Biomedical Text NER Tagging Tool with Web Interface for Generating BERT-Based Fine-Tuning Dataset
Abstract
1. Introduction
- Our web service is a biomedical text mining tool that uses deep learning based high-performance BioBERT NER and RE models;
- The NER result is displayed in a unique color, and the RE result is shown in a graph;
- We introduced a tagging tool system to our service so that users can tag the entities they desired;
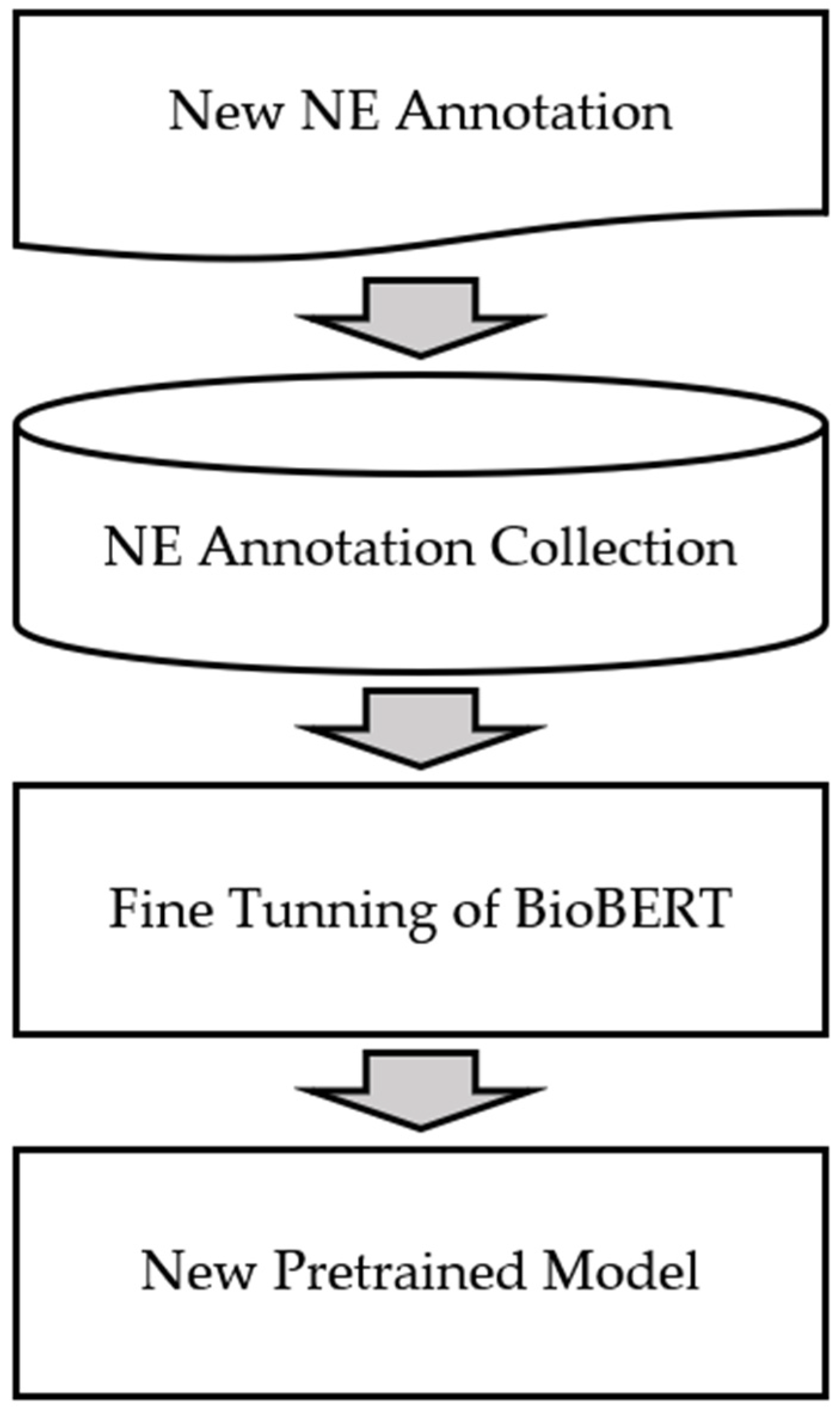
- Our web service can download newly tagged annotations and use it as a dataset for retraining. If this is used for retraining, better NER results can be used through the new pretrained model;
- This service is freely available at http://nertag.kw.ac.kr (accessed on 10 October 2022)
2. Related Work
2.1. Textpresso Central(TPC)
2.2. ezTag
2.3. TeamTat
3. Materials and Methods
4. Results
- Implementation Environments;
- Web Service Implementation;
- Model Retraining.
4.1. Implementation Environments
4.2. Web Service Implementation
4.3. Model Retraining
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ayush, S.; Simmons, M.; Lu, Z. Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PLoS Comput. Biol. 2016, 12, e1005017. [Google Scholar]
- Poux, S.; Arighi, C.N.; Magrane, M.; Bateman, A.; Wei, C.H.; Lu, Z.; Boutet, E.; Bye-A-Jee, H.; Famiglietti, M.L.; Roechert, B.; et al. On expert curation and scalability: UniProtKB/Swiss-Prot as a case study. Bioinformatics 2017, 33, 3454–3460. [Google Scholar] [CrossRef] [PubMed]
- Rak, R.; Batista-Navarro, R.T.; Rowley, A.; Carter, J.; Ananiadou, S. Text-mining-assisted biocuration workflows in Argo. Database 2014, 2014, bau070. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Kim, S.; Shin, S.Y.; Chatr-aryamontri, A.; Wilbur, W.J. Assisting manual literature curation for protein–protein interactions using BioQRator. Database 2014, 2014, bau067. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Lourenço, J.; Matos, S.; Oliveira, J.L. Egas: A collaborative and interactive document curation platform. Database 2014, 2014, bau048. [Google Scholar] [CrossRef]
- Pafilis, E.; Buttigieg, P.L.; Ferrell, B.; Pereira, E.; Schnetzer, J.; Arvanitidis, C.; Jensen, L.J. EXTRACT: Interactive extraction of environment metadata and term suggestion for metagenomic sample annotation. Database 2016, 2016, baw005. [Google Scholar] [CrossRef]
- Salgado, D.; Krallinger, M.; Depaule, M.; Drula, E.; Tendulkar, A.V.; Leitner, F.; Valencia, A.; Marcelle, C. MyMiner: A web application for computer-assisted biocuration and text annotation. Bioinformatics 2012, 28, 2285–2287. [Google Scholar] [CrossRef]
- Rinaldi, F.; Clematide, S.; Marques, H.; Ellendorff, T.; Romacker, M.; Rodriguez-Esteban, R. OntoGene web services for biomedical text mining. BMC Bioinform. 2014, 15, S6. [Google Scholar] [CrossRef]
- Wei, C.-H.; Kao, H.-Y.; Lu, Z. PubTator: A web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013, 41, W518–W522. [Google Scholar] [CrossRef]
- Cejuela, J.M.; McQuilton, P.; Ponting, L.; Marygold, S.J.; Stefancsik, R.; Millburn, G.H.; Rost, B.; FlyBase Consortium. tagtog: Interactive and text-mining-assisted annotation of gene mentions in PLOS full-text articles. Database 2014, 2014, bau033. [Google Scholar] [CrossRef]
- Rak, R.; Rowley, A.; Black, W.; Ananiadou, S. Argo: An integrative, interactive, text mining-based workbench supporting curation. Database 2012, 2012, bas010. [Google Scholar] [CrossRef] [PubMed]
- López-Fernández, H.; Reboiro-Jato, M.; Glez-Peña, D.; Aparicio, F.; Gachet, D.; Buenaga, M.; Fdez-Riverola, F. BioAnnote: A software platform for annotating biomedical documents with application in medical learning environments. Comput. Methods Programs Biomed. 2013, 111, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Bontcheva, K.; Cunningham, H.; Roberts, I.; Roberts, A.; Tablan, V.; Aswani, N.; Gorrell, G. GATE Teamware: A web-based, collaborative text annotation framework. Lang. Resour. Eval. 2013, 47, 1007–1029. [Google Scholar] [CrossRef]
- Pérez-Pérez, M.; Glez-Peña, D.; Fdez-Riverola, F.; Lourenço, A. Marky: A tool supporting annotation consistency in multi-user and iterative document annotation projects. Comput. Methods Programs Biomed. 2015, 118, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.; Glez-Peña, D.; Fdez-Riverola, F.; Lourenço, A. Marky: A lightweight web tracking tool for document annotation. In 8th International Conference on Practical Applications of Computational Biology & Bioinformatics (PACBB 2014); Springer: Cham, Switzerland, 2014. [Google Scholar]
- Hans-Michael, M.; Kenny, E.E.; Sternberg, P.W. Textpresso: An ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004, 2, e309. [Google Scholar]
- Müller, H.-M.; Van Auken, K.; Li, Y.; Sternberg, P.W. Textpresso Central: A customizable platform for searching, text mining, viewing, and curating biomedical literature. BMC Bioinform. 2018, 19, 94. [Google Scholar] [CrossRef]
- Frédérique, S. In Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics, Avignon, France, 23–27 April 2012.
- Ulf, L.; Hakenberg, J. What makes a gene name? Named entity recognition in the biomedical literature. Brief. Bioinform. 2005, 6, 357–369. [Google Scholar]
- David, C.; Matos, S.; Oliveira, J.L. Biomedical named entity recognition: A survey of machine-learning tools. Theory Appl. Adv. Text Min. 2012, 11, 175–195. [Google Scholar]
- Safaa, E.; Salim, N. Chemical named entities recognition: A review on approaches and applications. J. Cheminform. 2014, 6, 17. [Google Scholar]
- Islamaj, D.R.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar]
- Kwon, D.; Kim, S.; Wei, C.-H.; Leaman, R.; Lu, Z. ezTag: Tagging biomedical concepts via interactive learning. Nucleic Acids Res. 2018, 46, W523–W529. [Google Scholar] [CrossRef] [PubMed]
- Islamaj, R.; Kwon, D.; Kim, S.; Lu, Z. TeamTat: A collaborative text annotation tool. Nucleic Acids Res. 2020, 48, W5–W11. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Wei, C.-H.; Kao, H.-Y.; Lu, Z. GNormPlus: An integrative approach for tagging genes, gene families, and protein domains. BioMed Res. Int. 2015, 2015, 918710. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.-H.; Phan, L.; Feltz, J.; Maiti, R.; Hefferon, T.; Lu, Z. tmVar 2.0: Integrating genomic variant information from literature with dbSNP and ClinVar for precision medicine. Bioinformatics 2018, 34, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Robert, L.; Lu, Z. TaggerOne: Joint named entity recognition and normalization with semi-Markov Models. Bioinformatics 2016, 32, 2839–2846. [Google Scholar]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007, 36 (Suppl. 1), D13–D21. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 5998–6008. [Google Scholar]
- Smith, L.; Tanabe, L.K.; Ando, R.J.N.; Kuo, C.-J.; Chung, I.-F.; Hsu, C.-N.; Lin, Y.-S.; Klinger, R.; Friedrich, C.M.; Ganchev, K.; et al. Overview of BioCreative II gene mention recognition. Genome Biol. 2008, 9, S2. [Google Scholar] [CrossRef]
- Bravo, À.; Piñero, J.; Queralt-Rosinach, N.; Rautschka, M.; Furlong, L.I. Extraction of relations between genes and diseases from text and large-scale data analysis: Implications for translational research. BMC Bioinform. 2015, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Sachan, D.S.; Xie, P.; Sachan, M.; Xing, E.P. Effective use of bidirectional language modeling for transfer learning in biomedical named entity recognition. In Machine Learning for Healthcare Conference; PMLR: Westminster, UK, 2018. [Google Scholar]
- Yoon, W.; So, C.H.; Lee, J.; Kang, J. Collabonet: Collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinform. 2019, 20, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Habibi, M.; Weber, L.; Neves, M.; Wiegandt, D.L.; Leser, U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 2017, 33, i37–i48. [Google Scholar] [CrossRef] [PubMed]









| System Environments | |
|---|---|
| CPU | Inter® Core™ i9-10920X 12-core Processor |
| RAM | 96 GB |
| VGA | NVIDIA GeForce RTX 3090 |
| OS | Windows 10 |
| TOOL | Python 3.6.9 |
| Entity Types | Pretrained NER Models | Precision | Recall | F1-score |
|---|---|---|---|---|
| Disease | BioBERT [25] | 0.8904 | 0.8969 | 0.8936 |
| Sachan et al. [34] | 0.8641 | 0.8831 | 0.8734 | |
| CollaboNet [35] | 0.8548 | 0.8727 | 0.8636 | |
| LSTM-CRF (iii) of Habibi et al. [36] | 0.8531 | 0.8358 | 0.8444 | |
| Gene/Protein | BioBERT [25] | 0.8516 | 0.8365 | 0.8440 |
| Sachan et al. [34] | 0.8181 | 0.8157 | 0.8169 | |
| CollaboNet [35] | 0.8049 | 0.7899 | 0.7973 | |
| LSTM-CRF (iii) of Habibi et al. [36] | 0.7750 | 0.7813 | 0.7782 |
| Word | Tag |
|---|---|
| production | O |
| of | O |
| neurotoxic | B |
| amyloid | I |
| beta | I |
| (Aβ) | O |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, Y.-J.; Lee, M.-a.; Yang, G.-J.; Park, S.J.; Sohn, C.-B. Biomedical Text NER Tagging Tool with Web Interface for Generating BERT-Based Fine-Tuning Dataset. Appl. Sci. 2022, 12, 12012. https://doi.org/10.3390/app122312012
Park Y-J, Lee M-a, Yang G-J, Park SJ, Sohn C-B. Biomedical Text NER Tagging Tool with Web Interface for Generating BERT-Based Fine-Tuning Dataset. Applied Sciences. 2022; 12(23):12012. https://doi.org/10.3390/app122312012
Chicago/Turabian StylePark, Yeon-Ji, Min-a Lee, Geun-Je Yang, Soo Jun Park, and Chae-Bong Sohn. 2022. "Biomedical Text NER Tagging Tool with Web Interface for Generating BERT-Based Fine-Tuning Dataset" Applied Sciences 12, no. 23: 12012. https://doi.org/10.3390/app122312012
APA StylePark, Y.-J., Lee, M.-a., Yang, G.-J., Park, S. J., & Sohn, C.-B. (2022). Biomedical Text NER Tagging Tool with Web Interface for Generating BERT-Based Fine-Tuning Dataset. Applied Sciences, 12(23), 12012. https://doi.org/10.3390/app122312012








