Differential Expression of MicroRNAs in Dark-Cutting Meat from Beef Carcasses
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Tissue Resources
2.2. RNA Extraction & Quantification
2.3. Small RNA Sequencing
2.4. Bioinformatic and Statistical Analysis
2.5. Institutional Approvals
3. Results
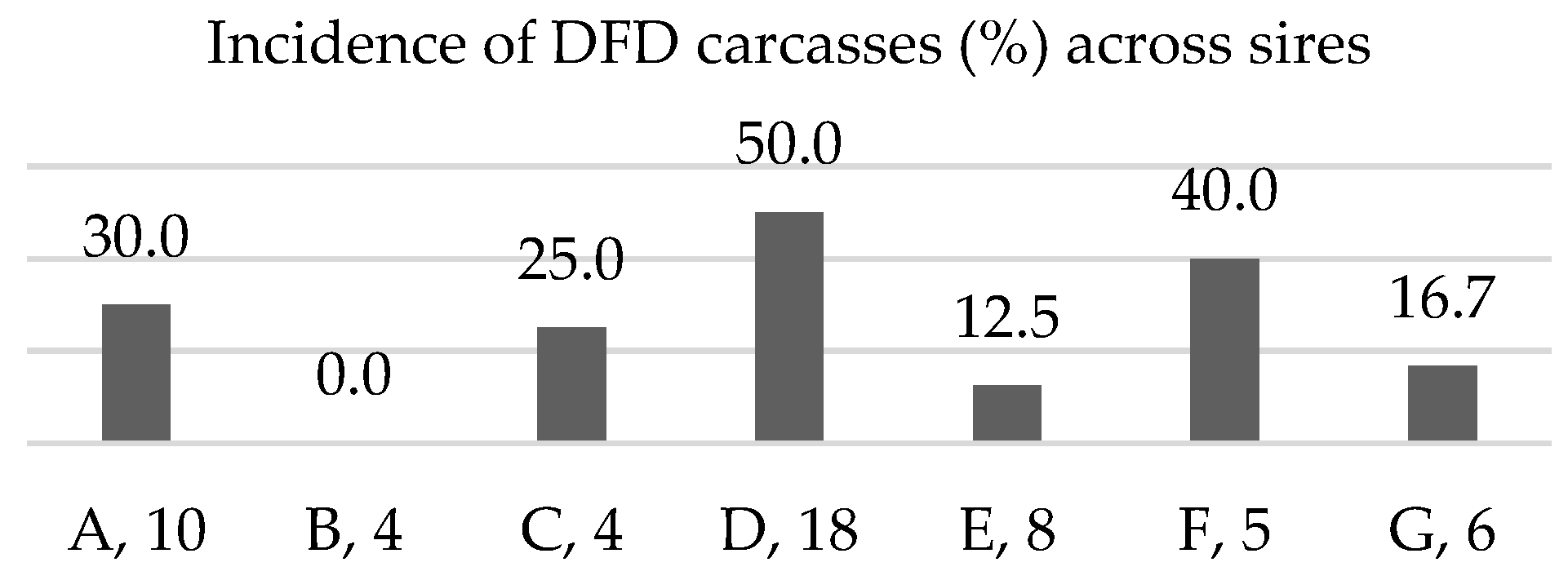
3.1. Family Distribution of Dark-Cutting Phenotype
3.2. miRNA Sequence Output
3.3. Identification of Differentially Expressed miRNAs
3.4. TargetScan Search for Putative Targets of bta-miR-2422
4. Discussion
4.1. MiRNA Role in Inflammation and Inflammatory Response
4.2. ThermomiRs and Stress Response
4.3. Muscle and MyomiRs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grandin, T. The effect of stress on livestock and meat quality prior to and during slaughter. Int. J. Study Anim. Probl. 1980, 5, 331–337. [Google Scholar]
- Lawrie, R.A. Physiological stress in relation to dark-cutting beef. J. Sci. Food Agric. 1958, 9, 721–727. [Google Scholar] [CrossRef]
- Ponnampalam, E.N.; Hopkins, D.L.; Bruce, H.; Li, D.; Baldi, G.; Bekhit, A.E.-D.A. Causes and Contributing Factors to “Dark Cutting” Meat: Current Trends and Future Directions: A Review. Compr. Rev. Food Sci. Food Saf. 2017, 16, 400–430. [Google Scholar] [CrossRef] [Green Version]
- Biggar, K.K.; Storey, K.B. Functional impact of microRNA regulation in models of extreme stress adaptation. J. Mol. Cell Biol. 2018, 10, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Hollins, S.; Cairns, M.J. MicroRNA: Small RNA mediators of the brains genomic response to environmental stress. Prog. Neurobiol. 2016, 143, 61–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bratzler, L.J. Dark cutting beef. MSC Vet. 1946, 6, 78–80. [Google Scholar]
- Cross, H.R.; Sorinmade, S.O.; Ono, K. Effect of Electrical Stimulation on Carcasses from Stressed and Unstressed Steers. J. Food Qual. 1983, 6, 73–79. [Google Scholar] [CrossRef]
- Tarrant, P.; Sherington, J. An investigation of ultimate pH in the muscles of commercial beef carcasses. Meat Sci. 1980, 4, 287–297. [Google Scholar] [CrossRef]
- Apaoblaza, A.; Gerrard, S.; Matarneh, S.; Wicks, J.; Kirkpatrick, L.; England, E.; Scheffler, T.; Duckett, S.; Shi, H.; Silva, S.; et al. Muscle from grass- and grain-fed cattle differs energetically. Meat Sci. 2020, 161, 107996. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Luo, X.; Yang, X.; Hopkins, D.L.; Mao, Y.; Zhang, Y. Understanding the development of color and color stability of dark cutting beef based on mitochondrial proteomics. Meat Sci. 2020, 163, 108046. [Google Scholar] [CrossRef]
- McGilchrist, P.; Perovic, J.L.; Gardner, G.E.; Pethick, D.W.; Jose, C.G. The incidence of dark cutting in southern Australian beef production systems fluctuates between months. Anim. Prod. Sci. 2014, 54, 1765–1769. [Google Scholar] [CrossRef] [Green Version]
- Boykin, C.A.; Eastwood, L.C.; Harris, M.K.; Hale, D.S.; Kerth, C.R.; Griffin, D.B.; Arnold, A.N.; Hasty, J.D.; Belk, K.E.; Woerner, D.R.; et al. National Beef Quality Audit–2016: In-plant survey of carcass characteristics related to quality, quantity, and value of fed steers and heifers1. J. Anim. Sci. 2017, 95, 2993–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, M.C.; Gray, G.D.; Hale, D.S.; Kerth, C.R.; Griffin, D.B.; Savell, J.W.; Raines, C.R.; Belk, K.E.; Woerner, D.R.; Tatum, J.D.; et al. National Beef Quality Audit–2011: In-plant survey of targeted carcass characteristics related to quality, quantity, value, and marketing of fed steers and heifers. J. Anim. Sci. 2012, 90, 5143–5151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, L.G.; Nicholson, K.L.; Hoffman, T.W.; Lawrence, T.E.; Hale, D.S.; Griffin, D.B.; Savell, J.W.; Vanoverbeke, D.L.; Morgan, J.B.; Belk, K.E.; et al. National Beef Quality Audit–2005: Survey of targeted cattle and carcass characteristics related to quality, quantity, and value of fed steers and heifers1. J. Anim. Sci. 2008, 86, 3533–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, D.R.; Roebert, D.L.; Bates, P.K.; Schmidt, T.B.; Hale, D.S.; Griffin, D.B.; Savell, J.W.; Brooks, J.C.; Morgan, J.B.; Montgomery, T.H.; et al. National Beef Quality Audit-2000: Survey of targeted cattle and carcass characteristics related to quality, quantity, and value of fed steers and heifers. J. Anim. Sci. 2002, 80, 1212–1222. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, C.D.; Fried, H.M.; Perkins, D.O. Additional layers of gene regulatory complexity from recently discovered microRNA mechanisms. Int. J. Biochem. Cell Biol. 2010, 42, 1236–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawes, M.; Kochan, K.J.; Riggs, P.K.; Lightfoot, J.T. Differential miRNA expression in inherently high- and low-active inbred mice. Physiol. Rep. 2015, 3, e12469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kochan, K.J.; Forman, S.E.; Hillhouse, A.E.; Cross, H.R.; Riggs, P.K. Methodology for quantification of circulating cell-free microRNA from bovine plasma for analysis of meat quality traits. In Proceedings of the 22nd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Townsville, QLD, Australia, 2–5 July 2017; Volume 22, p. 469. [Google Scholar]
- Riley, D.G.; Miller, R.K.; Nicholson, K.L.; Gill, C.A.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Savell, J.W.; Sanders, J.O. Genome association of carcass and palatability traits from Bos indicus-Bos taurus crossbred steers within electrical stimulation status and correspondence with steer temperament 1. Carcass. Livest. Sci. 2019, 229, 150–158. [Google Scholar] [CrossRef]
- Vaughn, R.N.; Kochan, K.J.; Torres, A.K.; Du, M.; Riley, D.G.; Gill, C.A.; Herring, A.D.; Sanders, J.O.; Riggs, P.K. Skeletal Muscle Expression of Actinin-3 (ACTN3) in Relation to Feed Efficiency Phenotype of F2Bos indicus-Bos taurus Steers. Front. Genet. 2022, 13, 796038. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ewing, B.; Hillier, L.; Wendl, M.C.; Green, P. Base-Calling of Automated Sequencer Traces UsingPhred. I. Accuracy Assessment. Genome Res. 1998, 8, 175–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC. 2017. Available online: https://github.com/s-andrews/FastQC (accessed on 27 February 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I. Statistical Modeling of High Dimensional Counts. In RNA Bioinformatics; Humana: New York, NY, USA, 2021; Volume 2284, pp. 97–134. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the falser discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Yekutieli, D.; Benjamini, Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Plan. Inference 1999, 82, 171–196. [Google Scholar] [CrossRef]
- Metpally, R.P.R.; Nasser, S.; Malenica, I.; Courtright, A.; Carlson, E.; Ghaffari, L.; Villa, S.; Tembe, W.; Van Keuren-Jensen, K. Comparison of Analysis Tools for miRNA High Throughput Sequencing Using Nerve Crush as a Model. Front. Genet. 2013, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, J.D.; Liu, G.; Luo, L.; Xiao, J.; Gerrein, J.; Juan-Guardela, B.; Tedrow, J.; Alekseyev, Y.O.; Yang, I.V.; Correll, M.; et al. Assessment of microRNA differential expression and detection in multiplexed small RNA sequencing data. RNA 2014, 21, 164–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giagnorio, E.; Malacarne, C.; Mantegazza, R.; Bonanno, S.; Marcuzzo, S. MyomiRs and their multifaceted regulatory roles in muscle homeostasis and amyotrophic lateral sclerosis. J. Cell Sci. 2021, 134, jcs258349. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Morenikeji, O.B.; Hawkes, M.E.; Hudson, A.O.; Thomas, B.N. Computational Network Analysis Identifies Evolutionarily Conserved miRNA Gene Interactions Potentially Regulating Immune Response in Bovine Trypanosomosis. Front. Microbiol. 2019, 10, 2010. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.-S.; Cha, H.-N.; Jo, H.J.; Song, I.-H.; Baek, S.-H.; Dan, J.-M.; Kim, Y.-W.; Kim, J.-Y.; Lee, I.-K.; Seo, J.-S.; et al. TLR2 deficiency attenuates skeletal muscle atrophy in mice. Biochem. Biophys. Res. Commun. 2015, 459, 534–540. [Google Scholar] [CrossRef]
- Li, R.; Zhang, C.-L.; Liao, X.-X.; Chen, D.; Wang, W.-Q.; Zhu, Y.-H.; Geng, X.-H.; Ji, D.-J.; Mao, Y.-J.; Gong, Y.-C.; et al. Transcriptome MicroRNA Profiling of Bovine Mammary Glands Infected with Staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef] [Green Version]
- Singh, J.; Dhanoa, J.K.; Choudhary, R.K.; Singh, A.; Sethi, R.S.; Kaur, S.; Mukhopadhyay, C.S. MicroRNA expression profiling in PBMCs of Indian water Buffalo (Bubalus bubalis) infected with Brucella and Johne’s disease. ExRNA 2020, 2, 8. [Google Scholar] [CrossRef]
- Ng, C.-T.; Li, J.J.; Balasubramanian, S.K.; You, F.; Yung, L.-Y.L.; Bay, B.-H. Inflammatory Changes in Lung Tissues Associated with Altered Inflammation-Related MicroRNA Expression after Intravenous Administration of Gold Nanoparticles in Vivo. ACS Biomater. Sci. Eng. 2016, 2, 1959–1967. [Google Scholar] [CrossRef]
- Welc, S.S.; Clanton, T.; Dineen, S.M.; Leon, L.R. Heat stroke activates a stress-induced cytokine response in skeletal muscle. J. Appl. Physiol. 2013, 115, 1126–1137. [Google Scholar] [CrossRef]
- Li, Q.; Yang, C.; Du, J.; Zhang, B.; He, Y.; Hu, Q.; Li, M.; Zhang, Y.; Wang, C.; Zhong, J. Characterization of miRNA profiles in the mammary tissue of dairy cattle in response to heat stress. BMC Genom. 2018, 19, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miretti, S.; Lecchi, C.; Ceciliani, F.; Baratta, M. MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock. Front. Veter. Sci. 2020, 7, 578193. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, L.H.; Zanon, N.M.; Garófalo, M.A.; Navegantes, L.C.C.; Kettelhut, I.C. Effect of short-term cold exposure on skeletal muscle protein breakdown in rats. J. Appl. Physiol. 2013, 115, 1496–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Bührer, C.; Wellmann, S. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. 2016, 73, 3839–3859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, P.; Hu, Y.; Cao, Y.; Lu, J.; Yang, Y.; Lv, H.; Lian, S.; Xu, B.; Li, S. Cold-Induced RNA-Binding Protein Promotes Glucose Metabolism and Reduces Apoptosis by Increasing AKT Phosphorylation in Mouse Skeletal Muscle Under Acute Cold Exposure. Front. Mol. Biosci. 2021, 8, 685993. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.J.-L.; Au, A.Y.; Gao, D.; Pinello, N.; Kwok, C.-T.; Thoeng, A.; Lau, K.A.; Gordon, J.E.; Schmitz, U.; Feng, Y.; et al. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 2016, 44, 2888–2897. [Google Scholar] [CrossRef] [Green Version]
- Fischer, C.; Seki, T.; Lim, S.; Nakamura, M.; Andersson, P.; Yang, Y.; Honek, J.; Wang, Y.; Gao, Y.; Chen, F.; et al. A miR-327–FGF10–FGFR2-mediated autocrine signaling mechanism controls white fat browning. Nat. Commun. 2017, 8, 2079. [Google Scholar] [CrossRef] [Green Version]
- Colitti, M.; Sgorlon, S.; Licastro, D.; Stefanon, B. Differential expression of miRNAs in milk exosomes of cows subjected to group relocation. Res. Veter. Sci. 2019, 122, 148–155. [Google Scholar] [CrossRef]
- Muroya, S.; Ogasawara, H.; Nohara, K.; Oe, M.; Ojima, K.; Hojito, M. Coordinated alteration of mRNA-microRNA transcriptomes associated with exosomes and fatty acid metabolism in adipose tissue and skeletal muscle in grazing cattle. Asian Australas. J. Anim. Sci. 2020, 33, 1824–1836. [Google Scholar] [CrossRef]
- Lee, J.; Heo, J.; Kang, H. miR-92b-3p-TSC1 axis is critical for mTOR signaling-mediated vascular smooth muscle cell proliferation induced by hypoxia. Cell Death Differ. 2019, 26, 1782–1795. [Google Scholar] [CrossRef] [Green Version]
- Seong, M.; Kang, H. Hypoxia-induced miR-1260b regulates vascular smooth muscle cell proliferation by targeting GDF11. BMB Rep. 2020, 53, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Yang, J.; Zhang, J.; Yang, C.; Fan, Z.; Li, Q.; Zhai, Y.; Liu, H.; Yang, J. Downregulated MicroRNA-327 Attenuates Oxidative Stress–Mediated Myocardial Ischemia Reperfusion Injury Through Regulating the FGF10/Akt/Nrf2 Signaling Pathway. Front. Pharmacol. 2021, 12, 669146. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Zhou, C.; Li, B.; Jiao, J.; Chen, L.; Ren, A.; Jie, H.; Tan, Z. Integrative plasma proteomic and microRNA analysis of Jersey cattle in response to high-altitude hypoxia. J. Dairy Sci. 2019, 102, 4606–4618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-S.; Kim, J.-M.; Lim, K.-S.; Hong, J.-S.; Hong, K.-C.; Lee, Y.S. Effects of polymorphisms in the porcine microRNAMIR206/MIR133Bcluster on muscle fiber and meat quality traits. Anim. Genet. 2012, 44, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Van de Worp, W.R.P.H.; Schols, A.M.W.J.; Dingemans, A.-M.C.; Op den Kamp, C.M.H.; Degens, J.H.R.J.; Kelders, M.C.J.M.; Coort, S.; Woodruff, H.C.; Kratassiouk, G.; Harel-Bellan, A.; et al. Identification of microRNAs in skeletal muscle associated with lung cancer cachexia. J. Cachexia Sarcopenia Muscle 2020, 11, 452–463. [Google Scholar] [CrossRef] [Green Version]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Chemello, F.; Grespi, F.; Zulian, A.; Cancellara, P.; Hebert-Chatelain, E.; Martini, P.; Bean, C.; Alessio, E.; Buson, L.; Bazzega, M.; et al. Transcriptomic Analysis of Single Isolated Myofibers Identifies miR-27a-3p and miR-142-3p as Regulators of Metabolism in Skeletal Muscle. Cell Rep. 2019, 26, 3784–3797.e8. [Google Scholar] [CrossRef] [Green Version]
- Maalouf, S.W.; Liu, W.-S.; Albert, I.; Pate, J.L. Regulating life or death: Potential role of microRNA in rescue of the corpus luteum. Mol. Cell. Endocrinol. 2014, 398, 78–88. [Google Scholar] [CrossRef]

| Cycle 1 | Cycle 2 | Cycle 3 |
|---|---|---|
| NA × NA | 4 crosses; combinations of NA and AN F1 parents | NA × NA |
| MicroRNA | Counts (Normalized) 1 | Fold Difference (DFD/NON) 2 | p Value (Wald Test) 3 | padj (FDR ≤ 0.05) 4 |
|---|---|---|---|---|
| bta-miR-2422 | 5.54 | 1.72 | 9.98 × 10−5 | 0.054073752 |
| bta-miR-10174-5p | 2.68 | 1.93 | 0.0018698 | 0.253362794 |
| bta-miR-1260b | 12.13 | 1.58 | 0.0009795 | 0.253362794 |
| bta-miR-144 | 25.26 | 0.56 | 0.0015518 | 0.253362794 |
| bta-miR-142-5p | 145.19 | 0.77 | 0.0045699 | 0.286059911 |
| bta-miR-2285at | 2.18 | 1.77 | 0.0043885 | 0.286059911 |
| bta-miR-2285e | 3.14 | 1.61 | 0.0036349 | 0.286059911 |
| bta-miR-3613a | 375.89 | 0.79 | 0.0053443 | 0.289662932 |
| bta-miR-493 | 20.74 | 1.26 | 0.006497 | 0.320127236 |
| bta-miR-27a-5p | 10.57 | 1.38 | 0.0074149 | 0.334904236 |
| bta-miR-136 | 8.86 | 0.68 | 0.0100435 | 0.400069983 |
| bta-miR-146a | 246.05 | 0.75 | 0.0110384 | 0.400069983 |
| bta-miR-23b-5p | 1.56 | 2.12 | 0.011072 | 0.400069983 |
| bta-miR-10162-5p | 1.32 | 1.84 | 0.0233158 | 0.554531616 |
| bta-miR-1307 | 12.53 | 1.29 | 0.0189402 | 0.554531616 |
| bta-miR-2285aa | 1.32 | 0.45 | 0.0198697 | 0.554531616 |
| bta-miR-2285r | 1.56 | 0.50 | 0.0189285 | 0.554531616 |
| bta-miR-31 | 4.48 | 0.63 | 0.0221755 | 0.554531616 |
| bta-miR-2284k | 1.44 | 0.54 | 0.0267659 | 0.580285276 |
| bta-miR-2299-5p | 1.89 | 0.56 | 0.0265474 | 0.580285276 |
| bta-miR-11994 | 6.87 | 1.28 | 0.0468123 | 0.604508006 |
| bta-miR-18b | 0.47 | 0.37 | 0.0462686 | 0.604508006 |
| bta-miR-2285q | 3.66 | 0.66 | 0.0304856 | 0.604508006 |
| bta-miR-2478 | 32.27 | 1.24 | 0.0307361 | 0.604508006 |
| bta-miR-431 | 1.26 | 2.08 | 0.0323255 | 0.604508006 |
| bta-miR-543 | 12.77 | 1.28 | 0.0328225 | 0.604508006 |
| bta-miR-6123 | 5.08 | 0.73 | 0.047955 | 0.604508006 |
| bta-miR-671 | 2.17 | 0.60 | 0.0348298 | 0.604508006 |
| MyomiRs 5 | ||||
| bta-miR-206 | 10,196.55 | 1.12 | 0.004750072 | 0.286059911 |
| bta-miR-208b | 236.105 | 0.80 | 0.063240802 | 0.604508006 |
| bta-miR-1 | 2,711,501.56 | 0.92 | 0.090611395 | 0.680638063 |
| bta-miR-499 | 16,961.84 | 0.93 | 0.258200039 | 0.904697654 |
| bta-miR-486 | 1758.97 | 1.06 | 0.333875388 | 0.973667764 |
| bta-miR-133a | 59,175.47 | 0.99 | 0.992791693 | 0.987194328 |
| bta-miR-133b | 833.48 | 1.04 | 0.440191294 | 0.992229627 |
| bta-miR-208a | 2.25 | 0.94 | 0.769097948 | 0.992229627 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riggs, P.K.; Therrien, D.A.; Vaughn, R.N.; Rotenberry, M.L.; Davis, B.W.; Herring, A.D.; Riley, D.G.; Cross, H.R. Differential Expression of MicroRNAs in Dark-Cutting Meat from Beef Carcasses. Appl. Sci. 2022, 12, 3555. https://doi.org/10.3390/app12073555
Riggs PK, Therrien DA, Vaughn RN, Rotenberry ML, Davis BW, Herring AD, Riley DG, Cross HR. Differential Expression of MicroRNAs in Dark-Cutting Meat from Beef Carcasses. Applied Sciences. 2022; 12(7):3555. https://doi.org/10.3390/app12073555
Chicago/Turabian StyleRiggs, Penny K., Dustin A. Therrien, Robert N. Vaughn, Marissa L. Rotenberry, Brian W. Davis, Andy D. Herring, David G. Riley, and H. Russell Cross. 2022. "Differential Expression of MicroRNAs in Dark-Cutting Meat from Beef Carcasses" Applied Sciences 12, no. 7: 3555. https://doi.org/10.3390/app12073555
APA StyleRiggs, P. K., Therrien, D. A., Vaughn, R. N., Rotenberry, M. L., Davis, B. W., Herring, A. D., Riley, D. G., & Cross, H. R. (2022). Differential Expression of MicroRNAs in Dark-Cutting Meat from Beef Carcasses. Applied Sciences, 12(7), 3555. https://doi.org/10.3390/app12073555








