Subtypes Classification of Pulmonary Ground Glass Nodules. Applied Sciences. 2022; 12(20):10587. https://doi.org/10.3390/app122010587
Chicago/Turabian StyleMa, Chenchen, Shihong Yue, and Chang Sun. 2022. "A Radiomics Approach Based on Follow-Up CT for Pathological Subtypes Classification of Pulmonary Ground Glass Nodules" Applied Sciences 12, no. 20: 10587. https://doi.org/10.3390/app122010587
APA StyleMa, C., Yue, S., & Sun, C. (2022). A Radiomics Approach Based on Follow-Up CT for Pathological Subtypes Classification of Pulmonary Ground Glass Nodules. Applied Sciences, 12(20), 10587. https://doi.org/10.3390/app122010587





