Cardiac Diffusion Tensor Biomarkers of Chronic Infarction Based on In Vivo Data
Abstract
:1. Introduction
2. Methods
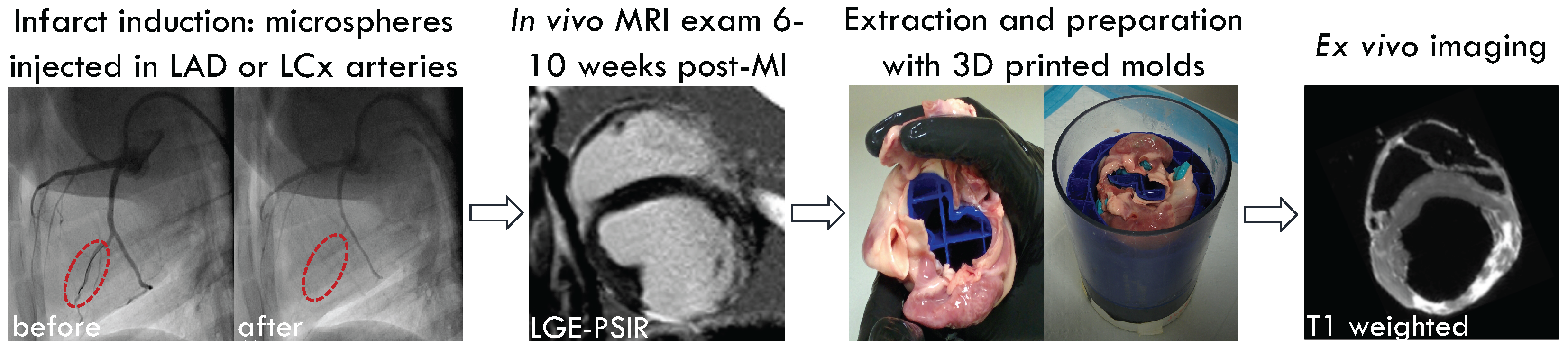
2.1. Animal Model and Infarct Induction
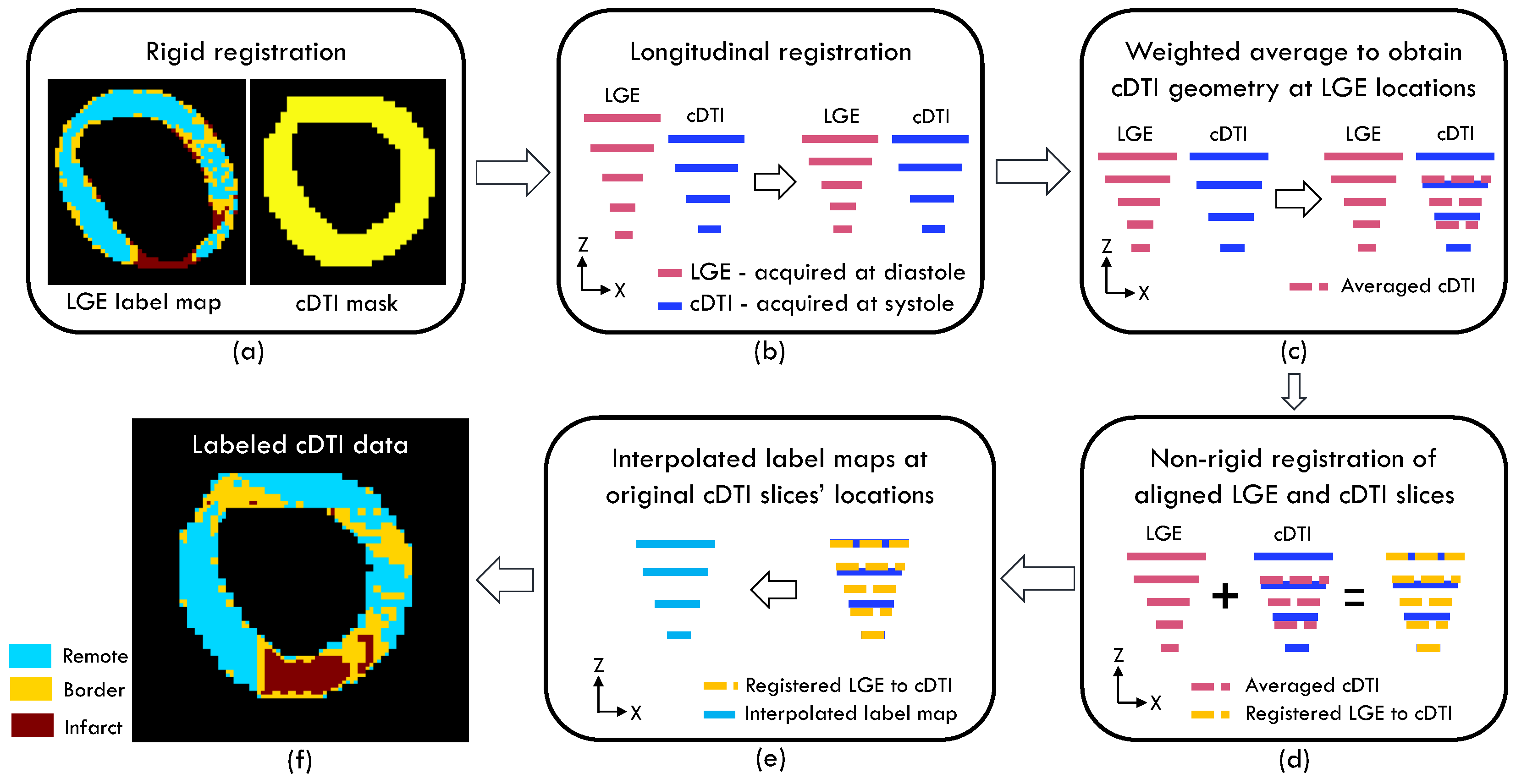
2.2. Regional Subdivision and Registration
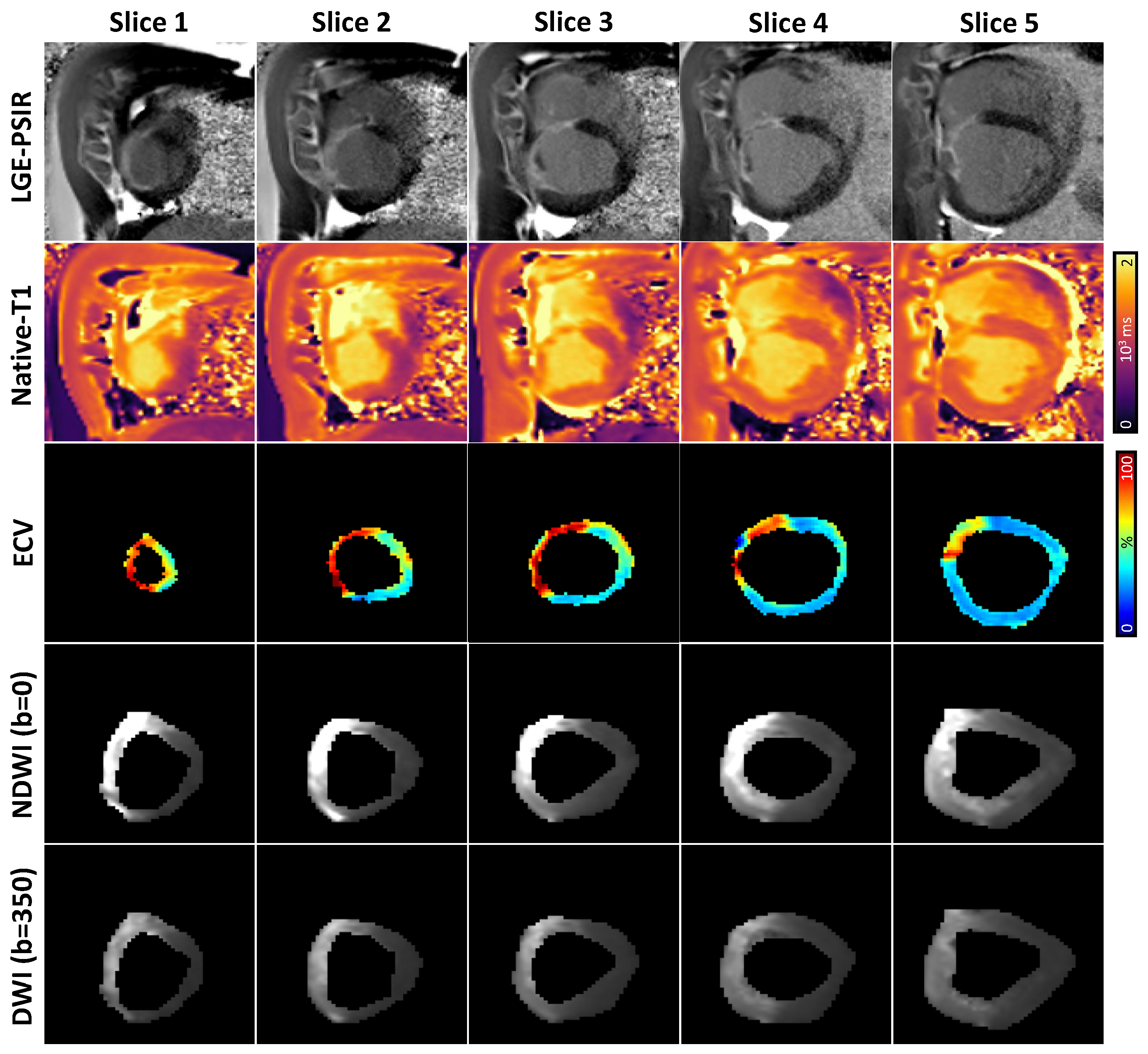
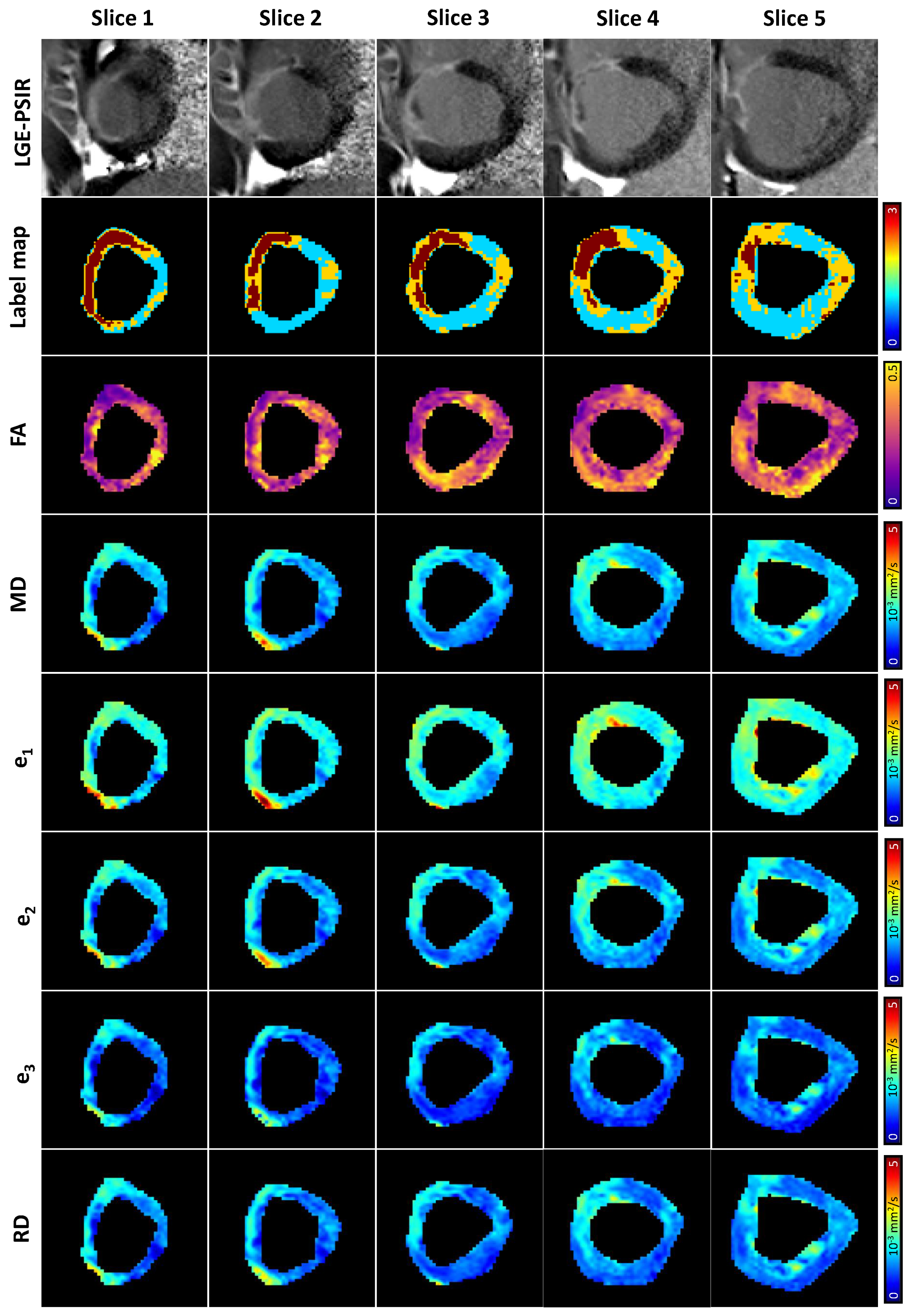
2.3. Data Analysis
2.4. Numerical Modeling
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Perotti, L.E.; Magrath, P.; Verzhbinsky, I.A.; Aliotta, E.; Moulin, K.; Ennis, D.B. Microstructurally anchored cardiac kinematics by combining in vivo DENSE MRI and cDTI. In Proceedings of the International Conference on Functional Imaging and Modeling of the Heart, Toronto, ON, Canada, 11–13 June 2017; pp. 381–391. [Google Scholar]
- Hooks, D.A.; Tomlinson, K.A.; Marsden, S.G.; LeGrice, I.J.; Smaill, B.H.; Pullan, A.J.; Hunter, P.J. Cardiac microstructure: Implications for electrical propagation and defibrillation in the heart. Circ. Res. 2002, 91, 331–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, P.F.; Kilner, P.J.; McGill, L.A.; Nielles-Vallespin, S.; Scott, A.D.; Ho, S.Y.; McCarthy, K.P.; Haba, M.M.; Ismail, T.F.; Gatehouse, P.D.; et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J. Cardiovasc. Magn. Reson. 2014, 16, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moulin, K.; Verzhbinsky, I.A.; Maforo, N.G.; Perotti, L.E.; Ennis, D.B. Probing cardiomyocyte mobility with multi-phase cardiac diffusion tensor MRI. PLoS ONE 2020, 15, e0241996. [Google Scholar] [CrossRef] [PubMed]
- Celle, T.D.; Cleutjens, J.P.; Blankesteijn, W.M.; Debets, J.J.; Smits, J.F.; Janssen, B.J. Long-term structural and functional consequences of cardiac ischaemia–reperfusion injury in vivo in mice. Exp. Physiol. 2004, 89, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Blankesteijn, W.; Creemers, E.; Lutgens, E.; Cleutjens, J.; Daemen, M.; Smits, J. Dynamics of cardiac wound healing following myocardial infarction: Observations in genetically altered mice. Acta Physiol. Scand. 2001, 173, 75–82. [Google Scholar] [CrossRef]
- Grieve, D.J.; Byrne, J.A.; Cave, A.C.; Shah, A.M. Role of oxidative stress in cardiac remodelling after myocardial infarction. Hear. Lung Circ. 2004, 13, 132–138. [Google Scholar] [CrossRef]
- Mehran, R.; Nikolsky, E. Contrast-induced nephropathy: Definition, epidemiology, and patients at risk. Kidney Int. 2006, 69, S11–S15. [Google Scholar] [CrossRef] [Green Version]
- Stoeck, C.T.; von Deuster, C.; Fuetterer, M.; Polacin, M.; Waschkies, C.F.; van Gorkum, R.J.; Kron, M.; Fleischmann, T.; Cesarovic, N.; Weisskopf, M.; et al. Cardiovascular magnetic resonance imaging of functional and microstructural changes of the heart in a longitudinal pig model of acute to chronic myocardial infarction. J. Cardiovasc. Magn. Reson. 2021, 23, 1–14. [Google Scholar] [CrossRef]
- Kung, G.L.; Vaseghi, M.; Gahm, J.K.; Shevtsov, J.; Garfinkel, A.; Shivkumar, K.; Ennis, D.B. Microstructural infarct border zone remodeling in the post-infarct swine heart measured by diffusion tensor MRI. Front. Physiol. 2018, 9, 826. [Google Scholar] [CrossRef]
- Wu, M.T.; Su, M.Y.M.; Huang, Y.L.; Chiou, K.R.; Yang, P.; Pan, H.B.; Reese, T.G.; Wedeen, V.J.; Tseng, W.Y.I. Sequential changes of myocardial microstructure in patients postmyocardial infarction by diffusion-tensor cardiac MR: Correlation with left ventricular structure and function. Circ. Cardiovasc. Imaging 2009, 2, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Scollan, D.F.; Holmes, A.; Winslow, R.; Forder, J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. Am. J. Physiol.-Heart Circ. Physiol. 1998, 275, H2308–H2318. [Google Scholar] [CrossRef]
- Scollan, D.; Holmes, A.; Zhang, J.; Winslow, R. Reconstruction of cardiac ventricular geometry and fiber orientation using magnetic resonance imaging. Ann. Biomed. Eng. 2000, 28, 934–944. [Google Scholar] [CrossRef]
- Rahman, T.; Moulin, K.; Ennis, D.B.; Perotti, L.E. Diffusion Biomarkers in Chronic Myocardial Infarction. In Proceedings of the International Conference on Functional Imaging and Modeling of the Heartr, Stanford, CA, USA, 21–25 June 2021; pp. 137–147. [Google Scholar]
- Verzhbinsky, I.A.; Perotti, L.E.; Moulin, K.; Cork, T.E.; Loecher, M.; Ennis, D.B. Estimating aggregate cardiomyocyte strain using Vivo Diffus. Displac. Encoded MRI. IEEE Trans. Med. Imaging 2019, 39, 656–667. [Google Scholar] [CrossRef]
- Perotti, L.E.; Ponnaluri, A.V.; Krishnamoorthi, S.; Balzani, D.; Ennis, D.B.; Klug, W.S. Method for the unique identification of hyperelastic material properties using full-field measures. Application to the passive myocardium material response. Int. J. Numer. Methods Biomed. Eng. 2017, 33, e2866. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Perotti, L.E.; Martinez, J.A.; Duarte-Vogel, S.M.; Ennis, D.B.; Wu, H.H. Real-time 3T MRI-guided cardiovascular catheterization in a porcine model using a glass-fiber epoxy-based guidewire. PLoS ONE 2020, 15, e0229711. [Google Scholar] [CrossRef] [Green Version]
- Stoeck, C.T.; Von Deuster, C.; Genet, M.; Atkinson, D.; Kozerke, S. Second-order motion-compensated spin echo diffusion tensor imaging of the human heart. Magn. Reson. Med. 2016, 75, 1669–1676. [Google Scholar] [CrossRef] [Green Version]
- Cork, T.E.; Perotti, L.E.; Verzhbinsky, I.A.; Loecher, M.; Ennis, D.B. High-Resolution Ex Vivo Microstructural MRI After Restoring Ventricular Geometry via 3D Printing. In Proceedings of the International Conference on Functional Imaging and Modeling of the Heart, Bordeaux, France, 6–8 June 2019; pp. 177–186. [Google Scholar]
- Porter, D.A.; Heidemann, R.M. High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2009, 62, 468–475. [Google Scholar] [CrossRef]
- Reese, T.G.; Heid, O.; Weisskoff, R.; Wedeen, V. Reduction of eddy-current-induced distortion in diffusion MRI using a twice-refocused spin echo. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2003, 49, 177–182. [Google Scholar] [CrossRef]
- Schelbert, E.B.; Hsu, L.Y.; Anderson, S.A.; Mohanty, B.D.; Karim, S.M.; Kellman, P.; Aletras, A.H.; Arai, A.E. Late gadolinium-enhancement cardiac magnetic resonance identifies postinfarction myocardial fibrosis and the border zone at the near cellular level in ex vivo rat heart. Circ. Cardiovasc. Imaging 2010, 3, 743–752. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Morgan, P.S.; Ashburner, J.; Smith, J.; Rorden, C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods 2016, 264, 47–56. [Google Scholar] [CrossRef]
- Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, PAMI-8, 679–698. [Google Scholar] [CrossRef]
- Thirion, J.P. Image matching as a diffusion process: An analogy with Maxwell’s demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef] [Green Version]
- Moulin, K. DiffusionRecon. Available online: https://github.com/KMoulin/DiffusionRecon (accessed on 24 January 2022).
- Haaf, P.; Garg, P.; Messroghli, D.R.; Broadbent, D.A.; Greenwood, J.P.; Plein, S. Cardiac T1 mapping and extracellular volume (ECV) in clinical practice: A comprehensive review. J. Cardiovasc. Magn. Reson. 2017, 18, 89. [Google Scholar] [CrossRef] [Green Version]
- Allen, M.; Poggiali, D.; Whitaker, K.; Marshall, T.R.; Kievit, R.A. Raincloud plots: A multi-platform tool for robust data visualization. Wellcome Open Res. 2019, 4, 63. [Google Scholar] [CrossRef] [Green Version]
- Balls, G.T.; Frank, L.R. A simulation environment for diffusion weighted MR experiments in complex media. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2009, 62, 771–778. [Google Scholar] [CrossRef] [Green Version]
- Berry, D.B.; Regner, B.; Galinsky, V.; Ward, S.R.; Frank, L.R. Relationships between tissue microstructure and the diffusion tensor in simulated skeletal muscle. Magn. Reson. Med. 2018, 80, 317–329. [Google Scholar] [CrossRef]
- Moulin, K.; Aliotta, E.; Ennis, D.B. Effect of flow-encoding strength on intravoxel incoherent motion in the liver. Magn. Reson. Med. 2019, 81, 1521–1533. [Google Scholar] [CrossRef]
- Moulin, K. DiffusionSimulation. Available online: https://github.com/KMoulin/DiffusionSimulation (accessed on 14 February 2022).
- Messroghli, D.R.; Nordmeyer, S.; Dietrich, T.; Dirsch, O.; Kaschina, E.; Savvatis, K.; O h Ici, D.; Klein, C.; Berger, F.; Kuehne, T. Assessment of diffuse myocardial fibrosis in rats using small-animal Look-Locker inversion recovery T1 mapping. Circ. Cardiovasc. Imaging 2011, 4, 636–640. [Google Scholar] [CrossRef] [Green Version]
- Messroghli, D.R.; Walters, K.; Plein, S.; Sparrow, P.; Friedrich, M.G.; Ridgway, J.P.; Sivananthan, M.U. Myocardial T1 mapping: Application to patients with acute and chronic myocardial infarction. Magn. Reson. Med. 2007, 58, 34–40. [Google Scholar] [CrossRef]
- Kali, A.; Choi, E.Y.; Sharif, B.; Kim, Y.J.; Bi, X.; Spottiswoode, B.; Cokic, I.; Yang, H.J.; Tighiouart, M.; Conte, A.H.; et al. Native T1 mapping by 3-T CMR imaging for characterization of chronic myocardial infarctions. JACC Cardiovasc. Imaging 2015, 8, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Arcari, L.; Engel, J.; Freiwald, T.; Zhou, H.; Zainal, H.; Gawor, M.; Buettner, S.; Geiger, H.; Hauser, I.; Nagel, E.; et al. Cardiac biomarkers in chronic kidney disease are independently associated with myocardial edema and diffuse fibrosis by cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2021, 23, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Camastra, G.; Arcari, L.; Ciolina, F.; Danti, M.; Cacciotti, L. Cardiac magnetic resonance imaging of transient myocardial dysfunction in a patient treated with checkpoint-targeted immunotherapy. Eur. J. Cancer 2021, 144, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Kolentinis, M.; Le, M.; Nagel, E.; Puntmann, V.O. Contemporary cardiac MRI in chronic coronary artery disease. Eur. Cardiol. Rev. 2020, 15, e50. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, S.K.; Liu, W.; McLean, M.; Allen, J.S.; Tan, J.; Wickline, S.A.; Yu, X. Remodeling of cardiac fiber structure after infarction in rats quantified with diffusion tensor MRI. Am. J. Physiol.-Heart Circ. Physiol. 2003, 285, H946–H954. [Google Scholar] [CrossRef] [Green Version]
- Pop, M.; Ghugre, N.R.; Ramanan, V.; Morikawa, L.; Stanisz, G.; Dick, A.J.; Wright, G.A. Quantification of fibrosis in infarcted swine hearts by ex vivo late gadolinium-enhancement and diffusion-weighted MRI methods. Phys. Med. Biol. 2013, 58, 5009. [Google Scholar] [CrossRef]
- Pashakhanloo, F.; Herzka, D.A.; Mori, S.; Zviman, M.; Halperin, H.; Gai, N.; Bluemke, D.A.; Trayanova, N.A.; McVeigh, E.R. Submillimeter diffusion tensor imaging and late gadolinium enhancement cardiovascular magnetic resonance of chronic myocardial infarction. J. Cardiovasc. Magn. Reson. 2017, 19, 9. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.N.; Nielles-Vallespin, S.; Ferreira, P.F.; Firmin, D.N.; Scott, A.D.; Doorly, D.J. Novel insights into in-vivo diffusion tensor cardiovascular magnetic resonance using computational modelling and a histology-based virtual microstructure. Magn. Reson. Med. 2019, 81, 2759–2773. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Kelly, C.; Teh, I.; Stoeck, C.T.; Kozerke, S.; Chowdhary, A.; Brown, L.A.; Saunderson, C.E.; Craven, T.P.; Chew, P.G.; et al. Acute Microstructural Changes after ST-Segment Elevation Myocardial Infarction Assessed with Diffusion Tensor Imaging. Radiology 2021, 299, 86–96. [Google Scholar] [CrossRef]
- Gottbrecht, M.; Kramer, C.M.; Salerno, M. Native T1 and extracellular volume measurements by cardiac MRI in healthy adults: A meta-analysis. Radiology 2019, 290, 317–326. [Google Scholar] [CrossRef]
- Risholm, P.; Pieper, S.; Samset, E.; Wells, W.M. Summarizing and visualizing uncertainty in non-rigid registration. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Beijing, China, 20–24 September 2010; pp. 554–561. [Google Scholar]

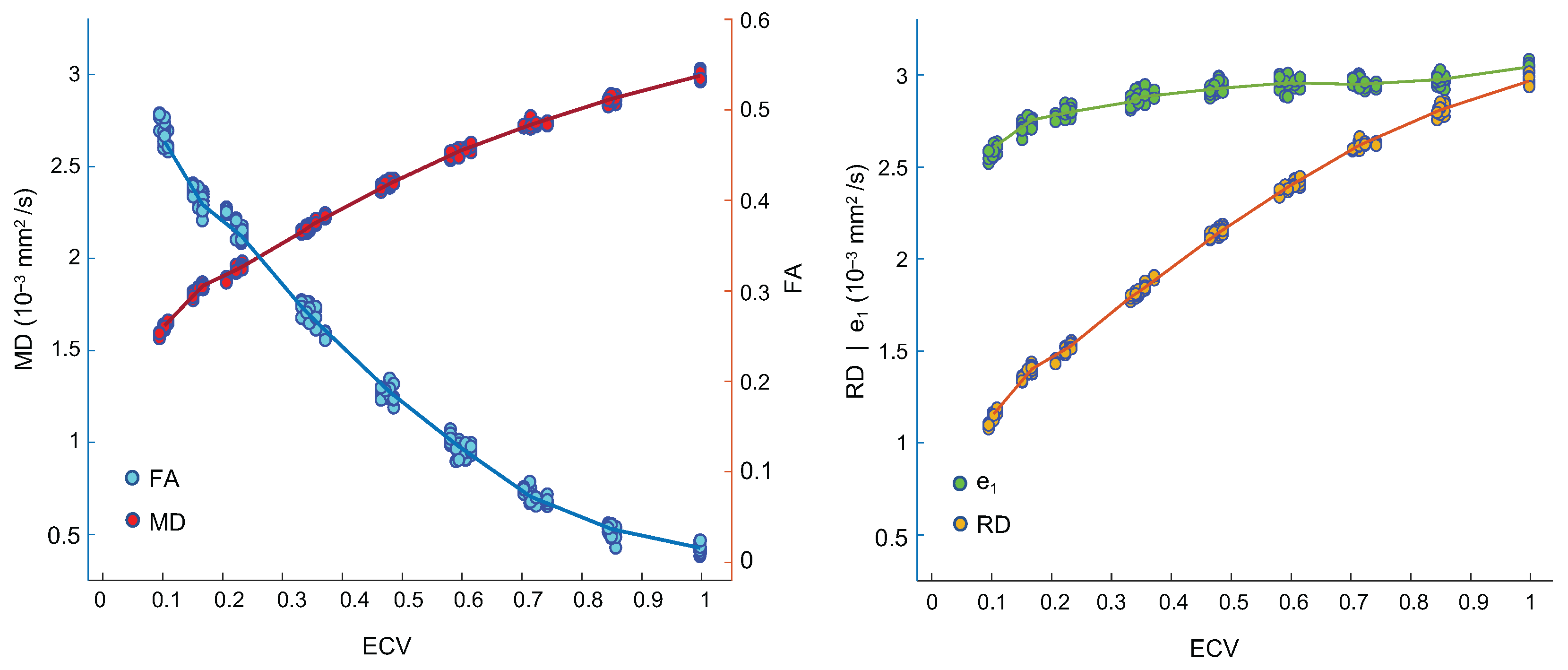
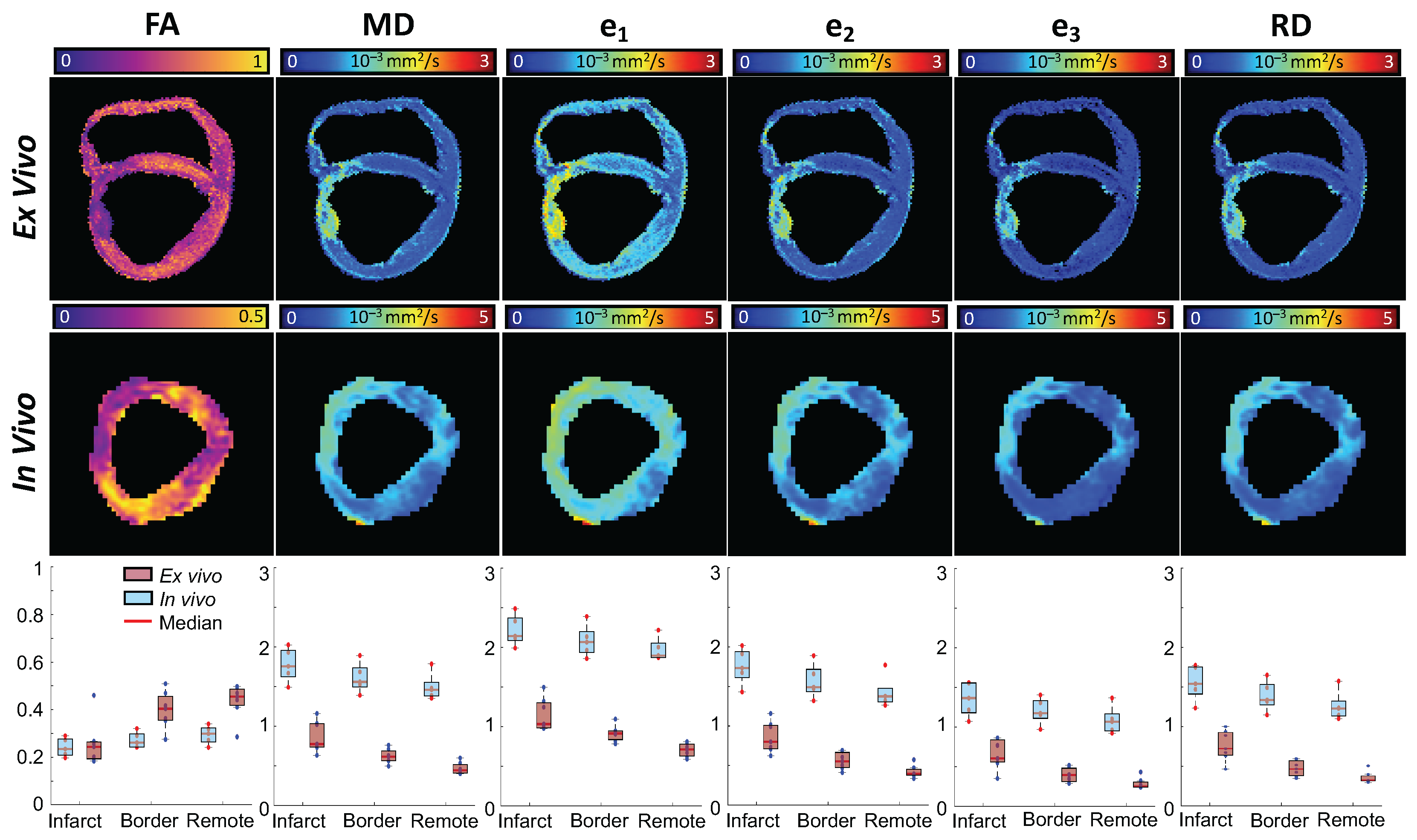
| Native T1 (ms) | ECV | |||||
|---|---|---|---|---|---|---|
| Infarct | Border | Remote | Infarct | Border | Remote | |
| Mean | 1546 | 1434 | 1344 | 0.49 | 0.39 | 0.32 |
| Median | 1554 | 1389 | 1282 | 0.47 | 0.38 | 0.31 |
| Q1, Q3 | 1363, 1700 | 1274, 1553 | 1210, 1442 | 0.36, 0.62 | 0.31, 0.46 | 0.27, 0.37 |
| FA | MD (1 × 10 mm/s) | |||||
| Infarct | Border | Remote | Infarct | Border | Remote | |
| Mean | 0.25 | 0.28 | 0.29 | 1.81 | 1.69 | 1.46 |
| Median | 0.23 | 0.27 | 0.29 | 1.84 | 1.68 | 1.46 |
| Q1, Q3 | 0.18, 0.30 | 0.21, 0.33 | 0.23, 0.35 | 1.52, 2.12 | 1.45, 1.92 | 1.32, 1.60 |
| e (1 × 10 mm/s) | e (1 × 10 mm/s) | |||||
| Infarct | Border | Remote | Infarct | Border | Remote | |
| Mean | 2.27 | 2.17 | 1.91 | 1.80 | 1.65 | 1.40 |
| Median | 2.28 | 2.14 | 1.94 | 1.83 | 1.65 | 1.38 |
| Q1, Q3 | 1.98, 2.55 | 1.90, 2.39 | 1.74, 2.10 | 1.48, 2.11 | 1.37, 1.92 | 1.22, 1.58 |
| e (1 × 10 mm/s) | RD (1 × 10 mm/s) | |||||
| Infarct | Border | Remote | Infarct | Border | Remote | |
| Mean | 1.38 | 1.25 | 1.06 | 1.59 | 1.45 | 1.23 |
| Median | 1.42 | 1.24 | 1.06 | 1.62 | 1.45 | 1.22 |
| Q1, Q3 | 1.09, 1.71 | 1.01, 1.48 | 0.91, 1.23 | 1.29, 1.90 | 1.20, 1.69 | 1.08, 1.39 |
| Native T1 | ECV | ||
|---|---|---|---|
| Border–Remote | Infarct–Remote | Border–Remote | Infarct–Remote |
| 8.4%, p < 0.01 | 21.2%, p < 0.01 | 22.6%, p < 0.01 | 51.6%, p < 0.01 |
| FA | MD | ||
| Border–Remote | Infarct–Remote | Border–Remote | Infarct–Remote |
| −7.6%, p < 0.01 | −19.8%, p < 0.01 | 14.7%, p < 0.01 | 26.1%, p < 0.01 |
| e | e | ||
| Border–Remote | Infarct–Remote | Border–Remote | Infarct–Remote |
| 10.4%, p < 0.01 | 17.5%, p < 0.01 | 19.2%, p < 0.01 | 31.9%, p < 0.01 |
| e | RD | ||
| Border–Remote | Infarct–Remote | Border–Remote | Infarct–Remote |
| 16.1%, p < 0.01 | 33.4%, p < 0.01 | 18.0%, p < 0.01 | 32.3%, p < 0.01 |
| Sub | FA | MD | ||||
|---|---|---|---|---|---|---|
| Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | |
| S1 | −18.4%, p < 0.01 | −24.0%, p < 0.01 | −38.0%, p < 0.01 | 14.6%, p < 0.01 | 21.3%, p < 0.01 | 39.0%, p < 0.01 |
| S2 | −6.5%, p = 0.25 | −2.7%, p < 0.01 | −9.0%, p < 0.01 | 7.0%, p = 0.02 | 5.4%, p < 0.01 | 12.8%, p < 0.01 |
| S3 | −11.8%, p < 0.01 | −11.1%, p < 0.01 | −21.6%, p < 0.01 | 15.0%, p < 0.01 | 4.6%, p < 0.01 | 20.3%, p < 0.01 |
| S4 | −9.5%, p < 0.01 | −5.7%, p = 0.1 | −14.6%, p = 0.02 | 7.3%, p < 0.01 | 2.9%, p < 0.01 | 10.3%, p < 0.01 |
| S5 | −10.2%, p < 0.01 | 8.3%, p < 0.01 | −2.7%, p < 0.01 | 7.1%, p < 0.01 | 6.0%, p < 0.01 | 13.5%, p < 0.01 |
| Sub | e | e | ||||
| Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | |
| S1 | 9.2%, p < 0.01 | 14.1%, p < 0.01 | 24.6%, p < 0.01 | 15.3%, p < 0.01 | 25.8%, p < 0.01 | 45.0%, p < 0.01 |
| S2 | 3.7%, p = 0.22 | 3.2%, p < 0.01 | 7.0%, p < 0.01 | 13.7%, p < 0.01 | 6.9%, p < 0.01 | 21.5%, p < 0.01 |
| S3 | 7.8%, p < 0.01 | 3.5%, p < 0.01 | 11.6%, p < 0.01 | 16.2%, p < 0.01 | 8.5%, p < 0.01 | 26.1%, p < 0.01 |
| S4 | 7.1%, p < 0.01 | −0.8%, p < 0.01 | 6.2%, p < 0.01 | 8.4%, p < 0.01 | 4.5%, p < 0.01 | 13.2%, p < 0.01 |
| S5 | 4.1%, p < 0.01 | 7.8%, p < 0.01 | 12.2%, p < 0.01 | 6.7%, p < 0.01 | 6.7%, p < 0.01 | 13.8%, p < 0.01 |
| Sub | e | RD | ||||
| Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | |
| S1 | 19.5%, p < 0.01 | 35.6%, p < 0.01 | 62.1%, p < 0.01 | 17.2%, p < 0.01 | 30.3%, p < 0.01 | 52.8%, p < 0.01 |
| S2 | 5.1%, p = 0.19 | 8.3%, p < 0.01 | 13.8%, p < 0.01 | 10.0%, p = 0.01 | 8.7%, p < 0.01 | 19.6%, p < 0.01 |
| S3 | 16.3%, p < 0.01 | 6.5%, p < 0.01 | 23.9%, p < 0.01 | 16.9%, p < 0.01 | 6.6%, p < 0.01 | 24.6%, p < 0.01 |
| S4 | 10.4%, p < 0.01 | 5.2%, p < 0.01 | 16.2%, p < 0.01 | 7.7%, p < 0.01 | 4.2%, p < 0.01 | 12.2%, p < 0.01 |
| S5 | 10.6%, p < 0.01 | 2.9%, p < 0.01 | 13.8%, p < 0.01 | 7.5%, p < 0.01 | 4.8%, p < 0.01 | 12.7%, p < 0.01 |
| Sub | Native T1 | ECV | ||||
| Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | Infarct vs. Border | Border vs. Remote | Infarct vs. Remote | |
| S1 | 16.7%, p = 0.06 | 4.0%, p = 0.88 | 21.3%, p < 0.01 | 23.8%, p < 0.01 | 2.1%, p < 0.01 | 26.4%, p < 0.01 |
| S2 | 18.7%, p = 0.66 | 14.1%, p < 0.01 | 35.4%, p < 0.01 | 61.7%, p < 0.01 | 29.2%, p < 0.01 | 108.9%, p < 0.01 |
| S3 | 7.1%, p = 0.11 | 14.2%, p < 0.01 | 22.3%, p < 0.01 | 12.1%, p < 0.01 | 36.8%, p < 0.01 | 53.3%, p < 0.01 |
| S4 | 2.0%, p = 0.3 | 4.5%, p < 0.01 | 6.6%, p = 0.018 | 10.1%, p < 0.01 | 11.3%, p < 0.01 | 22.6%, p < 0.01 |
| S5 | 9.3%, p < 0.01 | 1.4%, p = 0.1 | 10.8%, p < 0.01 | 13.9%, p < 0.01 | 8.3%, p < 0.01 | 23.3%, p < 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, T.; Moulin, K.; Perotti, L.E. Cardiac Diffusion Tensor Biomarkers of Chronic Infarction Based on In Vivo Data. Appl. Sci. 2022, 12, 3512. https://doi.org/10.3390/app12073512
Rahman T, Moulin K, Perotti LE. Cardiac Diffusion Tensor Biomarkers of Chronic Infarction Based on In Vivo Data. Applied Sciences. 2022; 12(7):3512. https://doi.org/10.3390/app12073512
Chicago/Turabian StyleRahman, Tanjib, Kévin Moulin, and Luigi E. Perotti. 2022. "Cardiac Diffusion Tensor Biomarkers of Chronic Infarction Based on In Vivo Data" Applied Sciences 12, no. 7: 3512. https://doi.org/10.3390/app12073512






