An Optimization-Based Diabetes Prediction Model Using CNN and Bi-Directional LSTM in Real-Time Environment
Abstract
:Featured Application
Abstract
1. Introduction
1.1. Diabetes Prediction in Real-Time Environment
1.2. Motivation
1.3. Major Contributions
- We used the best parameters to train our models. We ran a grid search algorithm that found the best values for parameters such as learning rate, epochs, optimizer, batch sizes, and hidden units;
- We split our dataset into test and training sets by using 10-fold cross-validation, and the precision of each model was improved. On the other hand, CNN-Bi-LSTM outperformed with 98% accuracy, 97% sensitivity, and 98% specificity;
- We proposed a framework to test our optimized models using a real-time dataset.
2. Related Work
3. Methodology
3.1. Availability of Real-Time Dataset for Training and Testing
3.2. Preprocessing of Real-Time Data
3.3. Feature Selection
3.4. Data Augmentation
3.5. Diabetes Prediction Models
3.5.1. Convolutional Neural Network
3.5.2. Architectural CNN-LSTM Model for Diabetes Prediction
3.5.3. CNN-Bi-LSTM: A Real-Time Framework
- I.
- CNN-Bi-LSTM Training Process: The main steps are explained by the activity diagram shown in Figure 7.
- Input data: The necessary data for CNN-Bi-LSTM training must be entered;
- Preprocessing of input data: The z-score standardization approach was used to normalize input data since there was a substantial gap in input data to fully train the algorithm, as indicated in Equation (4);
- Initialization of network: Here, we initialized weights and biases for each layer of CNN-Bi-LSTM;
- CNN Layer Calculation: Eight input features with an input shape of 6 × 1 were passed through a convolutionary layer, which is responsible for generating a feature map by striding filters of kernel size 1 at one step. We introduced non-linearity to the feature by using the ReLU function, which has a range of 0 and 1, i.e., it does not activate all neurons at the same time as it deactivates neurons whose values are less than zero. These function maps are then transferred into a batch normalization that regularises the meaning and prevents the over-fitting functions. Besides, these functions are transferred into the max-pooling layer, with pool size 1 being used for the downsampling of the function diagram. Down-sampled features are then passed through the flattening layer, which is responsible for translating these function matrices to 1D vectors.
- Bi-LSTM Estimation of the layer: Using Bi-LSTM [49,50], the output data of the CNN layer are determined. Bi-LSTM is a bidirectional RNN that consists of 32 hidden LSTM cells, so there are a total of 64 LSTM cells that have an additional peehole connection that prevents vanishing gradient problems and additional cell state memory that uses past and future knowledge to forecast output by using two separate hidden layers, such as forward state sequence, is represented by as shown in Equation (6) [43], the backward state sequence is shown in Equation (7) [43], and the output vector is represented by Equation (8)
- bh :the bias vector for hidden state vectors
- Dropout: 15% dropout was applied, which is used to reduce the overfitting in neural networks by dropping some of the random nodes during the network training process;
- Output values were transferred into a sigmoid function used for binary classification, which determines whether or not the input instance is diabetic [51];
- Calculation Error: The cost function assesses how effectively the neural network is equipped by describing the difference between the provided testing sample and the expected performance. The optimizer function was used to decrease the cost function. A cross-entropy function, which comes in a variety of forms and sizes, is commonly used in deep learning. Mathematically cost function is expressed as Equation (9) [35]:
- Evaluate if the prediction mechanism’s final criterion has been achieved: Effective completion of cycles depends on two factors: weights should not exceed a certain threshold, and the estimated error rate is below a specified threshold. If at least one of the graduation standards is fulfilled, the training is finished. Otherwise, the instruction would be resumed;
- Back Propagation Error: The calculated error is spread in the opposite direction, the weight and bias of each layer are modified, and then the process goes back to stage (4) to start network training.
- II.
- CNN-Bi-LSTM Prediction Process: The prediction of the CNN-Bi-LSTM model is explained by the activity diagram shown in Figure 7;
- Input Data: The essential data for CNN-Bi-LSTM predictions must be entered;
- Preprocessing of input data: This is performed through standardization through Equation (4);
- Process of prediction: Standardized data are fed into the CNN-Bi-LSTM, which is then used to calculate the output value;
- Output Result: Recovered results are provided to complete the prediction process. The model summary of CNN-Bi-LSTM is represented in Table 6.
3.6. Prototype Implementation and Testing of Proposed Model Using Real-Time Database
3.6.1. Sensing of Real-Time Data through a Proposed Framework
3.6.2. Prediction of Diabetes Using Proposed Model Using Real-Time Dataset
| Algorithm 1: Algorithm to fetch real-time data |
Require: initialization: Ensure: ,
if ≥ then while do Step1: Import chunk of size Step2: Perform testing using proposed model. Step3: Obtained result of chunk . Step4: Update previous results : ← ⋄ ← i+ 1 ← − 1 end while end if ← − = 0 |
- Notations used in Algorithm 1:
- Rc :Results of the model at an instance i
- Step1: Real-time dataset of size is imported from the cloud server in the form of chunks such as ;
- Step2: We check if the size of the dataset is greater than or equal to chunk size. Then, each chunk is tested over our optimized proposed model;
- Step3: For every instance, previous results were updated with a new result ;
- Step4: Lastly, we updated the size of the dataset ← − . If it was still greater, then the chunk size algorithm starts from step1 again.
4. Experimental Results and Its Analysis in Real-Time Environment
4.1. Real-Time Qualitative Analysis
- Learning Rate: states how many weights are modified in the loss gradient model;
- Batch Size: Specified are run through the model at any particular moment;
- Epochs: Defines the number of times the machine learning model is performed on the same dataset;
- Dropout: Is a method of regularization to reduce the issue of overfitting by dropping some of the random nodes during the training phase and improving generalization error in NN.
4.2. Real-Time Quantitative Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sneha, N.; Gangil, T. Analysis of diabetes mellitus for early prediction using optimal features selection. J. Big Data 2019, 6, 13. [Google Scholar] [CrossRef]
- Allam, F.; Nossai, Z.; Gomma, H.; Ibrahim, I.; Abdelsalam, M. A recurrent neural network approach for predicting glucose concentration in type-1 diabetic patients. In Engineering Applications of Neural Networks; Springer: Berlin/Heidelberg, Germany, 2011; pp. 254–259. [Google Scholar]
- Ashiquzzaman, A.; Tushar, A.K.; Islam, M.; Shon, D.; Im, K.; Park, J.H.; Lim, D.S.; Kim, J. Reduction of overfitting in diabetes prediction using deep learning neural network. In IT Convergence and Security 2017; Springer: Singapore, 2018; pp. 35–43. [Google Scholar]
- Metzger, B.E.; Coustan, D.R.; Trimble, E.R. Hyperglycemia and adverse pregnancy outcomes. N. Engl. J. Med. 2008, 358, 1991–2002. [Google Scholar]
- Care, D. Medical care in diabetes 2018. Diabet Care 2018, 41, S105–S118. [Google Scholar]
- Bruen, D.; Delaney, C.; Florea, L.; Diamond, D. Glucose sensing for diabetes monitoring: Recent developments. Sensors 2017, 17, 1866. [Google Scholar] [CrossRef] [Green Version]
- Acciaroli, G.; Vettoretti, M.; Facchinetti, A.; Sparacino, G. Calibration of minimally invasive continuous glucose monitoring sensors: State-of-the-art and current perspectives. Biosensors 2018, 8, 24. [Google Scholar] [CrossRef] [Green Version]
- Torres, I.; Baena, M.G.; Cayon, M.; Ortego-Rojo, J.; AguilarDiosdado, M. Use of sensors in the treatment and follow-up of patients with diabetes mellitus. Sensors 2010, 10, 7404–7420. [Google Scholar] [CrossRef]
- Rodríguez-Rodríguez, I.; Zamora-Izquierdo, M.Á.; Rodríguez, J.V. Towards an ict-based platform for type 1 diabetes mellitus management. Appl. Sci. 2018, 8, 511. [Google Scholar] [CrossRef] [Green Version]
- Nieminen, J.; Gomez, C.; Isomaki, M.; Savolainen, T.; Patil, B.; Shelby, Z.; Xi, M.; Oller, J. Networking solutions for connecting bluetooth low energy enabled machines to the internet of things. IEEE Netw. 2014, 28, 83–90. [Google Scholar] [CrossRef]
- Vhaduri, S.; Prioleau, T. Adherence to personal health devices: A case study in diabetes management. In Proceedings of the 14th EAI International Conference on Pervasive Computing Technologies for Healthcare, Atlanta, GA, USA, 18–20 May 2020; ACM Press: New York, NY, USA, 2020; pp. 62–72. [Google Scholar]
- Specifications—Samsung Galaxy Note8. The Official Samsung Galaxy Site. Available online: https://www.samsung.com/global/galaxy/galaxy-note8/specs/ (accessed on 7 July 2020).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Gomez, C.; Oller, J.; Paradells, J. Overview and evaluation of bluetooth low energy: An emerging low-power wireless technology. Sensors 2012, 12, 11734–11753. [Google Scholar] [CrossRef]
- Kumari, V.A.; Chitra, R. Classification of diabetes disease using support vector machine. Int. J. Eng. Res. Appl. 2013, 3, 1797–1801. [Google Scholar]
- Craven, M.W.; Shavlik, J.W. Using neural networks for data mining. Future Gener. Comput. Syst. 1997, 13, 211–229. [Google Scholar] [CrossRef]
- Radhimeenakshi, S. Classification and prediction of heart disease risk using data mining techniques of support vector machine and artificial neural networks. In Proceedings of the 2016 International Conference on Computing for Sustainable Global Development (INDIACom), New Delhi, India, 16–18 March 2016; pp. 3107–3111. [Google Scholar]
- Aslan, M.F.; Unlersen, M.F.; Sabanci, K.; Durdu, A. CNN-based transfer learning–BiLSTM network: A novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021, 98, 106912. [Google Scholar] [CrossRef]
- Dey, S.K.; Hossain, A.; Rahman, M.M. Implementation of a web application to predict diabetes disease: An approach using machine learning algorithm. In Proceedings of the 2018 21st International Conference of Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 21–23 December 2018; pp. 1–5. [Google Scholar]
- Srivastava, S.; Sharma, L.; Sharma, V.; Kumar, A.; Darbari, H. Prediction of diabetes using artificial neural network approach. In Engineering Vibration, Communication and Information Processing; Springer: Singapore, 2019; pp. 679–687. [Google Scholar]
- Radha, P.; Srinivasan, B. Predicting diabetes by cosequencing the various data mining classification techniques. Int. J. Innov. Sci. Eng. Technol. 2014, 1, 334–339. [Google Scholar]
- Karegowda, A.G.; Manjunath, A.S.; Jayaram, M.A. Application of genetic algorithm optimized neural network connection weights for medical diagnosis of pima Indians diabetes. Int. J. Soft Comput. 2011, 2, 15–23. [Google Scholar] [CrossRef]
- Zolfaghari, R. Diagnosis of diabetes in female population of pima indian heritage with ensemble of bp neural network and svm. Int. J. Comput. Eng. Manag. 2012, 15, 2230–7893. [Google Scholar]
- Sanakal, R.; Jayakumari, T. Prognosis of diabetes using data mining approach-fuzzy c means clustering and support vector machine. Int. J. Comput. Trends Technol. 2014, 11, 94–98. [Google Scholar] [CrossRef]
- Zhang, Y. Support vector machine classification algorithm and its application. In Proceedings of the International Conference on Information Computing and Applications, Chengdu, China, 14–16 September 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 179–186. [Google Scholar]
- Karatsiolis, S.; Schizas, C.N. Region based support vector machine algorithm for medical diagnosis on pima indian diabetes dataset. In Proceedings of the 12th International Conference on Bioinformatics & Bioengineering (BIBE), Larnaca, Cyprus, 11–13 November 2012; pp. 139–1442. [Google Scholar]
- Jarullah, A.; Asma, A. Decision tree discovery for the diagnosis of type II diabetes. In Proceedings of the 2011 International Conference on Innovations in Information Technology, Abu Dhabi, United Arab Emirates, 25–27 April 2011; pp. 303–307. [Google Scholar]
- Zou, Q.; Qu, K.; Luo, Y.; Yin, D.; Ju, Y.; Tang, H. Predicting diabetes mellitus with machine learning techniques. Front. Genet. 2018, 9, 515. [Google Scholar] [CrossRef]
- Saji, S.A.; Balachandran, K. Performance analysis of training algorithms of multilayer perceptrons in diabetes prediction. In Proceedings of the International Conference on Advances in Computer Engineering and Applications, Ghaziabad, India, 19–20 March 2015; pp. 201–206. [Google Scholar]
- Jahangir, M.; Afzal, H.; Ahmed, M.; Khurshid, K.; Nawaz, R. An expert system for diabetes prediction using auto tuned multi-layer perceptron. In Proceedings of the Intelligent Systems Conference (IntelliSys), London, UK, 7–8 September 2017; pp. 722–728. [Google Scholar]
- Kannadasan, K.; Edla, D.R.; Kuppili, V. Type 2 diabetes data classification using stacked autoencoders in deep neural networks. Clin. Epidemiol. Glob. Health 2019, 7, 530–535. [Google Scholar] [CrossRef] [Green Version]
- Apoorva, S.; Aditya, S.K.; Snigdha, P.; Darshini, P.; Sanjay, H.A. Prediction of diabetes mellitus type-2 using machine learning. In Proceedings of the International Conference on Computational Vision and Bio Inspired Computing, Coimbatore, India, 19–20 November 2019; Springer: Cham, Germany, 2019; pp. 364–370. [Google Scholar]
- Kamble, A.K.; Manza, R.R.; Rajput, Y.M. Review on diagnosis of diabetes in pima indians. Int. J. Comput. Appl. 2016, 975, 8887. [Google Scholar]
- Kotsiantis, S.B. Decision trees: A recent overview. Artif. Intell. Rev. 2013, 39, 261–283. [Google Scholar] [CrossRef]
- Swapna, G.; Vinayakumar, R.; Soman, K. Diabetes detection using deep learning algorithms. ICT Express 2018, 4, 243–246. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Massaro, A.; Maritati, V.; Giannone, D.; Convertini, D.; Galiano, A. LSTM DSS automatism and dataset optimization for diabetes prediction. Appl. Sci. 2019, 9, 3532. [Google Scholar] [CrossRef] [Green Version]
- Taspinar, Y.S.; Cinar, I.; Koklu, M. Classification by a stacking model using CNN features for COVID-19 infection diagnosis. J. X-ray Sci. Technol. 2021, 30, 73–88. [Google Scholar] [CrossRef]
- Rahman, M.; Islam, D.; Mukti, R.J.; Saha, I. A deep learning approach based on convolutional lstm for detecting diabetes. Comput. Biol. Chem. 2020, 88, 107329. [Google Scholar] [CrossRef]
- Rahman, M.M.; Roy, C.K.; Kula, R.G. Predicting usefulness of code review comments using textual features and developer experience. In Proceedings of the 2017 IEEE/ACM 14th International Conference on Mining Software Repositories (MSR), Buenos Aires, Argentina, 20–21 May 2017; pp. 215–226. [Google Scholar]
- Shetty, D.; Rit, K.; Shaikh, S.; Patil, N. March. Diabetes disease prediction using data mining. In Proceedings of the 2017 International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS), Coimbatore, India, 17–18 March 2017; pp. 1–5. [Google Scholar]
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; IEEE: Piscataway, NJ, USA, 1995; pp. 388–391. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Alghamdi, M.; Al-Mallah, M.; Keteyian, S.; Brawner, C.; Ehrman, J.; Sakr, S. Predicting diabetes mellitus using SMOTE and ensemble machine learning approach: The Henry Ford Exercise Testing (FIT) project. PLoS ONE 2017, 12, e017980. [Google Scholar] [CrossRef]
- Hemanth, D.J.; Deperlioglu, O.; Kose, U. An enhanced diabetic retinopathy detection and classification approach using deep convolutional neural network. Neural Comput. Appl. 2020, 32, 707–721. [Google Scholar] [CrossRef]
- Budreviciute, A.; Damiati, S.; Sabir, D.K.; Onder, K.; Schuller-Goetzburg, P.; Plakys, G.; Katileviciute, A.; Khoja, S.; Kodzius, R. Management and prevention strategies for non-communicable diseases (NCDs) and their risk factors. Front. Public Health 2020, 788. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 1 June 2015; pp. 448–456. [Google Scholar]
- Livieris, I.E.; Pintelas, E.; Pintelas, P. A cnn–lstm model for gold price time-series forecasting. Neural Comput. Appl. 2020, 32, 17351–17360. [Google Scholar] [CrossRef]
- Sun, Q.; Jankovic, M.V.; Bally, L.; Mougiakakou, S.G. Predicting blood glucose with an lstm and bi-lstm based deep neural network. In Proceedings of the 2018 14th Symposium on Neural Networks and Applications (NEUREL), Belgrade, Serbia, 20–21 November 2018; pp. 1–5. [Google Scholar]
- Orabi, K.M.; Kamal, Y.M.; Rabah, T.M. Early predictive system for diabetes mellitus disease. In Proceedings of the Industrial Conference on Data Mining, New York, NY, USA, 13–17 July 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 420–427. [Google Scholar]
- Rahman, M.; Siddiqui, F.H. An optimized abstractive text summarization model using peephole convolutional LSTM. Symmetry 2019, 11, 1290. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.; Vishwakarma, D.K. A deeply coupled convnet for human activity recognition using dynamic and rgb images. Neural Comput. Appl. 2021, 33, 469–485. [Google Scholar] [CrossRef]
- Rathor, S.; Agrawal, S. A robust model for domain recognition of acoustic communication using bidirectional lstm and deep neural network. Neural Comput. Appl. 2021, 33, 11223–11232. [Google Scholar] [CrossRef]
- Tama, B.A.; Lee, S. Comments on “stacking ensemble based deep neural networks modeling for effective epileptic seizure detection”. Expert Syst. Appl. 2021, 184, 115488. [Google Scholar] [CrossRef]
- Temurtas, H.; Yumusak, N.; Temurtas, F. A comparative study on diabetes disease diagnosis using neural networks. Expert Syst. Appl. 2009, 36, 8610–8615. [Google Scholar] [CrossRef]
- Gill, N.S.; Mittal, P. A computational hybrid model with two level classification using svm and neural network for predicting the diabetes disease. J. Theor. Appl. Inf. Technol. 2016, 87, 1–10. [Google Scholar]
- Yuvaraj, N.; SriPreethaa, K. Diabetes prediction in healthcare systems using machine learning algorithms on hadoop cluster. Clust. Comput. 2019, 22, 1–9. [Google Scholar] [CrossRef]
- Swapna, G.; Kp, S.; Vinayakumar, R. Automated detection of diabetes using CNN and CNN-LSTM network and heart rate signals. Procedia Comput. Sci. 2018, 132, 1253–1262. [Google Scholar]
- Lee, K.W.; Ching, S.M.; Ramachandran, V.; Yee, A.; Hoo, F.K.; Chia, Y.C.; Wan Sulaiman, W.A.; Suppiah, S.; Mohamed, M.H.; Veettil, S.K. Prevalence and risk factors of gestational diabetes mellitus in Asia: A systematic review and meta-analysis. BMC Pregnancy Childbirth 2018, 18, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Christobel, Y.A.; Sivaprakasam, P. A new classwise k nearest neighbor (CKNN) method for the classification of diabetes dataset. Int. J. Eng. Adv. Technol. 2013, 2, 396–400. [Google Scholar]
- George, G.; Lal, A.M.; Gayathri, P.; Mahendran, N. Comparative study of machine learning algorithms on prediction of diabetes mellitus disease. J. Comput. Theor. Nanosci. 2020, 17, 201–205. [Google Scholar] [CrossRef]
- Sivanesan, R.; Dhivya, K.D.R. A review on diabetes mellitus diagnoses using classification on Pima Indian diabetes data set. Int. J. Adv. Res. Comput. Sci. Manag. Stud. 2017, 5, 12–17. [Google Scholar]
- Naz, H.; Ahuja, S. Deep learning approach for diabetes prediction using PIMA Indian dataset. J. Diabetes Metab. Disord. 2020, 19, 391–403. [Google Scholar] [CrossRef]
- Polat, K.; Güneş, S. An expert system approach based on principal component analysis and adaptive neuro-fuzzy inference system to diagnosis of diabetes disease. Digit. Signal Process. 2007, 17, 702–710. [Google Scholar] [CrossRef]
- Haritha, R.; Babu, D.S.; Sammulal, P. A Hybrid Approach for Prediction of Type-1 and Type-2 Diabetes using Firefly and Cuckoo Search Algorithms. Int. J. Appl. Eng. Res. 2018, 13, 896–907. [Google Scholar]
- Mohammad, S.; Dadgar, H.; Kaardaan, M. A Hybrid Method of Feature Selection and Neural Network with Genetic Algorithm to Predict Diabetes. Int. J. Mechatron. Electr. Comput. Technol. 2017, 7, 3397–3404. [Google Scholar]
- Chen, W.; Chen, S.; Zhang, H.; Wu, T. A hybrid prediction model for type 2 diabetes using K-means and decision tree. In Proceedings of the IEEE International Conference on Software Engineering and Service Sciences, ICSESS, Beijing, China, 24–26 November 2017; pp. 386–390. [Google Scholar] [CrossRef]
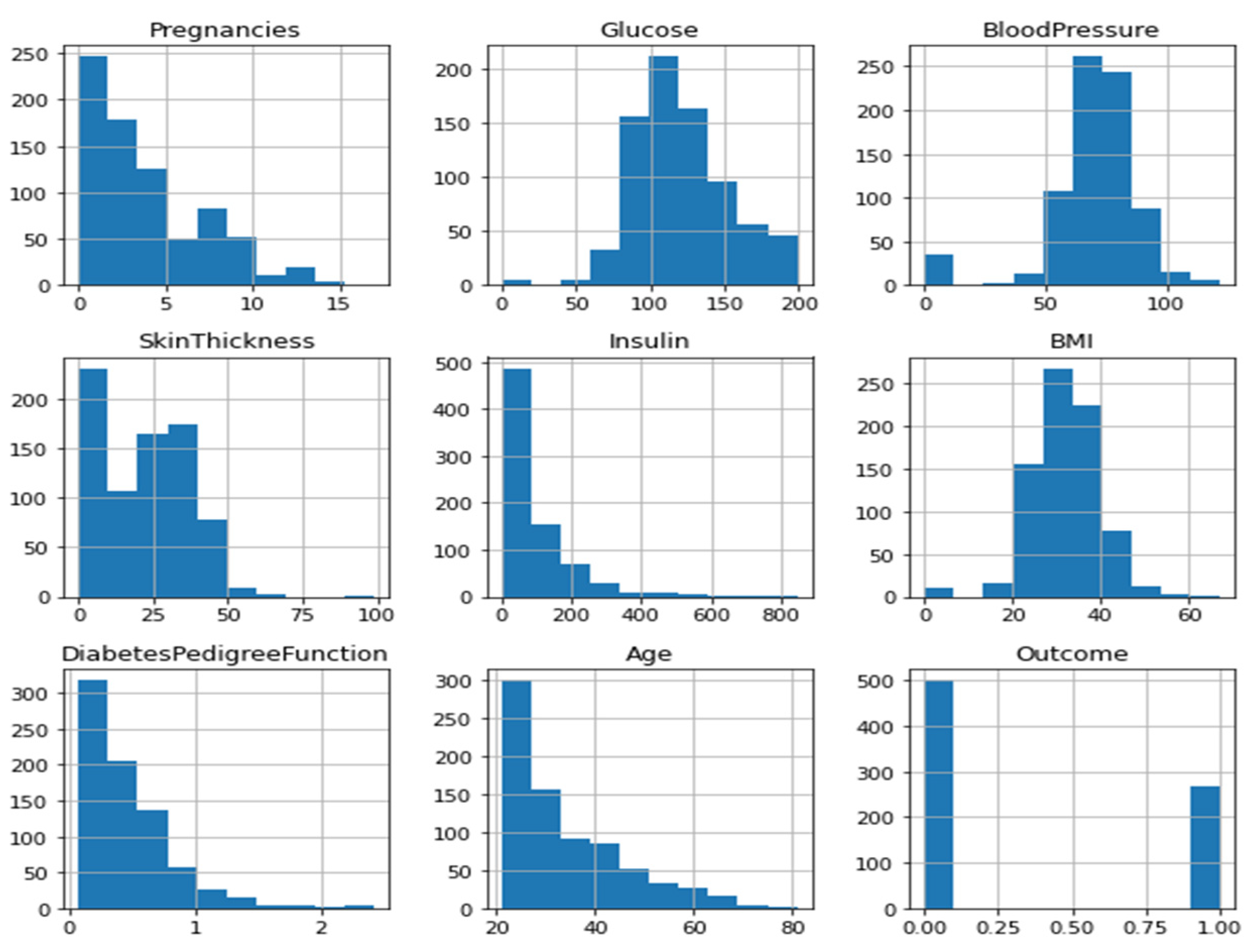











| S.no | Parameters | Description of Parameters | Range |
|---|---|---|---|
| 1. | Pregnancies | No. of times pregnant | 0–17 |
| 2. | Glucose | Plasma glucose 2 h in an oral glucose tolerance test (mg/dl) | 0–199 |
| 3. | Blood-pressure | Diastolic blood pressure (mm Hg) | 0–122 |
| 4. | Skin Thickness | Skin fold thickness (mm) | 0–99 |
| 5. | Insulin | 2-h serum insulin (mu U/mL) | 0–846 |
| 6. | BMI | (weight in kg/(height in m)2) | 0–67.1 |
| 7. | Diabetes-Pedigree | Diabetes pedigree function (weight in kg/(height in m)2) | 0.08–2.42 |
| 8. | Age | Age (years) | 21–81 |
| S.no | Attributes | Missing Values |
|---|---|---|
| 1. | Pregnancies | 0 |
| 2. | Glucose | 5 |
| 3. | Blood-pressure | 35 |
| 4. | Skin Thickness | 227 |
| 5. | Insulin | 374 |
| 6. | BMI | 11 |
| 7. | Diabetes-Pedigree | 0 |
| 8. | Age | 0 |
| S.no | Attributes | IQR Threshold Values |
|---|---|---|
| 1. | Pregnancies | 5 |
| 2. | Glucose | 41.25 |
| 3. | Blood-pressure | 18 |
| 4. | Skin Thickness | 32 |
| 5. | Insulin | 27.25 |
| 6. | BMI | 9.3 |
| 7. | Diabetes-Pedigree | 0.38 |
| 8. | Age | 17.0 |
| 9. | Outcome | 1 |
| S.no | Parameters | Mean | Standard Deviation | Minimum | Maximum |
|---|---|---|---|---|---|
| 1. | Pregnancies | 3.84 | 3.36 | 0 | 17 |
| 2. | Glucose | 121.6 | 30.46 | 44 | 199 |
| 3. | Blood-pressure | 74.8 | 16.68 | 24 | 142 |
| 4. | Skin Thickness | 56.89 | 44.51 | 7 | 142 |
| 5. | Insulin | 139.42 | 87.24 | 14 | 846 |
| 6. | BMI | 33.63 | 12.22 | 18 | 142 |
| 7. | Diabetes-Pedigree | 0.47 | 0.33 | 0 | 2.42 |
| 8. | Age | 33.24 | 11.76 | 21 | 81 |
| 9. | Outcome | 0.34 | 0.47 | 0 | 1 |
| S.no | Features | Chi-Squared Test | Extra Trees | LASSO |
|---|---|---|---|---|
| 1. | No of Pregnancies | 110.54 | 0.105 | 0.00 |
| 2. | Glucose | 1537.20 | 0.230 | 0.0065 |
| 3. | Blood-pressure | 54.26 | 0.089 | 0.0 |
| 4. | Skin Thickness | 145.20 | 0.090 | 0.0 |
| 5. | Insulin | 6779.24 | 0.147 | 0.0004 |
| 6. | BMI | 108.32 | 0.123 | 0.0006 |
| 7. | Age | 189.30 | 0.128 | 0.0002 |
| 8. | Diabetes-Pedigree | 4.30 | 0.128 | 0.0 |
| S.no | Layers (Type) | Filters | Output Shape | Param |
|---|---|---|---|---|
| 1. | Sequential model with input_shape (None, 1, 6) | - | - | - |
| 2. | CONV1D | F = 64, Kernel = [1 1] | (None, None, 1, 64) | 576 |
| 3. | BatchNormalization | - | (None, None, 1, 64) | 256 |
| 4. | MaxPooling | Filter_size = [1 1] | (None, None,1, 64) | 0 |
| 5. | Flattening | - | (None, None, 64) | 0 |
| 6. | Bi-LSTM | - | (None, 128) | 66,048 |
| 7. | Dropout | 50% | (None, 128) | 0 |
| 8. | Dense | (None, 1) | 129 | |
| 9. | Classification | - | - |
| Learning Rate | Batch Size | Hidden Units | Epochs | Mean Test Score |
|---|---|---|---|---|
| 0.01 | 32 | 32 | 250 | 90.38 |
| 0.03 | 64 | 64 | 300 | 85.58 |
| 0.05 | 128 | 128 | 500 | 79.49 |
| 0.09 | 64 | 64 | 300 | 89.0 |
| S.no | Parameters | Values |
|---|---|---|
| 1. | Regularization (dropouts) | 0.05 |
| 2. | Loss-function | Binary-crossentropy |
| 3. | Optimizer | Adam |
| 4. | Metrics | accuracy |
| 5. | Learning Rate | 0.01 |
| 6. | Batch-size | 32 |
| 7. | Epochs | 250 |
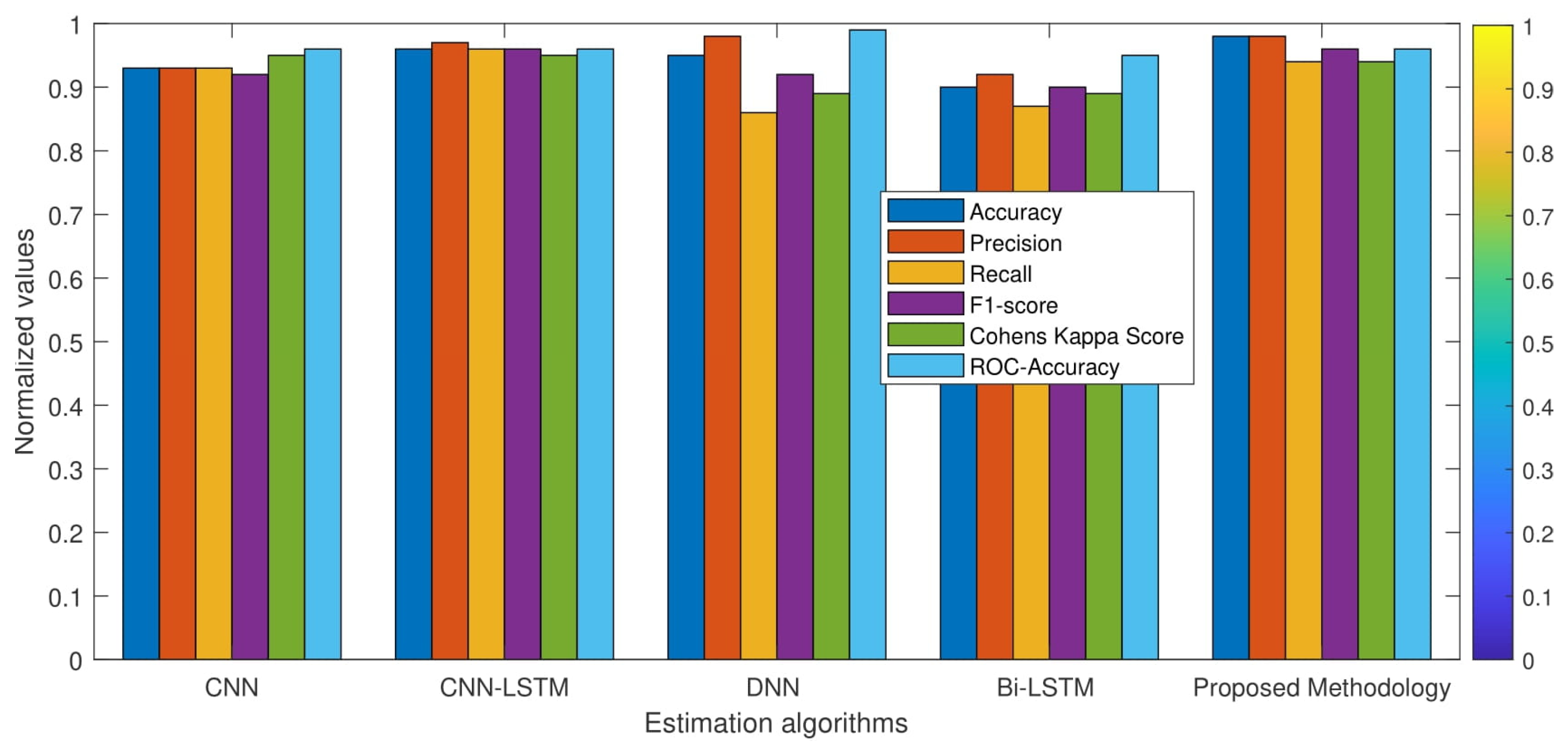
| S.no | Accuracy Measures | CNN-LSTM [26] | CNN [27] | Bi-LSTM [1] | Dense-NN [1] | Proposed Method |
|---|---|---|---|---|---|---|
| 1. | Accuracy | 90 | 82 | 85 | 87 | 88.37 |
| 2. | Precision | 0.84 | 0.72 | 0.82 | 0.84 | 0.85 |
| 3. | Recall | 0.79 | 0.77 | 0.70 | 0.75 | 0.79 |
| 4. | F1-score | 0.83 | 0.74 | 0.75 | 0.79 | 0.81 |
| 5. | Cohens Kappa Score | 0.76 | 0.61 | 0.64 | 0.70 | 0.73 |
| 6. | ROC-Accuracy | 0.89 | 0.87 | 0.87 | 0.91 | 0.90 |
| S.no | Accuracy Measures | CNN-LSTM [26] | CNN [27] | Bi-LSTM [1] | Dense-NN [1] | Proposed Method |
|---|---|---|---|---|---|---|
| 1. | Accuracy | 93 | 96 | 95 | 90 | 98.85 |
| 2. | Precision | 0.93 | 0.97 | 0.97 | 0.92 | 0.98 |
| 3. | Recall | 0.93 | 0.96 | 0.86 | 0.87 | 0.94 |
| 4. | F1-score | 0.92 | 0.96 | 0.92 | 0.90 | 0.96 |
| 5. | Cohens Kappa Score | 0.95 | 0.95 | 0.89 | 0.89 | 0.94 |
| 6. | ROC-Accuracy | 0.96 | 0.96 | 0.99 | 0.95 | 0.96 |
| Precision | Recall | F1-Score | |
|---|---|---|---|
| 0 | 0.83 | 0.88 | 0.85 |
| 1 | 0.75 | 0.65 | 0.70 |
| Accuracy | 80% |
| Precision | Recall | F1-Score | |
|---|---|---|---|
| 0 | 0.81 | 0.87 | 0.83 |
| 1 | 0.72 | 0.61 | 0.70 |
| Accuracy | 79% |
| S.no | Accuracy Measures | Mean-Test Score 85.58% | Mean-Test Score 79.49% | Mean-Test Score 89% |
|---|---|---|---|---|
| 1. | Accuracy | 91 | 78 | 67 |
| 2. | Precision | 0.87 | 0.67 | 0 |
| 3. | Recall | 0.82 | 0.67 | 0 |
| 4. | F1-Score | 0.85 | 0.67 | 0 |
| 5. | Cohens Kappa Score | 0.78 | 0.51 | 0 |
| 6. | ROC-Accuracy | 0.92 | 0.83 | 0.28 |
| 7. | Sensitivity | 91% | 84% | 2 |
| 8. | Specificity | 0.87% | 67% | 0 |
| S.no | Author | Dataset | Validation Technique | Algorithm Used | Accuracy |
|---|---|---|---|---|---|
| 1. | Ashiquzzaman (2017) et al. [3] | PIMA Dataset | 0.1 split validation | Deep learning architecture | 88.41% |
| 2. | Kumari et al. [15] | PIMA Dataset | Divide hyperplane | SVM | 77% |
| 3. | K.Kannadasan (2019) et al. [31] | PIMA Dataset | 70% Training, 30% Testing | DNN with auto-encoders | 86% |
| 4. | Massaro (2019) et al. [36] | PIMA Dataset | The training and validation sets are split in an 80/20 ratio | LSTM | 86% |
| 5. | Patil et al. [41] | PIMA Dataset | K-fold cross-validation | PCA, K-Means Algorithm | 73% |
| 6. | Gill and Mittal (2016) et al. [54] | PIMA Dataset | 70% Training, 30% Testing | SVM and NN | 96.09% |
| 7. | Yuvaraj andSriPreethaa (2019) et al. [57] | PIMA Dataset | 70% Training, 30% Testing | RF | 94% |
| 8. | Naz and Ahuja (2017) et al. [63] | PIMA Dataset | The training and validation sets are split in an 80/20 ratio. | ANN | 90.34% |
| 9. | Kemal Polat et al. [64] | PIMA Dataset | K-fold cross-validation | neuro-fuzzy-inference system (ANFIS) | 89.47% |
| 10. | Haritha et al. [65] | PIMA Dataset | 70% Training, 30% Testing | Firefly and Cuckoo Search Algorithms | 80% |
| 11. | Mohammad et al. [66] | PIMA Dataset | 70% Training, 30% Testing | NN with Genetic Algorithm | 86.78% |
| 12. | Chen et al. [67] | PIMA Dataset | 10 fold cross validation | K-means and DT | 91.23% |
| 13. | Proposed Method | PIMA Dataset | K-fold cross-validation | CNN-BI-LSTM | 98.85% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madan, P.; Singh, V.; Chaudhari, V.; Albagory, Y.; Dumka, A.; Singh, R.; Gehlot, A.; Rashid, M.; Alshamrani, S.S.; AlGhamdi, A.S. An Optimization-Based Diabetes Prediction Model Using CNN and Bi-Directional LSTM in Real-Time Environment. Appl. Sci. 2022, 12, 3989. https://doi.org/10.3390/app12083989
Madan P, Singh V, Chaudhari V, Albagory Y, Dumka A, Singh R, Gehlot A, Rashid M, Alshamrani SS, AlGhamdi AS. An Optimization-Based Diabetes Prediction Model Using CNN and Bi-Directional LSTM in Real-Time Environment. Applied Sciences. 2022; 12(8):3989. https://doi.org/10.3390/app12083989
Chicago/Turabian StyleMadan, Parul, Vijay Singh, Vaibhav Chaudhari, Yasser Albagory, Ankur Dumka, Rajesh Singh, Anita Gehlot, Mamoon Rashid, Sultan S. Alshamrani, and Ahmed Saeed AlGhamdi. 2022. "An Optimization-Based Diabetes Prediction Model Using CNN and Bi-Directional LSTM in Real-Time Environment" Applied Sciences 12, no. 8: 3989. https://doi.org/10.3390/app12083989
APA StyleMadan, P., Singh, V., Chaudhari, V., Albagory, Y., Dumka, A., Singh, R., Gehlot, A., Rashid, M., Alshamrani, S. S., & AlGhamdi, A. S. (2022). An Optimization-Based Diabetes Prediction Model Using CNN and Bi-Directional LSTM in Real-Time Environment. Applied Sciences, 12(8), 3989. https://doi.org/10.3390/app12083989










