Artificial Intelligence Mortality Prediction Model for Gastric Cancer Surgery Based on Body Morphometry, Nutritional, and Surgical Information: Feasibility Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Datasets
2.2. Clinical Information
2.3. Feature Selection and Preprocessing
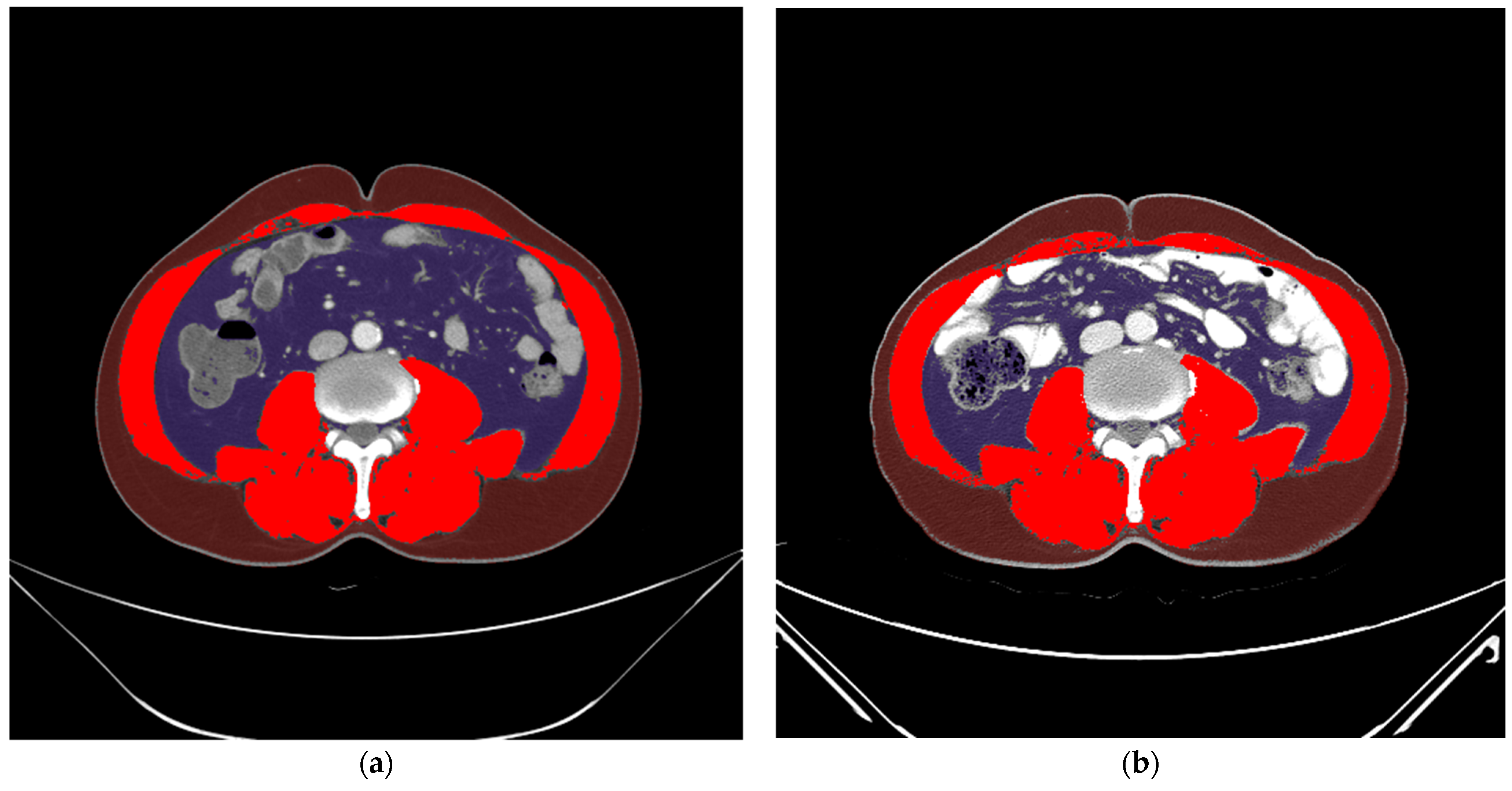
2.4. Processing of Data
2.5. Development of a CNN Model
2.6. Implementation
2.7. Performance Evaluation of the CNN Model
3. Results
3.1. Feature Importance and Preprocessing Results
- -
- Continuous features: age (year), height (cm), ΔBMIstandard, Δcholesterolstandard, Δhemoglobinstandard, Δalbuminstandard, ΔNRIstandard, ΔSFAstandard, ΔVFAstandard, ΔSMA/height2standard, and ΔSMA/BMIstandard;
- -
- Categorical features: sex, type of operation, type of anastomosis, tumor, nodes, and metastases (TNM) stage.
3.2. Learning Curves
3.3. Performance
3.4. Web Application of the AI Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.J.; Lin, J.C.; Tu, S.P. Etiology and prevention of gastric cancer. Gastrointest. Tumors 2016, 3, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Tegels, J.J.; van Vugt, J.L.; Reisinger, K.W.; Hulsewé, K.W.; Hoofwijk, A.G.; Derikx, J.P.; Stoot, J.H. Sarcopenia is highly prevalent in patients undergoing surgery for gastric cancer but not associated with worse outcomes. J. Surg. Oncol. 2015, 112, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Lee, K.; Lee, J.B.; Park, T.; Khang, S.; Jeong, H.; Ko, C.S.; Yook, J.H.; Kim, B.S.; Lee, I.S. Preoperative nutritional risk index and postoperative one-year skeletal muscle loss can predict the prognosis of patients with gastric adenocarcinoma: A registry-based study. BMC Cancer 2021, 21, 157. [Google Scholar] [CrossRef]
- Asplund, J.; Gottlieb-Vedi, E.; Leijonmarck, W.; Mattsson, F.; Lagergren, J. Prognosis after surgery for gastric adenocarcinoma in the Swedish Gastric Cancer Surgery Study (SWEGASS). Acta Oncol. 2021, 60, 513–520. [Google Scholar] [CrossRef]
- Hu, D.; Peng, F.; Lin, X.; Chen, G.; Zhang, H.; Liang, B.; Ji, K.; Lin, J.; Chen, L.F.; Zheng, X.; et al. Preoperative metabolic syndrome is predictive of significant gastric cancer mortality after gastrectomy: The Fujian prospective investigation of cancer (FIESTA) study. EBioMedicine 2017, 15, 73–80. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, E.S.; Kim, B.J.; Kim, W.S.; Park, J.Y.; Kim, J.G.; Park, J.M.; Kim, J.W.; Chi, K.C.; Kang, H. Risk factors and clinical outcomes of postgastrectomy sarcopenia newly developed after curative resection for gastric cancer. Medicine 2022, 101, e28699. [Google Scholar] [CrossRef]
- Lee, K.; Kim, K.W.; Lee, J.B.; Shin, Y.; Jang, J.K.; Yook, J.H.; Kim, B.S.; Lee, I.S. Impact of remnant stomach volume and anastomosis on nutrition and body composition in gastric cancer patients. Surg. Oncol. 2019, 31, 75–82. [Google Scholar] [CrossRef]
- Kuwada, K.; Kuroda, S.; Kikuchi, S.; Yoshida, R.; Nishizaki, M.; Kagawa, S.; Fujiwara, T. Clinical Impact of Sarcopenia on Gastric Cancer. Anticancer Res. 2019, 39, 2241–2249. [Google Scholar] [CrossRef] [Green Version]
- Karimi, P.; Islami, F.; Anandasabapathy, S.; Freedman, N.D.; Kamangar, F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomark. Prev. 2014, 23, 700–713. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, X.; Dang, Y.; Zhang, F.; Huang, Q. Low serum albumin correlates with poor survival in gastric cancer patients. Clin. Lab. 2018, 64, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bai, B.; Xie, F.; Zhao, Q. Distal versus total gastrectomy for middle and lower-third gastric cancer: A systematic review and meta-analysis. Int. J. Surg. 2018, 53, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wu, X.; Gao, X.; Shan, F.; Ying, X.; Zhang, Y.; Ji, J. Development and validation of an artificial neural network prognostic model after gastrectomy for gastric carcinoma: An international multicenter cohort study. Cancer Med. 2020, 9, 6205–6215. [Google Scholar] [CrossRef] [PubMed]
- Niu, P.H.; Zhao, L.L.; Wu, H.L.; Zhao, D.B.; Chen, Y.T. Artificial intelligence in gastric cancer: Application and future perspectives. World J. Gastroenterol. 2020, 26, 5408–5419. [Google Scholar] [CrossRef]
- Jin, P.; Ji, X.; Kang, W.; Li, Y.; Liu, H.; Ma, F.; Ma, S.; Hu, H.; Li, W.; Tian, Y. Artificial intelligence in gastric cancer: A systematic review. J. Cancer Res. Clin. Oncol. 2020, 146, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, P.C.; Krause, J.; Peng, L. How to read articles that use machine learning: Users’ guides to the medical literature. JAMA 2019, 322, 1806–1816. [Google Scholar] [CrossRef]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An a attempt at h Histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef]
- Veterans Affairs Total Parenteral Nutrition Cooperative Study Group. Perioperative total parenteral nutrition in surgical patients. N. Engl. J. Med. 1991, 325, 525–532. [Google Scholar] [CrossRef]
- Park, H.J.; Shin, Y.; Park, J.; Kim, H.; Lee, I.S.; Seo, D.W.; Huh, J.; Lee, T.Y.; Park, T.; Lee, J.; et al. Development and validation of a deep learning system for segmentation of abdominal muscle and fat on computed tomography. Korean J. Radiol. 2020, 21, 88–100. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Lin, W.-C.; Tsai, C.-F.; Hu, Y.-H.; Jhang, J.-S. Clustering-based undersampling in class-imbalanced data. Inf. Sci. 2017, 409–410, 17–26. [Google Scholar] [CrossRef]
- Arnold, T.B. kerasR: R interface to the keras deep learning library. J. Open Source Softw. 2017, 2, 296. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Rahman, S.A.; Maynard, N.; Trudgill, N.; Crosby, T.; Park, M.; Wahedally, H.; Underwood, T.J.; Cromwell, D.A. Prediction of long-term survival after gastrectomy using random survival forests. Br. J. Surg. 2021, 108, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Luo, W.; Su, M.; Wei, H.; Wei, J.; Zhang, X.; Zou, C. Comparison between artificial neural network and Cox regression model in predicting the survival rate of gastric cancer patients. Biomed. Rep. 2013, 1, 757–760. [Google Scholar] [CrossRef] [Green Version]
- Biglarian, A.; Hajizadeh, E.; Kazemnejad, A.; Zali, M. Application of artificial neural network in predicting the survival rate of gastric cancer patients. Iran. J. Public Health 2011, 40, 80–86. [Google Scholar]
- Suo, Q.; Ma, F.; Yuan, Y.; Huai, M.; Zhong, W.; Zhang, A.; Gao, J. Personalized disease prediction using a cnn-based similarity learning method. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 811–816. [Google Scholar]
- Zhu, Z.; Yin, C.; Qian, B.; Cheng, Y.; Wei, J.; Wang, F. Measuring patient similarities via a deep architecture with medical concept embedding. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining (ICDM), Barcelona, Spain, 12–15 December 2016; pp. 749–758. [Google Scholar]
- Korhani Kangi, A.; Bahrampour, A. Predicting the survival of gastric cancer patients using artificial and bayesian neural networks. Asian Pac. J. Cancer Prev. 2018, 19, 487–490. [Google Scholar] [CrossRef]
- Biglarian, A.; Hajizadeh, E.; Kazemnejad, A.; Zayeri, F. Determining of prognostic factors in gastric cancer patients using artificial neural networks. Asian Pac. J. Cancer Prev. 2010, 11, 533–536. [Google Scholar]
- Ha, J.; Park, T.; Kim, H.K.; Shin, Y.; Ko, Y.; Kim, D.W.; Sung, Y.S.; Lee, J.; Ham, S.J.; Khang, S.; et al. Development of a fully automatic deep learning system for L3 selection and body composition assessment on computed tomography. Sci. Rep. 2021, 11, 21656. [Google Scholar] [CrossRef]





| Characteristics | AMC (n = 621) | AUH (n = 33) | Total (n = 654) | ||
|---|---|---|---|---|---|
| Clinicopathologic | |||||
| Age (year) | 55.7 ± 11.6 | 60.0 ± 12.0 | 55.9 ± 11.6 | ||
| Sex | |||||
| Male | 388 (62.5%) | 20 (60.6%) | 408 (62.4%) | ||
| Female | 233 (37.5%) | 13 (39.4%) | 246 (37.6%) | ||
| Height (cm) | 162.95 ± 8.35 | 162.55 ± 8.12 | 162.93 ± 8.34 | ||
| Weight (kg) | 63.1 ± 10.3 | 61.5 ± 11.4 | 63.0 ± 10.4 | ||
| BMI (kg/m2) | 23.7 ± 2.9 | 23.2 ± 3.2 | 23.7 ± 2.9 | ||
| Type of operation | |||||
| Distal gastrectomy | 396 (63.8%) | 25 (75.8%) | 421 (64.4%) | ||
| Total gastrectomy | 225 (36.2%) | 8 (24.2%) | 233 (35.6%) | ||
| TNM stage 1 | |||||
| 1A | 78 (12.6%) | 18 (54.5%) | 96 (14.7%) | ||
| 1B | 21 (3.4%) | 3 (9.1%) | 24 (3.7%) | ||
| 2A | 138 (22.2%) | 2 (6.1%) | 140 (21.4%) | ||
| 2B | 135 (21.7%) | 1 (3.0%) | 136 (20.8%) | ||
| 3A | 110 (17.7%) | 3 (9.1%) | 113 (17.3%) | ||
| 3B | 93 (15.0%) | 3 (9.1%) | 96 (14.7%) | ||
| 3C | 42 (6.8%) | 3 (9.1%) | 45 (6.9%) | ||
| 4 | 4 (0.6%) | 0 (0%) | 4 (0.6%) | ||
| Preoperative Body/Nutrition | |||||
| SMA (cm2) | 124.2 ± 30.0 | 125.8 ± 31.7 | 124.3 ± 30.0 | ||
| SFA (cm2) | 118.0 ± 55.2 | 118.0 ± 55.2 | 118.1 ± 55.5 | ||
| VFA (cm2) | 97.7 ± 56.6 | 100.3 ± 72.1 | 97.9 ± 57.4 | ||
| NRI | 101.1 ± 6.4 | 103.9 ± 7.6 | 101.3 ± 6.5 | ||
| Features | Based on XGBoost (n = 20) | Based on Experts (n = 13) | Selected for AI Model (n = 15) |
|---|---|---|---|
| Age | O | O | O |
| Sex | O | O | O |
| Height | O | X | O |
| BMI | O | O | O |
| SFA | O | O | O |
| VFA | O | O | O |
| SMA/height2 | O | O | O |
| SMA/BMI | O | O | O |
| NRI | O | O | O |
| Cholesterol | O | O | O |
| Hemoglobin | O | O | O |
| Albumin | O | X | O |
| Protein | O | X | X |
| Type of operation (total vs. distal gastrectomy) | O | O | O |
| Type of anastomosis | O | O | O |
| TNM stage 1 | O | O | O |
| Distal resection margin | O | X | X |
| Proximal resection margin | O | X | X |
| Number of metastatic lymph nodes | O | X | X |
| Number of retrieved lymph nodes | O | X | X |
| Performance | All Validation Sets (n = 53) | Internal Validation Set (n = 20) | External Validation Set (n = 33) |
|---|---|---|---|
| Accuracy | 0.887 | 0.900 | 0.879 |
| F1 score | 0.889 | 0.909 | 0.882 |
| AUC | 0.887 | 0.900 | 0.881 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, Y.; Shin, H.; Shin, J.; Hur, H.; Huh, J.; Park, T.; Kim, K.W.; Lee, I.-S. Artificial Intelligence Mortality Prediction Model for Gastric Cancer Surgery Based on Body Morphometry, Nutritional, and Surgical Information: Feasibility Study. Appl. Sci. 2022, 12, 3873. https://doi.org/10.3390/app12083873
Ko Y, Shin H, Shin J, Hur H, Huh J, Park T, Kim KW, Lee I-S. Artificial Intelligence Mortality Prediction Model for Gastric Cancer Surgery Based on Body Morphometry, Nutritional, and Surgical Information: Feasibility Study. Applied Sciences. 2022; 12(8):3873. https://doi.org/10.3390/app12083873
Chicago/Turabian StyleKo, Yousun, Hooyoung Shin, Juneseuk Shin, Hoon Hur, Jimi Huh, Taeyong Park, Kyung Won Kim, and In-Seob Lee. 2022. "Artificial Intelligence Mortality Prediction Model for Gastric Cancer Surgery Based on Body Morphometry, Nutritional, and Surgical Information: Feasibility Study" Applied Sciences 12, no. 8: 3873. https://doi.org/10.3390/app12083873
APA StyleKo, Y., Shin, H., Shin, J., Hur, H., Huh, J., Park, T., Kim, K. W., & Lee, I.-S. (2022). Artificial Intelligence Mortality Prediction Model for Gastric Cancer Surgery Based on Body Morphometry, Nutritional, and Surgical Information: Feasibility Study. Applied Sciences, 12(8), 3873. https://doi.org/10.3390/app12083873







