Design and Experimental Evaluation of a New RNA-FISH Probe to Detect and Identify Paenibacillus sp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Design and In Silico Evaluation of a DNA-FISH Probe for Detection and Identification of Paenibacillus spp.
2.2. Experimental Assessment of the Efficiency and Specificity of the Designed Probe
2.2.1. Microorganisms and Growth Conditions
2.2.2. RNA-FISH Procedure
3. Results and Discussion
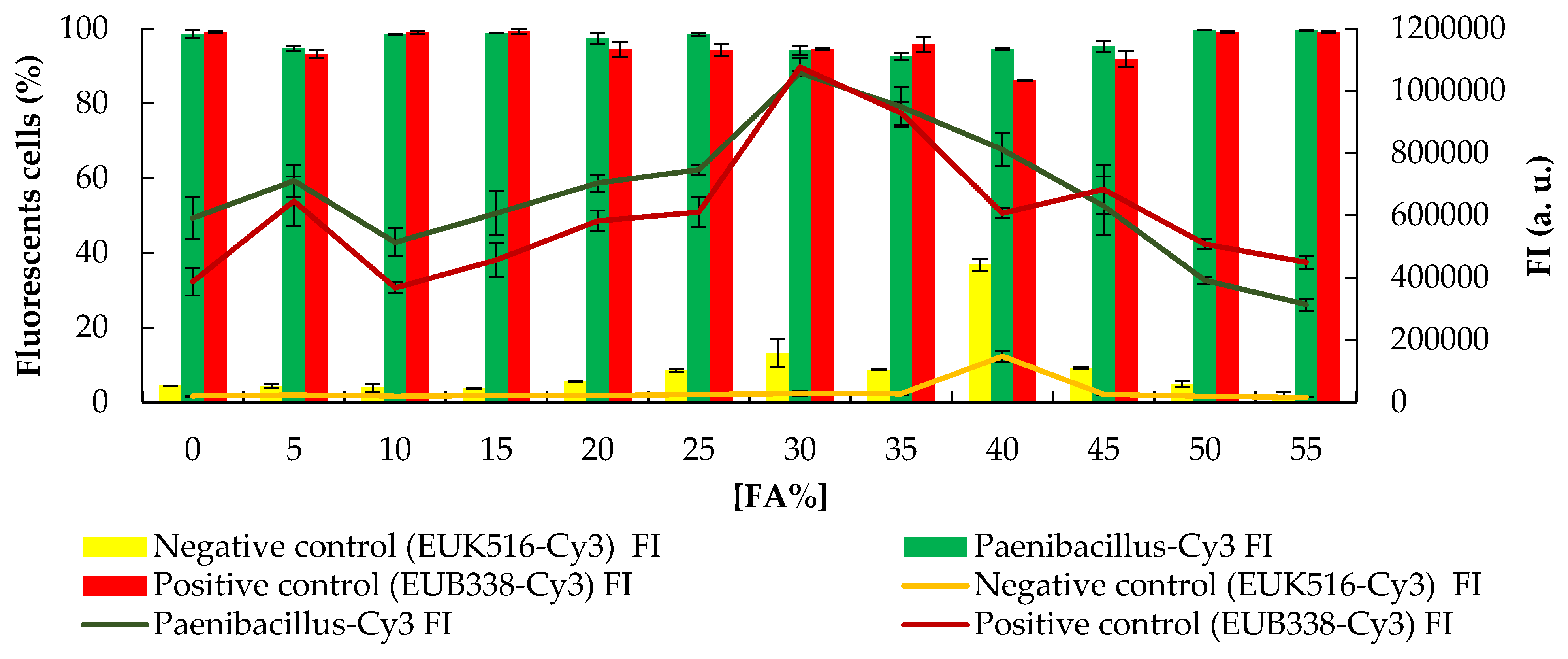
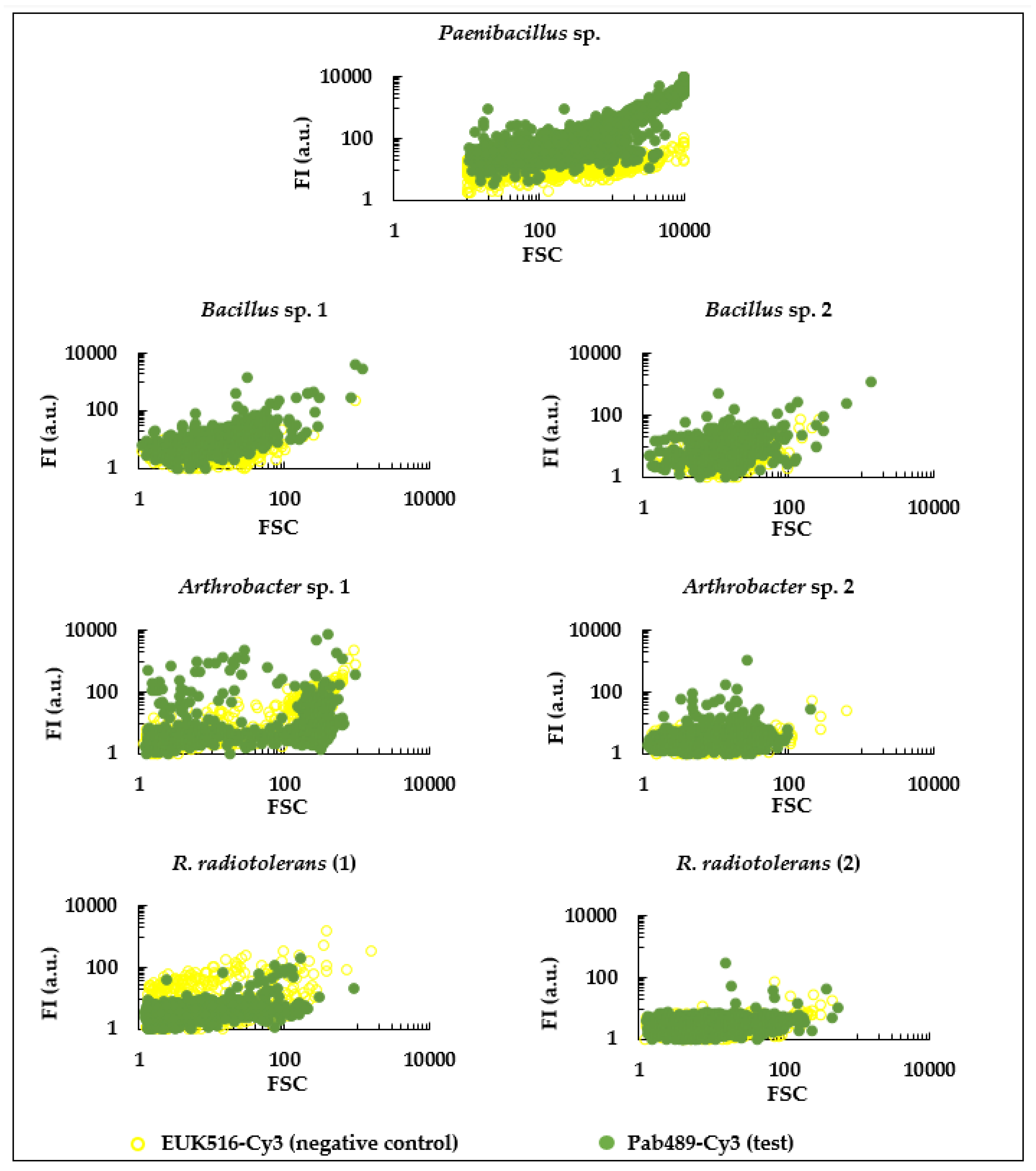
3.1. Design and In Silico Evaluation of a Paenibacillus sp. Specific Probe
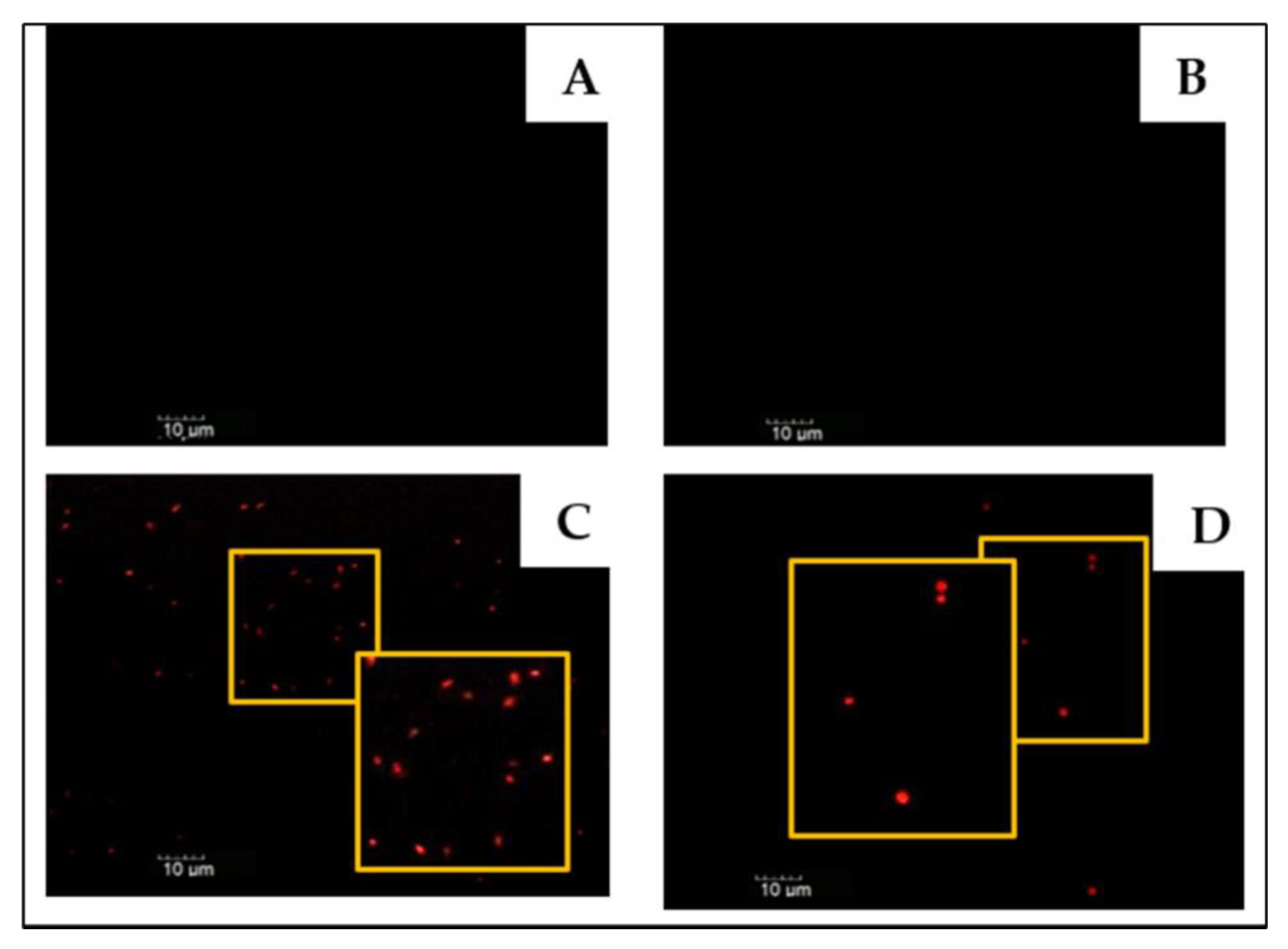
3.2. Experimental Evaluation of a Paenibacillus sp. Specific Probe
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ash, C.; Priest, F.G.; Collins, M.D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Leeuwenhoek 1993, 64, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Saez-Nieto, J.A.; Medina-Pascual, M.J.; Carrasco, G.; Garrido, N.; Fernandez-Torres, M.A.; Villalon, P.; Valdezate, S. Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes New Infect. 2017, 19, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Prasad, V.; Lata, C. Bacillus: Plant Growth Promoting Bacteria for Sustainable Agriculture and Environment. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 43–55. [Google Scholar]
- Rafigh, S.M.; Yazdi, A.V.; Vossoughi, M.; Safekordi, A.A.; Ardjmand, M. Optimization of culture medium and modeling of curdlan production from Paenibacillus polymyxa by RSM and ANN. Int. J. Biol. Macromol. 2014, 70, 463–473. [Google Scholar] [CrossRef]
- Son, J.S.; Kang, H.U.; Ghim, S.Y. Paenibacillus dongdonensis sp. nov., isolated from rhizospheric soil of Elymus tsukushiensis. Int. J. Syst. Evol. Microbiol. 2014, 64, 2865–2870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.K.; Kim, E.; Yi, H. Paenibacillus crassostreae sp. nov., isolated from the Pacific oyster Crassostrea gigas. Int. J. Syst. Evol. Microbiol. 2018, 68, 58–63. [Google Scholar] [CrossRef]
- Kong, B.H.; Liu, Q.F.; Liu, M.; Liu, Y.; Liu, L.; Li, C.L.; Yu, R.; Li, Y.H. Paenibacillus typhae sp. nov., isolated from roots of Typha angustifolia L. Int. J. Syst. Evol. Microbiol. 2013, 63, 1037–1044. [Google Scholar] [CrossRef] [Green Version]
- Smerda, J.; Sedlacek, I.; Pacova, Z.; Krejci, E.; Havel, L. Paenibacillus sepulcri sp. nov., isolated from biodeteriorated mural paintings in the Servilia tomb. Int. J. Syst. Evol. Microbiol. 2006, 56, 2341–2344. [Google Scholar] [CrossRef] [Green Version]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [Green Version]
- Roux, V.; Fenner, L.; Raoult, D. Paenibacillus provencensis sp. nov., isolated from human cerebrospinal fluid, and Paenibacillus urinalis sp. nov., isolated from human urine. Int. J. Syst. Evol. Microbiol. 2008, 58, 682–687. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, L.K. Paenibacillus apiarius sp. nov. Int. J. Syst. Bacteriol. 1996, 46, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Chung, Y.R.; Kim, C.H.; Hwang, I.; Chun, J. Paenibacillus koreensis sp. nov., a new species that produces an iturin-like antifungal compound. Int. J. Syst. Evol. Microbiol. 2000, 50, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Lee, K.C.; Chang, Y.H.; Hong, S.G.; Oh, H.W.; Pyun, Y.R.; Bae, K.S. Paenibacillus daejeonensis sp. nov., a novel alkaliphilic bacterium from soil. Int. J. Syst. Evol. Microbiol. 2002, 52, 2107–2111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, M.; Kamagata, Y.; Shinmaru, S.; Nishiyama, T.; Koizumi, J. Paenibacillus koleovorans sp. nov., able to grow on the sheath of Sphaerotilus natans. Int. J. Syst. Evol. Microbiol. 2002, 52, 1597–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, N.A.; De Clerck, E.; Lebbe, L.; Verhelst, A.; Goris, J.; Forsyth, G.; Rodriguez-Diaz, M.; Heyndrickx, M.; De Vos, P. Paenibacillus cineris sp. nov. and Paenibacillus cookii sp. nov., from Antarctic volcanic soils and a gelatin-processing plant. Int. J. Syst. Evol. Microbiol. 2004, 54, 1071–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Zhao, F.; Li, Y.H. Isolation of Paenibacillus tumbae sp. nov., from the tomb of the emperor Yang of the Sui dynasty, and emended description of the genus Paenibacillus. Antonie Leeuwenhoek 2017, 110, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Jurado, V.; Gonzalez-Pimentel, J.L.; Hermosin, B.; Saiz-Jimenez, C. Biodeterioration of Salón de Reinos, Museo Nacional del Prado, Madrid, Spain. Appl. Sci. 2021, 11, 8858. [Google Scholar] [CrossRef]
- Rojas, T.I.; Aira, M.J.; Batista, A.; Cruz, I.L.; González, S. Fungal biodeterioration in historic buildings of Havana (Cuba). Grana 2012, 51, 44–51. [Google Scholar] [CrossRef]
- Sterflinger, K. Fungi: Their role in deterioration of cultural heritage. Fungal Biol. Rev. 2010, 24, 47–55. [Google Scholar] [CrossRef]
- Rosado, T.; Gil, M.; Mirão, J.; Candeias, A.; Caldeira, A.T. Oxalate biofilm formation in mural paintings due to microorganisms –A comprehensive study. Int. Biodeterior. Biodegrad. 2013, 85, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Amann, R.; Fuchs, B.M.; Behrens, S. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 2001, 12, 231–236. [Google Scholar] [CrossRef]
- Bottari, B.; Ercolini, D.; Gatti, M.; Neviani, E. Application of FISH technology for microbiological analysis: Current state and prospects. Appl. Microbiol. Biotechnol. 2006, 73, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.V.B.; Junior, R.M.D.S.; Koshikene, D.; Silva, E.S. Applications of fluorescent in situ hybridization (FISH) in environmental microbiology. J. Food Agric. Environ. 2007, 5, 408–411. [Google Scholar]
- Franco-Duarte, R.; Cernakova, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stepien, K.; Leszczewicz, M.; et al. Advances in Chemical and Biological Methods to Identify Microorganisms-From Past to Present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kempf, V.A.; Mandle, T.; Schumacher, U.; Schafer, A.; Autenrieth, I.B. Rapid detection and identification of pathogens in blood cultures by fluorescence in situ hybridization and flow cytometry. Int. J. Med. Microbiol. 2005, 295, 47–55. [Google Scholar] [CrossRef]
- Branco, P.; Candeias, A.; Caldeira, A.T.; Gonzalez-Perez, M. A simple procedure for detecting Dekkera bruxellensis in wine environment by RNA-FISH using a novel probe. Int. J. Food Microbiol. 2020, 314, 108415. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Wright, E. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35, W43–W46. [Google Scholar] [CrossRef]
- Yilmaz, L.S.; Parnerkar, S.; Noguera, D.R. MathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl. Environ. Microbiol. 2011, 77, 1118–1122. [Google Scholar] [CrossRef] [Green Version]
- González-Pérez, M.; Brinco, C.; Vieira, R.; Rosado, T.; Mauran, G.; Pereira, A.; Candeias, A.; Caldeira, A.T. Dual phylogenetic staining protocol for simultaneous analysis of yeast and bacteria in artworks. Appl. Phys. A 2017, 123, 142. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, T.; Del Giorgio, P.A. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): A quantitative review of published reports. FEMS Microbiol. Ecol. 2003, 44, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Keller, G.H.; Cumming, C.U.; Huang, D.P.; Manak, M.M.; Ting, R. A chemical method for introducing haptens onto DNA probes. Anal. Biochem. 1988, 170, 441–450. [Google Scholar] [CrossRef]
- Owczarzy, R.; Vallone, P.M.; Gallo, F.J.; Paner, T.M.; Lane, M.J.; Benight, A.S. Predicting sequence-dependent melting stability of short duplex DNA oligomers. Biopolymers 1997, 44, 217–239. [Google Scholar] [CrossRef]
- Henriques, A.; Cereija, T.; Machado, A.; Cerca, N. In silico vs. in vitro analysis of primer specificity for the detection of Gardnerella vaginalis, Atopobium vaginae and Lactobacillus spp. BMC Res. Notes 2012, 5, 637. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, L.S.; Loy, A.; Wright, E.S.; Wagner, M.; Noguera, D.R. Modeling formamide denaturation of probe-target hybrids for improved microarray probe design in microbial diagnostics. PLoS ONE 2012, 7, e43862. [Google Scholar] [CrossRef]
- Icgen, B.; Yilmaz, F. Design a cadA-targeted DNA probe for screening of potential bacterial cadmium biosorbents. Environ. Sci. Pollut. Res. Int. 2016, 23, 5743–5752. [Google Scholar] [CrossRef]
- Daims, H.; Stoecker, K.; Wagner, M. Fluorescence In Situ Hybridization for the Detection of Prokaryotes. In Advanced Methods in Molecular Microbial Ecology; Osborn, A.M., Smith, C.J., Eds.; Bios-Garland: Abingdon, UK, 2005; pp. 213–239. [Google Scholar]
- Fuchs, B.M.; Glockner, F.O.; Wulf, J.; Amann, R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 2000, 66, 3603–3607. [Google Scholar] [CrossRef] [Green Version]
- Program, N.T. Toxicology and Carcinogenesis Studies of Formamide (Cas No. 75-12-7) in F344/N Rats and B6C3F1 Mice (Gavage Studies); 0888-8051; National Toxicology Program Technical Report Series; National Toxicology Program: Springfield, IL, USA, 2008; pp. 1–192.
- George, J.D.; Price, C.J.; Marr, M.C.; Myers, C.B.; Jahnke, G.D. Evaluation of the developmental toxicity of formamide in Sprague-Dawley (CD) rats. Toxicol. Sci. 2000, 57, 284–291. [Google Scholar] [CrossRef] [Green Version]
- IARC. Formaldehyde, 2-Butoxyethanol and 1-tert-butoxypropan-2-ol, 2007/03/21 ed.; IARC: Lyon, France, 2006; Volume 88, pp. 1–478.
- Wei, S.; Jiang, Z.; Liu, H.; Zhou, D.; Sanchez-Silva, M. Microbiologically induced deterioration of concrete—A review. Braz. J. Microbiol. 2013, 44, 1001–1007. [Google Scholar] [CrossRef]
- Urzi, C.; La Cono, V.; Stackebrandt, E. Design and application of two oligonucleotide probes for the identification of Geodermatophilaceae strains using fluorescence in situ hybridization (FISH). Environ. Microbiol. 2004, 6, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Moter, A.; Gobel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- Palkova, Z. Multicellular microorganisms: Laboratory versus nature. EMBO Rep. 2004, 5, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Lukumbuzya, M.; Schmid, M.; Pjevac, P.; Daims, H. A Multicolor Fluorescence in situ Hybridization Approach Using an Extended Set of Fluorophores to Visualize Microorganisms. Front. Microbiol. 2019, 10, 1383. [Google Scholar] [CrossRef] [PubMed]
- Shakoori, A.R. Fluorescence In Situ Hybridization (FISH) and Its Applications. In Chromosome Structure and Aberrations; Springer: Berlin/Heidelberg, Germany, 2017; pp. 343–367. [Google Scholar]
- Urzì, C.; La Cono, V.; De Leo, F.; Donato, P. Fluorescent In Situ Hybridization (FISH) to Study Biodeterioration in Cultural Heritage. In Molecular Biology and Cultural Heritage; Routledge: London, UK, 2017; pp. 55–60. [Google Scholar]
- González, M.; Vieira, R.; Nunes, P.; Rosado, T.; Martins, S.; Candeias, A.; Pereira, A.; Caldeira, A.T. Fluorescence In Situ Hybridization: A Potentially Useful Technique for Detection of Microorganisms on Mortars. e-conservation J. 2014, 2, 45–52. [Google Scholar] [CrossRef]
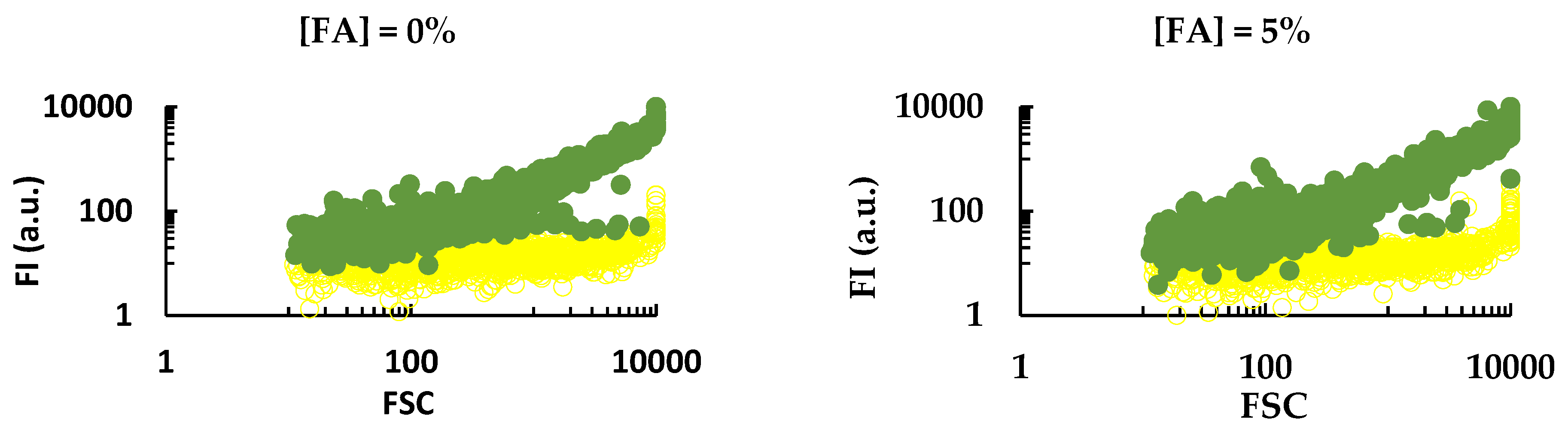

| Name | Sequence (5′–3′) | Score a | Efficiency b | GC (%) | Length | Hybridization Efficiency c (%) | Hairpin | Self-Dimer | Target Microorganisms d | Non-Target Microorganisms d |
|---|---|---|---|---|---|---|---|---|---|---|
| Pab489 | CCGGGGCTTTCTTCTCA | 0 | 0.717 | 58.80 | 17 | 99.99 | none | none | 444 | 0 |
| Pab482 | GCTTTCTTCTCAGGTACCG | 0 | 0.731 | 52.60 | 19 | 99.99 | none | none | 394 | 1 |
| Pab516 | TACCGCGGCTGCTGGC | 0 | 0.772 | 75.00 | 16 | 99.99 | none | none | 2 | 0 |
| Pab492 | AGCCGGGGCTTTCTTC | 0 | 0.642 | 62.50 | 16 | 99.99 | none | none | 168 | 0 |
| Pab533 | CCCTACGTATTACCGCGG | 0 | 70.81 | 61.00 | 18 | 99.86 | none | none | 17 | 10 |
| Pab435 | GCAACAGAGCTTTACGATCC | 0 | 0.584 | 50.00 | 20 | 98.71 | none | none | 426 | 0 |
| Pab433 | GCATTCTTCCCTGGCAAC | 0 | 0.713 | 55.60 | 18 | 98.06 | none | none | 238 | 1 |
| Pab525 | GCGGCTGCTGGCACGT | 0 | 0.792 | 80.00 | 15 | 88.12 | none | none | 13 | 3 |
| Pab564 | GCGCGCTTTACGCCCA | 0 | 0.779 | 68.80 | 16 | 56.26 | none | none | 2 | 1 |
| Pab428 | CCCTGGCAACAGAGCTTTA | 0 | 0.717 | 52.60 | 19 | 54.91 | none | none | 396 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arantes, S.; Branco, P.; Caldeira, A.T. Design and Experimental Evaluation of a New RNA-FISH Probe to Detect and Identify Paenibacillus sp. Appl. Sci. 2022, 12, 2348. https://doi.org/10.3390/app12052348
Arantes S, Branco P, Caldeira AT. Design and Experimental Evaluation of a New RNA-FISH Probe to Detect and Identify Paenibacillus sp. Applied Sciences. 2022; 12(5):2348. https://doi.org/10.3390/app12052348
Chicago/Turabian StyleArantes, Sílvia, Patrícia Branco, and Ana Teresa Caldeira. 2022. "Design and Experimental Evaluation of a New RNA-FISH Probe to Detect and Identify Paenibacillus sp." Applied Sciences 12, no. 5: 2348. https://doi.org/10.3390/app12052348
APA StyleArantes, S., Branco, P., & Caldeira, A. T. (2022). Design and Experimental Evaluation of a New RNA-FISH Probe to Detect and Identify Paenibacillus sp. Applied Sciences, 12(5), 2348. https://doi.org/10.3390/app12052348







