Using Simulated Pest Models and Biological Clustering Validation to Improve Zoning Methods in Site-Specific Pest Management
Abstract
1. Introduction
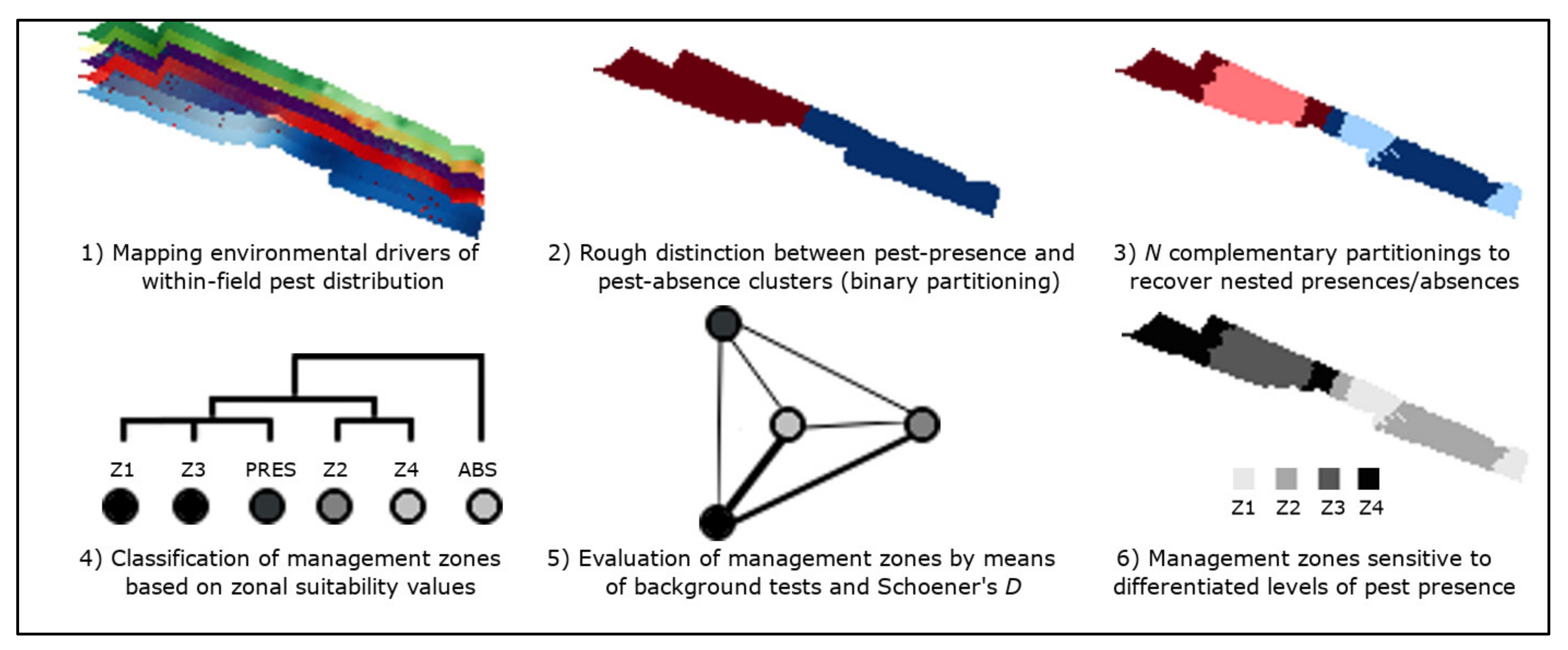
2. Materials and Methods
2.1. Summary
2.2. Study Site
2.3. Data Sets
2.4. Environmental Predictors of Virtual Pests
2.5. Within-Field Distribution of Virtual Pests
2.6. Nested Field Partitioning Essays
2.7. Validation of Field Partition Models
2.8. Classification of Management Zones
2.9. Validation of Management Zones
3. Results
3.1. Nested Field Partitioning Essays (Binary)
3.2. Nested Field Partitioning Essays (Complementary, Presence-Only)
3.3. Nested Field Partitioning Essays (Complementary, Absence-Only)
3.4. Classified Management Zones
3.5. Validated Management Zones
4. Discussion
4.1. Nested Field Partitioning Essays in eSSPM
4.2. Performance of MC Algorithms within the Context of eSSPM
4.3. Validation of Field Partition Models Using Biologically Meaningful CVI
4.4. SDM-Based Validation of Management Zones
4.5. New Workflow for MZ Delineation in eSSPM
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Abbreviation | Meaning | Class |
|---|---|---|
| AL | average linkage | clustering algorithm |
| CL | complete linkage | clustering algorithm |
| CLA | clustering large applications | clustering algorithm |
| DIA | divisive analysis | clustering algorithm |
| FNY | fuzzy analysis | clustering algorithm |
| MCL | model-based clustering | clustering algorithm |
| PAM | partitioning around medioids | clustering algorithm |
| SL | single linkage | clustering algorithm |
| SOM | self-organizing maps | clustering algorithm |
| WL | Ward’s linkage | clustering algorithm |
| SSPM | site-specific pest management | discipline |
| eSSPM | ecological site-specific pest management | discipline |
| IPM | integrated pest management | discipline |
| PA | precision agriculture | discipline |
| SDM | species distribution modeling | discipline |
| SSIPM | site-specific insect pest management | discipline |
| aFRot | active foot rot | pest driver |
| cropFVC | fractional vegetation cover of the research orchard | pest driver |
| cropHeight | height of trees included in the research orchard | pest driver |
| cropNDVI | normalized differences vegetation index of the research orchard | pest driver |
| DSM | digital surface model | pest driver |
| DTM | digital terrain model | pest driver |
| flowAccum | flow accumulation | pest driver |
| flowDir | flow direction | pest driver |
| FVC | fractional vegetation cover | pest driver |
| iFRot | inactive foot rot | pest driver |
| maxTemp | maximum ambient temperature | pest driver |
| minTemp | minimum ambient temperature | pest driver |
| NDVI | normalized differences vegetation index | pest driver |
| relHum | relative humidity | pest driver |
| SI-NDVI | single-image normalized differences vegetation index | pest driver |
| soilEC | soil electrical conductivity | pest driver |
| soilPH | soil potential of hydrogen | pest driver |
| sunRad | sun radiation | pest driver |
| TDS | total dissolved solids | pest driver |
| TRI | topographic roughness index | pest driver |
| VPD | vapor pressure deficit | pest driver |
| FPS | frames per second | precision agriculture tool |
| GIS | geographic information system | precision agriculture tool |
| GPS | global positioning system | precision agriculture tool |
| MZ | management zones | precision agriculture tool |
| UAS | unmanned aerial system | precision agriculture tool |
| ANOVA | analysis of variance | statistical method |
| BHI | biological homogeneity index | statistical method |
| BSI | biological stability index | statistical method |
| CVI | classification validation index | statistical method |
| D | Schoener’s D | statistical method |
| IDW | inverse distance weights | statistical method |
| MC | multivariate clustering (algorithm) | statistical method |
| MLM | mixed linear models | statistical method |
| PAST | presence–absence suitability threshold | statistical method |
| pD | probability of D | statistical method |
| S2T | total within-field suitability variance | statistical method |
- DIANA (Divisive analysis [52]) is an algorithm that initially starts with all observations in a single cluster, and successively divides the clusters until each one contains a single observation; thus, hierarchies are built in n − 1 steps. During each step, the cluster C with the largest diameter is selected based on the following equation:Assuming diam(C) > 0, we then split up C into two clusters A and B, according to a variant of the method of Macnaughton-Smith et al. [80]. At first A := C and B := θ, later one object is moved from A to B and then other objects are moved from A to B.
- 2.
- PAM (Partitioning around medioids [52]), similar to “k-means”, the number of clusters (i.e., k) is fixed in advance and an initial set of cluster centers (i.e., “medioids”, in contrast to “means” used in k-means) is required to start the algorithm. PAM is considered more robust than k-means because it admits the use of other dissimilarities besides Euclidean distance. The implementation of PAM clustering was based on the equation:where TD is the total deviation, defined as the sum of dissimilarities of each point Xj ∈ C1 to medioid mi of its cluster.
- 3.
- CLARA (Clustering large applications [52]), a sampling-based algorithm that implements PAM on a number of sub-datasets, which allows for faster running times when a number of observations is relatively large. CLARA complies with the following algorithm:
- Create randomly, from the original dataset, multiple subsets with fixed size (sampsize).
- Compute PAM algorithm on each subset and choose the corresponding k representative objects (medioids). Assign each observation of the entire data set to the closest medioid.
- Calculate the mean (or the sum) of the dissimilarities of the observations to their closest medioid. This is used as a measure of the goodness of the clustering.
- Retain the sub-dataset for which the mean (or sum) is minimal. A further analysis is carried out on the final partition.
- 4.
- FANNY (Fuzzy analysis [52]), this algorithm performs fuzzy clustering, where each observation can have partial membership in each cluster. Thus, each observation has a vector that gives the partial membership to each of the clusters. A hard cluster can be produced by assigning each observation to the cluster where it has the highest membership. FANNY clustering is based on the equation:where uiv is the membership of element i in relation to group v, n is the number of elements that form the data set, k is the number of groups to be formed, r corresponds to a pertinent exponent, and d(i, j) is the distance between elements i and j.
- 5.
- SOM (Self-organizing maps [54]), an unsupervised learning technique based on neural networks that is popular among computational biologists and machine learning researchers. SOM is a concept of competition network that tries to find the most similar distance between the input vector and neuron with weight vector . SOM always consist of both input vector x and output vector y. At the start of the learning, all the weights () are initialized to small random numbers. The set of weights forms a vector where is the row number and is the column number. Euclidian distance between the input vector and the neuron with weight vector of the given neuron is computed by:where t is an integer. Next, SOM will search for the winner neuron using the minimum distance (best matching unit, BMU). BMU is calculated as follows:To increase the similarity with the input vector, weights are adjusted after obtaining the winning neuron. The rule for updating the weight vector is given by:where wi(t + 1) is the updated weight vector, xj is the input record, hc(i)(t) is the neighborhood function related to the winning unit ci at step t, and S is the number of input samples. The neighborhood function (usually assumed as Gaussian) determines the rate of change of the neighborhood around the winner neuron as in equation:where rc(i) and ri are, respectively, the positions on the map of the winning neuron and of the generic unit i; σ(t) is the neighborhood radius at the iteration t of the training process and corresponds to the width of the neighborhood function at step t. Initially, σ(t) can be as large as the size of the map and then, to guarantee convergence and stability, it decreases linearly with time till one during the process.
- 6.
- MCL (Model-based clustering [53]) operates on the assumption that the analyzed data originate from a finite mixture of underlying probability distributions [83]. Each mixture component represents a cluster, and the mixture components and group memberships are estimated using maximum likelihood (EM algorithm). MCL usually assumes a normal or Gaussian mixture model as in the following equation:where G is the number of components, x represents the data, are the density and parameters of the kth component in the mixture, (mean vector) and (covariance matrix) are parameters to model each component k by the multivariate distribution, τk is the probability that an observation belongs to the kth component, and:
References
- Strickland, R.M.; Ess, D.R.; Parsons, S.D. Precision farming and precision pest management: The power of new crop production technologies. J. Nematol. 1998, 30, 431–435. [Google Scholar]
- Ye, X.; Sakai, K.; Manago, M.; Asada, S.; Sasao, A. Prediction of citrus yield from airborne hyperspectral imagery. Precis. Agric. 2007, 8, 111–125. [Google Scholar] [CrossRef]
- Park, Y.; Krell, R.K.; Carroll, M. Theory, technology, and practice of site-specific insect pest management. J. Asia Pac. Entomol. 2007, 10, 89–101. [Google Scholar] [CrossRef]
- Weisz, R.; Fleischer, S.; Smilowitz, S. Map generation in high-value horticultural integrated pest management: Appropriate interpolation methods for site-specific pest management of Colorado potato beetle (Coleoptera: Chrysomelidae). J. Econ. Entomol. 1995, 88, 1650–1657. [Google Scholar] [CrossRef]
- Weisz, R. Site-specific integrated pest management for high-value crops: Impact on potato pest management. J. Econ. Entomol. 1996, 89, 501–509. [Google Scholar] [CrossRef]
- Park, Y.; Tollefson, J.J. Spatial prediction of corn rootworm (Coleoptera: Chrysomelidae) adult emergence in Iowa cornfields. J. Econ. Entomol. 2005, 98, 8. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Perring, T.M.; Farrar, C.A.; Gispert, C. Spatial and temporal distributions of two sympatric Homalodisca spp. (Hemiptera: Cicadellidae): Implications for areawide pest management. Agric. Ecosyst. Environ. 2006, 113, 168–174. [Google Scholar] [CrossRef]
- Méndez-Vázquez, L.J.; Lira-Noriega, A.; Lasa-Covarrubias, R.; Cerdeira-Estrada, S. Delineation of site-specific management zones for pest control purposes: Exploring precision agriculture and species distribution modeling approaches. Comput. Electron. Agric. 2019, 167, 105101. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Gavioli, A.; Godoy de Souza, E.; Bazzi, C.L.; Schenatto, K.; Betzek, N.M. Identification of management zones in precision agriculture: An evaluation of alternative cluster analysis methods. Biosyst. Eng. 2019, 181, 86–102. [Google Scholar] [CrossRef]
- Córdoba, M.A.; Bruno, C.I.; Costa, J.L.; Peralta, N.R.; Balzarini, M.G. Protocol for multivariate homogeneous zone delineation in precision agriculture. Biosyst. Eng. 2016, 143, 95–107. [Google Scholar] [CrossRef]
- Hirzel, A.H.; Helfer, V.; Metral, F. Assessing habitat-suitability models with a virtual species. Ecol. Model. 2001, 145, 111–121. [Google Scholar] [CrossRef]
- Zurell, D.; Berger, U.; Cabral, J.S.; Jeltsch, F.; Meynard, C.N.; Münkemüller, T.; Nehrbass, N.; Pagel, J.; Reineking, B.; Schröder, B.; et al. The virtual ecologist approach: Simulating data and observers. Oikos 2010, 119, 622–635. [Google Scholar] [CrossRef]
- Miller, J.A. Virtual species distribution models: Using simulated data to evaluate aspects of model performance. Prog. Phys. Geogr. Earth Environ. 2014, 38, 117–128. [Google Scholar] [CrossRef]
- Hernández-Landa, L.; López-Collado, J.; Nava-Tablada, M.E.; García-García, C.G. Percepción de la problemática del Huanglongbing por agentes relevantes en zonas urbanas. Rev. Mex. Cien. Agrícolas 2017, 8, 993–1000. [Google Scholar] [CrossRef]
- Bautista-Zúñiga, F.; Solórzano, H.R.; Durán de Bazúa, C. Caracterización y clasificación de suelos con fines productivos en Córdoba, Veracruz, México. Investig. Geogr. 1998, 1, 21–33. [Google Scholar] [CrossRef]
- Moreno-Casasola, P.; Paradowska, K. Especies útiles de la selva baja caducifolia en las dunas costeras del centro de Veracruz. Madera Bosques 2009, 15, 21–44. [Google Scholar] [CrossRef][Green Version]
- Beisel, N.S.; Callaham, J.B.; Sng, N.J.; Taylor, D.J.; Paul, A.; Ferl, R.J. Utilization of single-image normalized difference vegetation index (SI-NDVI) for early plant stress detection. Appl. Plant Sci. 2018, 6, e01186. [Google Scholar] [CrossRef]
- Malone, M.; Foster, E. A mixed-methods approach to determine how conservation management programs and techniques have affected herbicide use and distribution in the environment over time. Sci. Total Environ. 2019, 660, 145–157. [Google Scholar] [CrossRef]
- Greenspan, S.E.; Morris, W.; Warburton, R.; Edwards, L.; Duffy, R.; Pike, D.A.; Schwarzkopf, L.; Alford, R.A. Low-cost fluctuating-temperature chamber for experimental ecology. Methods Ecol. Evol. 2016, 7, 1567–1574. [Google Scholar] [CrossRef]
- Virk, G.S.; Nagpal, A. Citrus diseases caused by Phytophthora species. GERF Bull. Biosci. 2012, 3, 18–27. [Google Scholar]
- Colauto-Stenzel, N.M.; Janeiro-Neves, C.S.V. Rootstocks for ’Tahiti’ lime. Sci. Agric. 2004, 61, 151–155. [Google Scholar] [CrossRef][Green Version]
- Ouédraogo, M.M.; Degré, A.; Debouche, C.; Lisein, J. The evaluation of unmanned aerial system-based photogrammetry and terrestrial laser scanning to generate DEMs of agricultural watersheds. Geomorphology 2014, 214, 339–355. [Google Scholar] [CrossRef]
- Lum, C.; Mackenzie, M.; Shaw-Feather, C.; Luker, E.; Dunbabin, M. Multi-spectral imaging and elevation mapping from an unmanned aerial system for precision agriculture applications. In Proceedings of the 13th International Conference on Precision Agriculture, St. Louis, MO, USA, 31 July–4 August 2016. [Google Scholar]
- Thorp, K.R.; Hunsaker, D.J.; French, A.N. Assimilating leaf area index estimates from remote sensing into the simulations of a cropping systems model. Trans. ASABE 2010, 53, 251–262. [Google Scholar] [CrossRef]
- Wiegand, C.L.; Richardson, A.J.; Kanemasu, E.T. Leaf area index estimates for wheat from LANDSAT and their implications for evapotranspiration and crop modeling. Agron. J. 1979, 71, 336. [Google Scholar] [CrossRef]
- Thorp, K.R.; Wang, G.; West, A.L.; Moran, M.S.; Bronson, K.F.; White, J.W.; Mon, J. Estimating crop biophysical properties from remote sensing data by inverting linked radiative transfer and ecophysiological models. Remote Sens. Environ. 2012, 124, 224–233. [Google Scholar] [CrossRef]
- Arge, L.; Toma, L.; Vitter, J.S. I/O-Efficient Algorithms for Problems on Grid-Based Terrains. Journal of Experimental Algorithmics. J. Exp. Algorithmics 2001, 6, 1. [Google Scholar] [CrossRef]
- Riley, S.J.; DeGloria, S.D.; Elliiot, R.A. Terrain Ruggedness Index That Quantifies Topographic Heterogeneity. Int. J. Soil Sci. 1999, 5, 23–27. [Google Scholar]
- Neteler, M.; Bowman, M.H.; Landa, M.; Metz, M. GRASS GIS: A multi-purpose open source GIS. Environ. Model. Softw. 2012, 31, 124–130. [Google Scholar] [CrossRef]
- Makori, D.; Fombong, A.; Abdel-Rahman, E.; Nkoba, K.; Ongus, J.; Irungu, J.; Mosomtai, G.; Makau, S.; Mutanga, O.; Odindi, J.; et al. Predicting spatial distribution of key honeybee pests in Kenya using remotely sensed and bioclimatic variables: Key honeybee pests distribution models. ISPRS Int. J. Geo Inf. 2017, 6, 66. [Google Scholar] [CrossRef]
- Ficklin, D.L.; Novick, K.A. Historic and projected changes in vapor pressure deficit suggest a continental-scale drying of the United States atmosphere: Increasing U.S. vapor pressure deficit. J. Geophys. Res. Atmos. 2017, 122, 2061–2079. [Google Scholar] [CrossRef]
- Allen, R.G.; Pereira, L.S.; Raes, D.; Smith, M. Crop evapotranspiration: Guidelines for computing crop water requirements. FAO Irrig. Drain. Pap. 1998, 56, 15. [Google Scholar]
- Hartkamp, A.D.; De Beurs, K.; Stein, A.; White, J.W. Interpolation Techniques for Climate Variables; CIMMYT: Mexico City, Mexico, 1999. [Google Scholar]
- Beckler, A.A.; French, B.W.; Chandler, L.D. Using GIS in areawide pest management: A case study in South Dakota. Trans. GIS 2005, 9, 109–127. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, L.; Qui, X.; Qui, J.; Wang, J.; Zhu, Y. An improved method of delineating rectangular management zones using a semivariogram-based technique. Comput. Electron. Agric. 2016, 121, 74–83. [Google Scholar] [CrossRef]
- Bazzi, C.L.; Schenatto, K.; Betzek, N.M.; Gavioli, A. A Software for the delineation of crop management zones (SDUM). Aust. J. Crop Sci. 2019, 13, 26–34. [Google Scholar] [CrossRef]
- Shipp, J.L.; Zhang, Y.; Hunt, D.W.A.; Ferguson, G. Influence of humidity and greenhouse microclimate on the efficacy of Beauveria bassiana (Balsamo) for control of greenhouse arthropod pests. Environ. Entomol. 2003, 32, 1154–1163. [Google Scholar] [CrossRef]
- Classen, A.T.; Hart, S.C.; Whitman, T.G.; Cobb, N.S.; Koch, G.W. Insect infestations linked to shifts in microclimate: Important climate change implications. Soil Sci. Soc. Am. J. 2006, 70, 305. [Google Scholar] [CrossRef]
- Gogo, E.O.; Saidi, M.; Ochieng, J.M.; Martin, T.; Baird, V.; Ngouajio, M. Microclimate modification and insect pest exclusion using agronet improve pod yield and quality of French bean. HortScience 2014, 49, 1298–1304. [Google Scholar] [CrossRef]
- Corwin, D.L.; Lesch, S.M. Characterizing soil spatial variability with apparent soil electrical conductivity. Comput. Electron. Agric. 2005, 46, 135–152. [Google Scholar] [CrossRef]
- Chaudhari, P.R.; Ahire, D.B.; Chkravarty, M.; Maity, S. Electrical conductivity as a tool for determining the physical properties of Indian soils. Int. J. Sci. Res. Publ. 2014, 4, 1–4. [Google Scholar]
- Chakraborty, K.; Mistri, B. Soil pH as a master variable of agricultural productivity in burdwan-I C.d. block, Barddhaman, West Bengal. Indian J. Spat. Sci. 2016, 11, 55–64. [Google Scholar]
- Philar, U.B. Holistic, Cost Effective Method for Management of Huang Long Bing (HLB), Phytophthora gummosis, Asian Citrus Psyllid and Other Serious Infestations in Citrus and Other Crops. U.S. Patent 10,264,792B2, 23 April 2019. [Google Scholar]
- Leroy, B.; Meynard, C.N.; Bellard, C.; Courchamp, F. Virtualspecies, an R package to generate virtual species distributions. Ecography 2015, 39, 599–607. [Google Scholar] [CrossRef]
- O’Bannon, J.H.; Radewald, J.D.; Tomerlin, A.T. Population fluctuation of three parasitic nematodes in Florida citrus. J. Nematol. 1972, 4, 6. [Google Scholar]
- Graham, J.H.; Gottwald, T.R.; Cubero, J.; Achor, D.S. Xanthomonas axonopodis pv. Citri: Factors affecting successful eradication of citrus canker. Mol. Plant Pathol. 2004, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hailnu, N.; Fininsa, C.; Mamo, G. Effects of temperature and moisture on growth of common bean and its resistance reaction against common bacterial blight (Xanthomonas axonopodis pv. phaseoli strains). J. Plant Pathol. Microbiol. 2017, 8, 419. [Google Scholar] [CrossRef]
- Marutani-Hert, M.; Hunter, W.B.; Morgan, J.K. Associated bacteria of Asian citrus psyllid (Hemiptera: Psyllidae: Diaphorina citri). Southwest. Entomol. 2011, 36, 323–330. [Google Scholar] [CrossRef]
- Brock, G.; Pihur, V.; Datta, S.; Datta, S. ClValid: An R package for cluster validation. J. Stat. Softw. 2008, 25, 1–22. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C. Algorithms for Clustering Data; Michigan State University; Prentice Hall: Englewood Cliffs, NJ, USA, 1988. [Google Scholar]
- Kaufman, L.; Rousseeuw, P.J. Finding Groups in Data; John Wiley & Sons Inc.: Hoboken, NJ, USA, 1990. [Google Scholar]
- Tipping, M.E. Deriving cluster analytic distance functions from Gaussian mixture models. In Proceedings of the 9th International Conference on Artificial Neural Networks: ICANN ’99, Edinburgh, UK, 7–10 September 1999; pp. 815–820. [Google Scholar] [CrossRef]
- Kohonen, T. Exploration of very large databases by self-organizing maps. Proceedings of International Conference on Neural Networks (ICNN’97), Houston, TX, USA, 12 June 1997; Volume 1, pp. PL1–PL6. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Datta, S.; Datta, S. Methods for evaluating clustering algorithms for gene expression data using a reference set of functional classes. BMC Bioinform. 2006, 7, 397. [Google Scholar] [CrossRef]
- Schoener, T.W.; Schoener, A. Distribution of vertebrates on some very small islands. I. Occurrence sequences of individual species. J. Anim. Ecol. 1983, 52, 209. [Google Scholar] [CrossRef]
- Warren, D.L.; Glor, R.E.; Turelli, M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution 2008, 62, 2868–2883. [Google Scholar] [CrossRef]
- Warren, D.L. ENMTools: A Toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Schapire, R.E. A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, 83, ICML’04, Banff, AB, Canada, 4–8 July 2004. [Google Scholar] [CrossRef]
- Fridgen, J.J.; Fraisse, C.W.; Kitchen, N.R.; Sudduth, K.A. Delineation and analysis of site-specific management zones. In Proceedings of the Second International Conference on Geospatial Information in Agriculture and Forestry, Lake Buena Vista, FL, USA, 10–12 January 2000; Volume 685, pp. 10–12. [Google Scholar]
- Hornung, A.; Khosla, R.; Reich, R.; Inman, D.; Westfall, D.G. Comparison of site-specific management zones. Agron. J. 2006, 98, 407. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Z.; Li, F.; Li, H. Delineation of site-specific management zones using fuzzy clustering analysis in a coastal saline land. Comput. Electron. Agric. 2007, 56, 174–186. [Google Scholar] [CrossRef]
- Gavioli, A.; Godoy de Souza, E.; Bazzi, C.L.; Carvalho-Guedes, L.P.; Schenatto, K. Optimization of management zone delineation by using spatial principal components. Comput. Electron. Agric. 2016, 127, 302–310. [Google Scholar] [CrossRef]
- Szeto, L.; Liew, A.W.; Yan, H.; Tang, S. Gene expression data clustering and visualization based on a binary hierarchical clustering framework. J. Vis. Lang. Comput. 2003, 14, 341–362. [Google Scholar] [CrossRef]
- Tamasauskas, D.; Sakalauskas, V.; Kriksciuniene, D. Evaluation framework of hierarchical clustering methods for binary data. In Proceedings of the 12 International Conference on Hybrid Intelligent Systems (HIS), Pune, India, 4–7 December 2012. [Google Scholar] [CrossRef]
- Löhr, S.C.; Grigorescu, M.; Hodgkinson, J.H.; Cox, M.E.; Fraser, S.J. Iron occurrence in soils and sediments of a coastal catchment. Geoderma 2010, 156, 253–266. [Google Scholar] [CrossRef]
- Sharma, A.; López, Y.; Tsunoda, T. Divisive hierarchical maximum likelihood clustering. BMC Bioinform. 2017, 18, 546. [Google Scholar] [CrossRef]
- Damian, J.M.; de Castro Pias, O.H.; Cherubin, M.R.; da Fonseca, A.Z.; Fornari, E.Z.; Santi, A.L. Applying the NDVI from satellite images in delimiting management zones for annual crops. Sci. Agric. 2020, 77, 1–11. [Google Scholar] [CrossRef]
- Arbelaitz, O.; Gurrutxaga, I.; Muguerza, J.; Pérez, J.M.; Perona, I. An extensive comparative study of cluster validity indices. Pattern Recognit. 2013, 46, 243–256. [Google Scholar] [CrossRef]
- Wilmer, P.G. Microclimate and the environmental physiology of insects. In Advances in Insect Physiology; Berridge, M.J., Treherne, J.E., Wiglessworth, V.B., Eds.; Elsevier: New York, NY, USA, 1982. [Google Scholar]
- Sgrò, C.M.; Terblanche, J.F.; Hoffmann, A.A. What can plasticity contribute to insect responses to climate change? Annu. Rev. Entomol. 2016, 61, 433–451. [Google Scholar] [CrossRef]
- Ladányi, M.; Horváth, L. A Review of the potential climate change impact on insect populations: General and agricultural aspects. Appl. Ecol. Environ. Res. 2010, 8, 143–152. [Google Scholar] [CrossRef]
- Kingsolver, J.G.; Woods, H.A.; Buckley, L.B.; Potter, K.A.; MacLean, H.J.; Higgins, J.K. Complex life cycles and the responses of insects to climate change. Integr. Comp. Biol. 2011, 51, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Andrew, N.; Hill, S.J. Effect of climate change on insect pest management. In Environmental Pest Management: Challenges for Agronomists, Ecologists, Economists and Policymakers; Coll, M., Wajnberg, E., Eds.; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Mitchell, M.S.; Powell, R.A. A mechanistic home range model for optimal use of spatially distributed resources. Ecol. Model. 2004, 177, 209–232. [Google Scholar] [CrossRef]
- Johnson, L.F.; Bosch, D.F.; Williams, D.C.; Lobitz, B.M. Remote sensing of vineyard management zones: Implications for wine quality. Appl. Eng. Agric. 2001, 17, 557–560. [Google Scholar] [CrossRef]
- Gardner, A.S.; Maclean, I.M.D.; Gaston, K.G.; Bütikofer, L. Forecasting future crop suitability with microclimate data. Agric. Syst. 2001, 190, 103084. [Google Scholar] [CrossRef]
- Soberón, J. Grinnellian and Eltonian niches and geographic distributions of species. Ecol. Lett. 2007, 10, 1115–1123. [Google Scholar] [CrossRef]
- Macnaughton-Smith, P.; Williams, W.T.; Dale, M.B.; Mockett, L.G. Dissimilarity analysis: A new technique of hierarchical sub-division. Nature 1964, 202, 1031–1035. [Google Scholar] [CrossRef]
- Chollet, F.; Allaire, J.J. Deep Learning with R; Manning Publications Co.: Shelter Island, NY, USA, 2017. [Google Scholar]
- Meila, M.; Heckerman, D. An experimental comparison of model-based clustering methods. Mach. Learn. 2001, 42, 9–29. [Google Scholar] [CrossRef]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. MCL 5: Clustering, classification and density estimation using Gaussian finite mixture models. R J. 2016, 8, 289. [Google Scholar] [CrossRef]















| Code | Variable | Estimation Method |
|---|---|---|
| aFRot | active citrus foot rot | IDW interpolation of presence-absence data |
| flowAccum | flow accumulation | “r.terraflow” function of GRASS GIS 7 |
| cropFVC | fractional vegetation cover | FVC = (1 + NDVI)/(1 − NDVI) × NDVI^0.5 |
| iFRot | inactive citrus foot rot | IDW interpolation of presence-absence data |
| relHum | mean relative humidity | IDW interpolation of data logs |
| sunRad | mean sub-canopy radiation | IDW interpolation of data logs |
| cropNDVI | single image NDVI | SI-NDVI = (NIR − BLUE)/(NIR + BLUE) |
| soilEC | soil electrical conductivity | IDW interpolation of soil samples |
| soiPH | soil pH | IDW interpolation of soil samples |
| TRI | topographic roughness index | “r.tri function” of GRASS GIS 7 |
| VPD | vapor-pressure deficit | VPD = esm − ea |
| Pest | Variable 1 | Fun. Var 1 | Range Var 1 | Variable 2 | Fun. Var 2 | Range Var 2 |
|---|---|---|---|---|---|---|
| 1 | VPD | normal | m = 0.55, sd = 0.25 | TRI | normal | m = 0.2, sd = 0.15 |
| 2 | flowDir | quadratic | a = 3, b = 1, c = 0.25 | sunRad | custom | m = 195, diff = 55, prob = 0.95 |
| 3 | relHum | quadratic | a = 3, b = 1, c = 0.25 | aFRot | logistic | beta = 0.3, alpha = 0.25 |
| 4 | soilPH | logistic | beta = 10, alpha = 1 | cropNDVI | normal | m = 0.05, sd = 0.1 |
| 5 | cropHeight | normal | m = 1.5, sd = 0.1 | ambTemp | quadratic | a = 3, b = 1, c = 0.25 |
| 6 | iFRot | logistic | beta = 0.75, alpha = 0.05 | soilEC | normal | m = 155, sd = 35 |
| Method | Acronym | Class | Reference | Package |
|---|---|---|---|---|
| Average linkage | AL | hierarchical | [51] | fastcluster |
| Clustering large applications | CLA | partitioning | [52] | cluster |
| Complete linkage | CL | hierarchical | [51] | fastcluster |
| Divisive analysis | DIA | hierarchical | [52] | cluster |
| Fuzzy analysis | FNY | partitioning | [52] | cluster |
| Model-based clustering | MCL | model-based | [53] | mclust |
| Partitioning around medioids | PAM | partitioning | [52] | cluster |
| Self-organizing maps | SOM | machine learning | [54] | kohonen |
| Single linkage | SL | hierarchical | [51] | fastcluster |
| Ward’s linkage | WL | hierarchical | [55] | fastcluster |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méndez-Vázquez, L.J.; Lasa-Covarrubias, R.; Cerdeira-Estrada, S.; Lira-Noriega, A. Using Simulated Pest Models and Biological Clustering Validation to Improve Zoning Methods in Site-Specific Pest Management. Appl. Sci. 2022, 12, 1900. https://doi.org/10.3390/app12041900
Méndez-Vázquez LJ, Lasa-Covarrubias R, Cerdeira-Estrada S, Lira-Noriega A. Using Simulated Pest Models and Biological Clustering Validation to Improve Zoning Methods in Site-Specific Pest Management. Applied Sciences. 2022; 12(4):1900. https://doi.org/10.3390/app12041900
Chicago/Turabian StyleMéndez-Vázquez, Luis Josué, Rodrigo Lasa-Covarrubias, Sergio Cerdeira-Estrada, and Andrés Lira-Noriega. 2022. "Using Simulated Pest Models and Biological Clustering Validation to Improve Zoning Methods in Site-Specific Pest Management" Applied Sciences 12, no. 4: 1900. https://doi.org/10.3390/app12041900
APA StyleMéndez-Vázquez, L. J., Lasa-Covarrubias, R., Cerdeira-Estrada, S., & Lira-Noriega, A. (2022). Using Simulated Pest Models and Biological Clustering Validation to Improve Zoning Methods in Site-Specific Pest Management. Applied Sciences, 12(4), 1900. https://doi.org/10.3390/app12041900







