Abstract
(1) Background: Clinical information modeling tools are software instruments designed to support the definition of semantic structures able to be implemented in health information systems. Based on the analysis of existing tools, this research developed a tool that proposes new approaches to promoting clinician involvement and supporting information modeling processes through mechanisms that ensure governance, information consistency and consensus building. (2) Method: This research developed the MedicalForms system, which is based on the requirements identified in both a Delphi study about tool requirements and the ISO/TS 13972 specifications. (3) Results: This system allows the management of projects, information structures and implementable forms related to clinical documentation. Users can easily define clinical documents in collaboration with the rest of the professionals in their team by being able to reuse previously defined forms, terminologies and information structures. The system is able to export the defined forms as interoperable specifications or as several implementable form formats compatible with multiple open source EHR systems and research platforms. End user perception of this tool was evaluated through the Technology Acceptance Questionnaire with satisfactory results. Finally, the system was applied to develop 12 research registries and 2 clinical trial research forms, 3 mobile applications and 1 decision support system.
1. Introduction
Semantic interoperability in health informatics is a field where multiple technologies and specifications have been defined. Multiple standard development organizations (HL7, ISO and CEN) and other international initiatives (IHE, CDISC, openEHR) work towards defining how information should be structured to allow computer consistent processing and interpretation in well-defined scenarios. As long as Electronic Health Record (EHR) systems are becoming more mature, they are advancing towards supporting patient information flow associated with continuity of care and reusing healthcare data scenarios such as research, epidemiological analysis, business analytics and decision support. As part of this research, we use the term clinical information model (CIM) to describe any semantic structure able to represent how clinical concepts are interrelated, including terminology bindings, to be able to provide the full context to safely process this information in multiple EHR systems or research platforms.
This paper explains the development and validation of a new tool focused on promoting clinician involvement in processes associated with the defined implementable forms and semantic structures for EHR systems through mechanisms that ensure governance, information consistency and consensus building. As a result, improvements in management of semantic relationships between EHR concepts could contribute towards the reuse of clinical data for multiple use cases such as clinical decision support integrated with biomedical knowledge [1], advanced analytics and wearable technologies [2].
1.1. Quality Standards for Developing Clinical Information Models
The clinical information modeling process (CIMP) could be described as an iterative process that includes the analysis of the domain and requirements, designing, implementing, validating and maintaining CIMs. The analysis of how the multiple interoperability resources are defined shows that there are no particular differences on the processes applied for modeling CIMs without regard for the adopted specification [3]. On the other hand, the process carried out for modeling clinical information is identified as a relevant factor to determine the quality of the semantic interoperability resources [4].
In the clinical information modeling field, ISO 13972 Technical Specification describes how to implement quality processes that lead to the adoption of best practices based on the definition of CIMs [5]. Moreover, the standard details a set of testable quality attributes for these resulting models and how to implement a Quality Management System for the CIMP. The implementation of a Quality Management System allows a continuous improvement cycle to be established. The definition of a “Plan, Do, Check, Act” (PDCA) cycle [6] promotes the continuous adaptation of processes and measurements within each of the steps to obtain improved quality in the final product. As a result, a software-based methodology based on the ISO 13972 specification could guide the participation of multiple domain experts in tasks associated with definition, validation and maintenance of CIMs in compliance with defined best practice and support the adoption of a continuous improvement cycle associated with information management.
1.2. Interoperability Specifications and International Initiatives
Some of the most relevant international initiatives are openEHR [7] and the HL7 CIMI working groups [8], which are focused on defining CIMs. These initiatives are focused on obtaining a consistent semantic definition of CIMs able to be implemented in multiple scenarios. They are working on defining CIMs compatible with Archetype Description Language, each of them on their own specific reference model. CIMI is working on the definition of CIMs able to be the CIMI Reference Model Specification and define HL7 FHIR profiles. On the other hand, the openEHR CIMs are known as archetypes based on the openEHR reference models and they are able to be transferred as EHR extracts [7]. In addition, they are working on supporting mappings to HL7 FHIR resources given the increased acceptance of EHR vendors.
There are additional specifications that aim to define how clinical information is structured in order to be transferred between EHR systems. Some of the most relevant specifications are the Clinical Information Model [5], HL7 CDA templates [9] and ISO 13606 [10]. There are national and regional Ministries of Health working on each of these specifications. The CDISC consortium developed several specifications to define how information associated with clinical trials should be structured. They have several standards which are able to define how to satisfy regulatory submission to the Food and Drug Administration (FDA) such as the Operational Data Model (ODM) [11]. Some of their standards are harmonizing element names, definitions and metadata to establish a standardized data collection baseline across all submissions through Clinical Data Acquisition Standards Harmonization (CDASH) [12].
1.3. Clinical Information Modeling Process and Tools
Clinical information modeling tools are software platforms designed to support the processes associated with the definition of CIMs, as well as establish governance for the multiple CIMs applicable within an infrastructure or domain. Currently there are multiple tools such as the openEHR suite, DCM suite and LinkEHR that provide the mechanisms for defining and managing CIMs. A previous evaluation of existing modeling tools showed that they generally have a good adoption of functionalities related to the management EHR specifications, data types, terminology binding and CIM metadata. However, the need to improve tool support for information modeling and software development processes has been identified, especially in those areas related to governance, clinician involvement and optimizing the technical validation of testing processes [13]. In addition, several experts pointed out the need to improve modeling tools to provide better support for the modeling process in an international survey about CIMP [14].
1.4. Implementation as Medical Form for Clinical Care or Research
Instead of focusing only on producing semantic structures addressing the needs defined by existing health informatics specifications, our approach aims to also produce semantic structures able to be widely implemented for use in systems. There are multiple open source initiatives for EHR systems and research platforms that provide mechanisms to define implementable forms. They are able to import and process the defined forms from an external file that specifies how to display multiple items on a screen, including presentation and user interaction functionalities. This research study has identified the following systems as relevant initiatives able to test the implementation of semantic structures for data collection or analytics tasks:
- Diraya is an open source EHR system developed by the Andalusian Health Service able to be shared with any public administrations in all the primary care centers of the region. This system has been deployed to provide care for 8 million people through the full network of 29 hospitals and 1500 Primary care centers distributed throughout the region [15,16].
- RedCap and OpenClinica are the most widespread open source Clinical Trial Management Systems (CMTS). OpenClinica is an open source software that has been used in 3 million clinical trials in 100 different countries [17]. REDCap is an open source software that has been used by 2800 institutions in 120 countries in 500,000 projects [18]. These systems allow the documentation of patients’ clinical information in accordance with FDA regulations.
- TranSmart and i2b2 research platforms are patient cohort analysis systems designed to analyze data obtained through conventional care while ensuring compliance with patient privacy requirements. This system supports the anonymized analysis of specific patient cohorts. As a consequence, it facilitates the generation of new research hypotheses based on the analysis of the patient population treated at their centers [19,20].
- The CroniCare system is a mobile health platform that is focused on establishing mechanisms for an agile and customized development of interventions based on the use of mobile technologies. The system allows users to configure the information, interactions, questionnaires and alarms that each patient will access through the mobile app in order to improve the capacity for self-management, empowerment and control of chronic and multimorbidity patients [21,22].
2. Materials and Methods
2.1. Project Objective
This project aimed to develop and validate a system for information management in the healthcare and the research field. This system should enable the participation of clinicians in CIMP to produce standards-based specifications that support the interoperability of health information systems, as well as implementable forms and semantic structures for EHR systems and research platforms.
We aimed to advance knowledge in areas related to promoting clinician involvement and support CIMP through mechanisms that ensure governance, information consistency and consensus building.
2.2. Requirement Definition
The definition of requirements was based on: (i) CIMP identified as part of the systematic literature review about papers talking about semantic interoperability in EHR systems [3]; (ii) metrics defined as part of the ISO 13972 standard for implementing a Quality Management System [6]; and (iii) essential requirements for modeling tools [23].
2.3. Use Cases and Validation
The MedicalForm system has designed a set of use cases to validate its proper management of semantics for:
- Representing international datasets, structures and forms: This use case will be validated through the definition of semantic structures and forms specified as part of the European Heart Failure Summary defined by SemanticHealthNet project [24].
- Representing datasets, structures and forms for large healthcare providers: This use case will be validated through the definition of semantic structures and forms according to the Andalusian Stroke Integrated Care process EHR [25].
- Export semantic structures as compliance instances with multiple relevant health informatics specifications. This use case will be validated by exporting CIMs as an implementable interoperability specification such as ISO 13606 and CDISC.
- Export semantic structures with W3C semantic web technology stack. This use case will be validated exporting CIMs according to the Web Ontology Language (OWL).
- Define implementable forms for research platforms, EHR systems and mHealth solutions: This use case will be validated by implementing a questionnaires in platforms (ITCBio, openClinica and Redcap), Diraya EHR system and CroniCare mobile app for patient follow-up from hospital [21,22].
- Define a mechanism for reusing collected data in data analytics tools: This use case will be validated with the database configuration in TranSmart and i2b2 analytics tool. As a result, it will be required to be translated to define a semantic structure in the analytics tools in the form of a tree of concepts consistent with the MedicalForm definition in order to ensure the appropriate management of semantics in the performed analysis.
2.4. End User Evaluation
There was a defined questionnaire based on the Technology Acceptance Model [26] addressing user perception through 14 questions about perceived usefulness (U), ease of use, attitude toward using (AU) and behavioral intention to use (IU). The answers for each dimension were collected through a 5-point Likert Scale (from 1—high level of disagreement to 5—high level of agreement). This questionnaire was applied to collect the perceptions from members of the hospital committee for clinical documentation and healthcare professionals who required the definition of the EHR model or research registries in our hospital.
- Group of Members of the Clinical Documentation Committee: The system was presented to a group of end users in a 45-min session in order to obtain a functional evaluation of the system by healthcare professionals, experts in clinical documentation and EHR systems. At the end of the session, the professionals filled in the TAM questionnaire on a voluntary basis.
- Group of Principal Investigators: These users received a 45-min training session and used the tool to define the forms to create a research record on the patients they cared for during their daily practice. As described in the piloting section for the definition, several revisions were made to the forms until reaching the consensus version that would finally be implemented in the registry. After finalizing the definition of the forms, users completed the TAM questionnaire on a voluntary basis.
2.5. Implementation in Real Use
The system was required to be applied in multiple clinical domains to support the specification of how information is structured for research and healthcare scenarios at the Virgen Macarena University Hospital.
3. Results
3.1. Technological Infrastructure
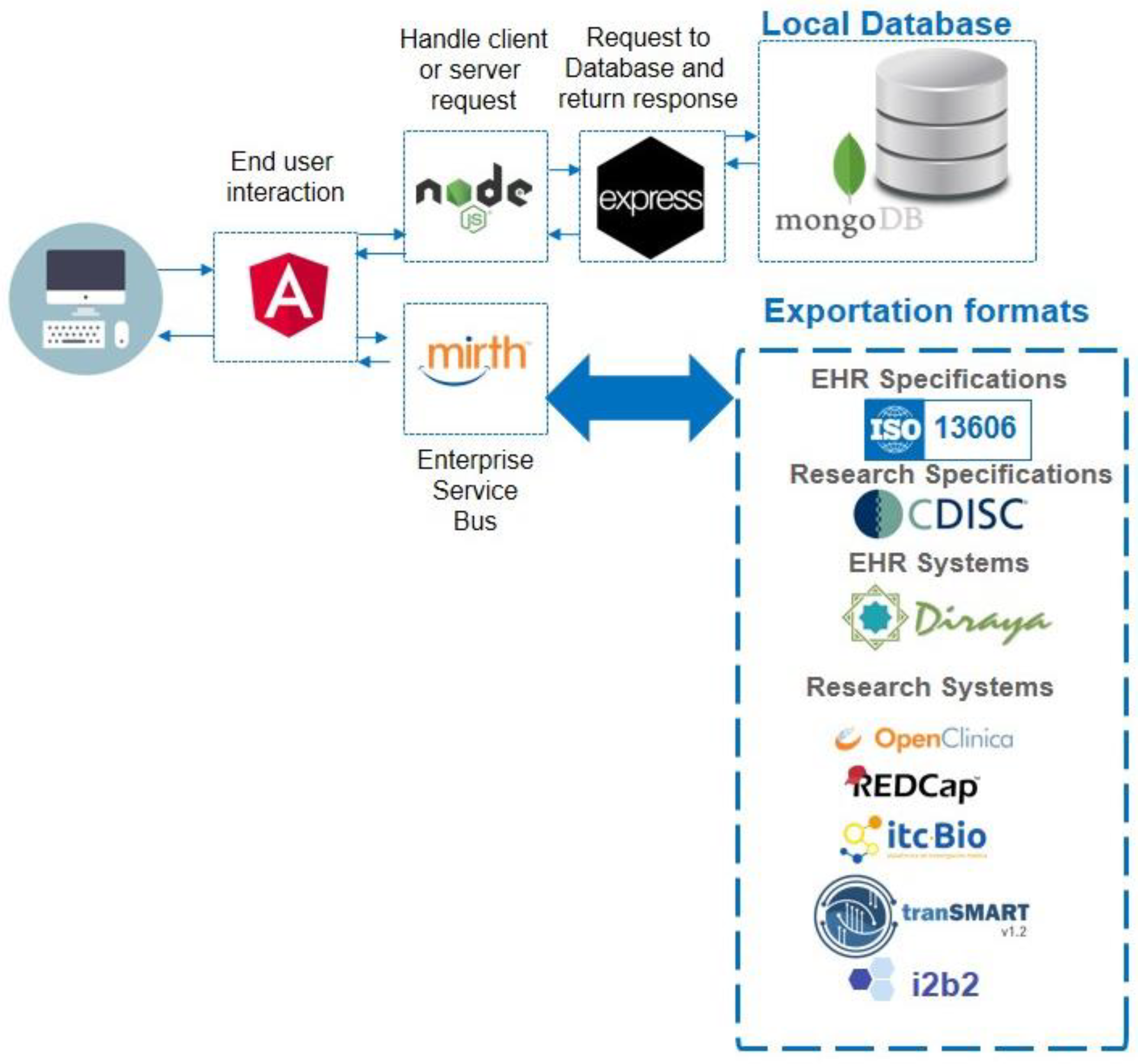
The MedicalForms system was developed on a Linux Centos server with AngularJS framework (frontend), NodeJS (BackEnd) and MongoDB. The system includes an Enterprise Service Bus (MirthConnect) to support model and form exportation to external systems. Figure 1 details the system architecture and exporting formats. The whole system was included as a docker container; this makes it easily deployed. The MedicalForms website provides additional information about how to access and test it [27].
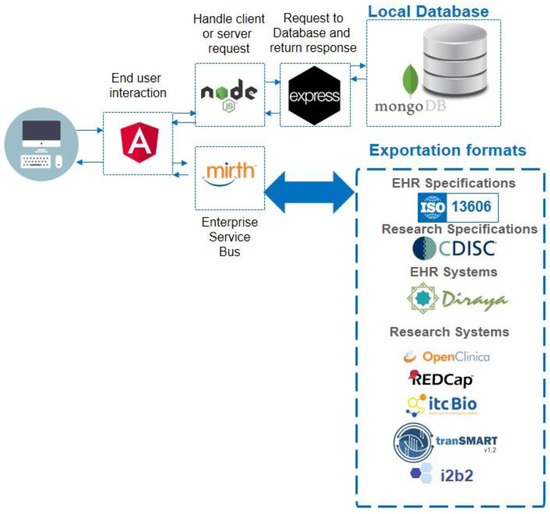
Figure 1.
MedicalForms system architecture, including multiple components for user interaction, handling request, local database and Enterprise Service Bus mappings to produce results compliant with interoperability specifications and relevant opensource systems.
3.2. Requirements Definition
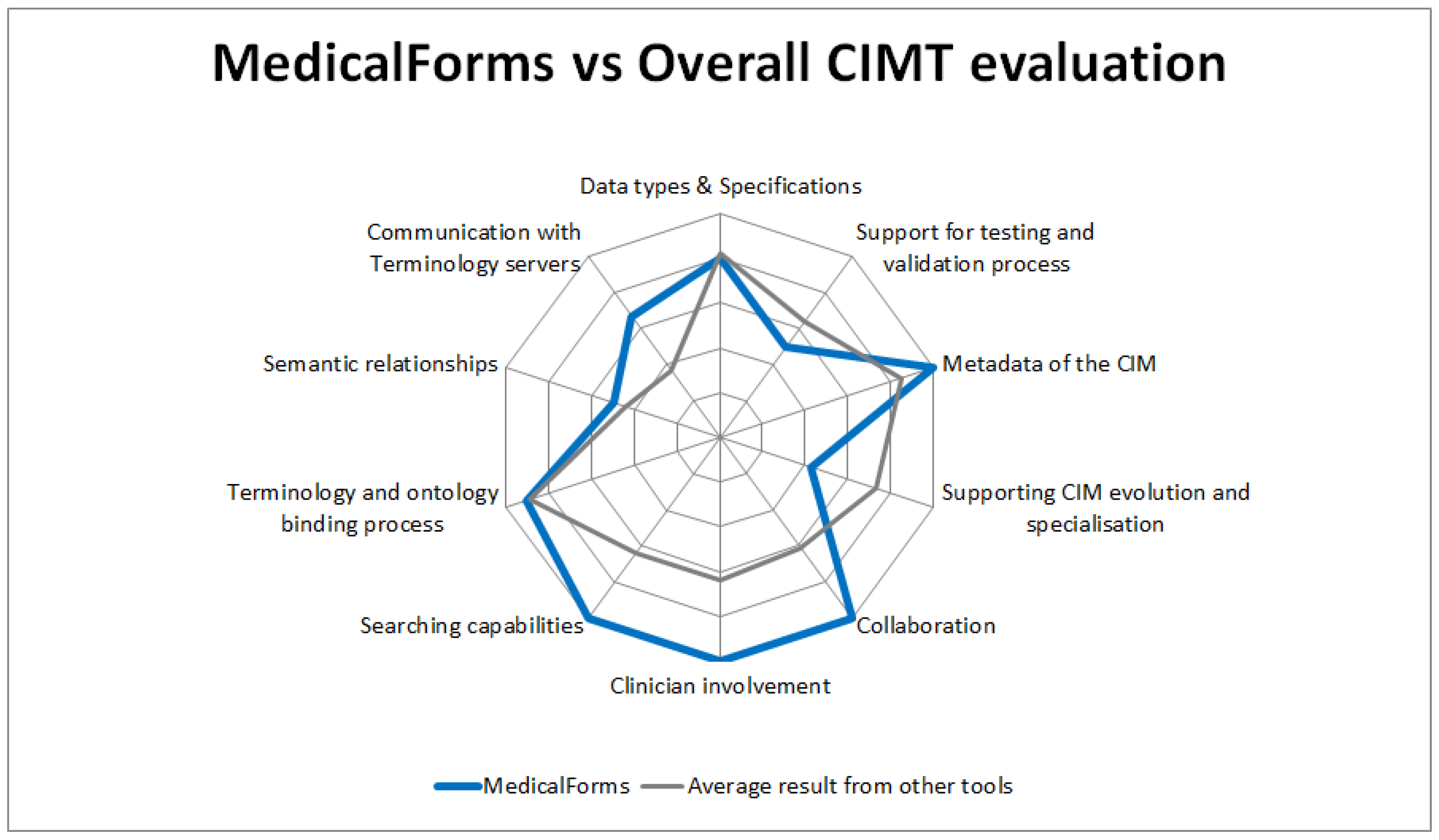
The current version of the MedicalForms system was evaluated against the requirements defined as part of the Delphi study about tool requirements [23] and compared with obtained results from 11 CIMT identified in a previous study [13]. The evaluation presented in Figure 2 shows that the MedicalForms system has:

Figure 2.
Comparison between the evaluation of the MedicalForms system against the requirements for CIMTs defined in a previous Delphi study, and average results from tools included in that study.
- An excellent level of fulfillment: in areas related to clinician involvement, collaboration, searching capabilities and metadata of the CIMs.
- Above average level of compliance: in areas related to communication with terminology servers.
- Average level of compliance: for supporting the management of semantics, as well as the ontology and terminology binding process.
- Below average level of compliance: in areas related to supporting CIM evolution and specialization and support of the testing and validation processes.
3.3. Clinical Information Process Support
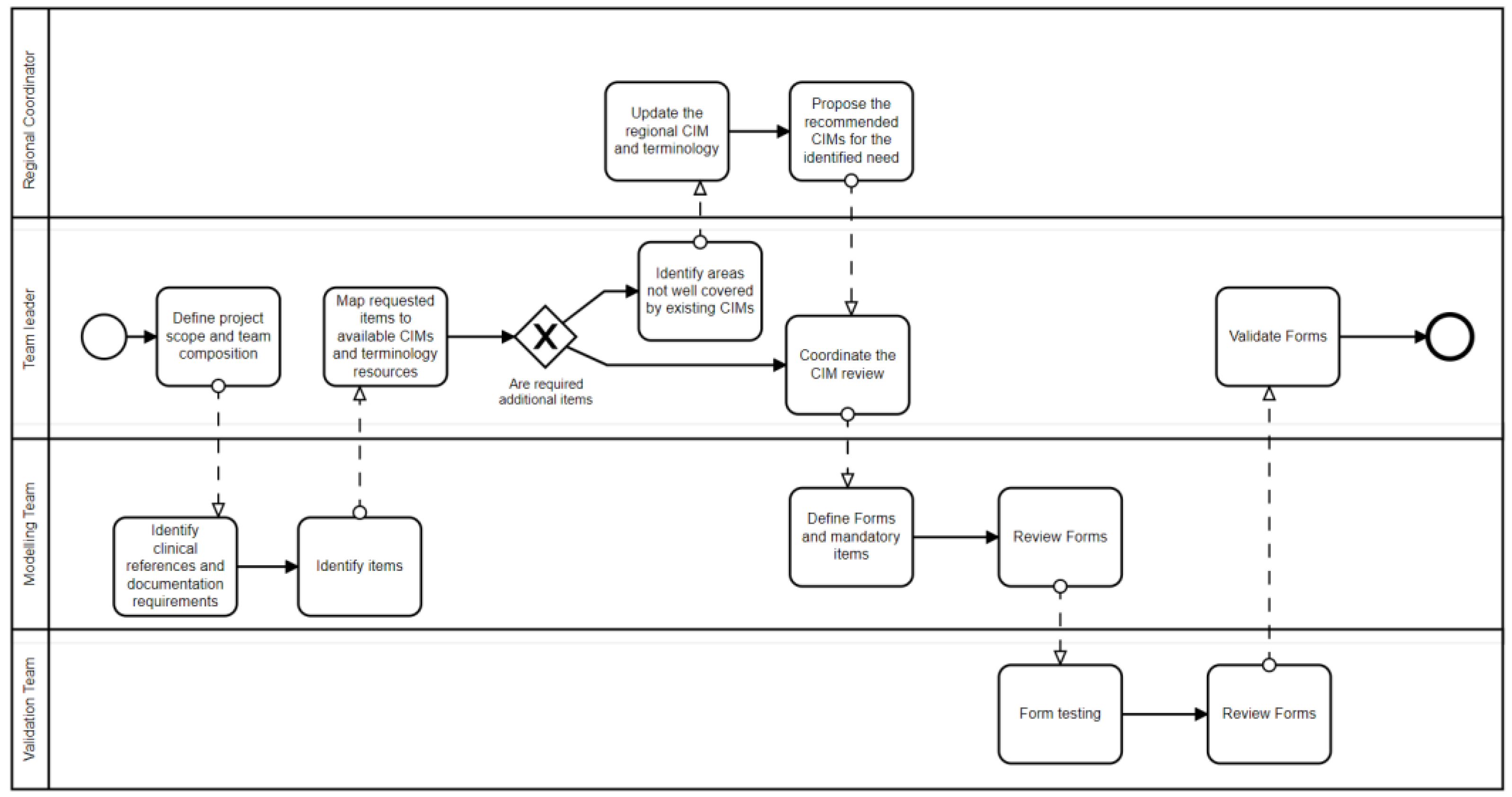
The MedicalForms system was designed as a software instrument to support the coordination of those healthcare professionals involved in the modeling process. Figure 2 details how multiple professionals involved in multiple tasks associated with CIMP could benefit from system functionalities. This system includes roles to classify users up to four levels to support the establishment of governance in large healthcare organizations or international modeling initiatives.
The first level includes the coordinators that ensure the establishment of the information governance process. They coordinate and manage the definition of resources applicable for the multiple healthcare domains in the generic CIM form. The second level includes a project leader that will define the project scope and guide clinicians participating in the CIMP. The third level includes a core team of multidisciplinary experts who work in depth on the detailed clinical and technical needs that the system and CIMs will need to satisfy. The fourth level comprises a larger group of domain experts responsible for validating the proposed clinical document or EHR form. Figure 3 details how multiple roles interact as part of this process.
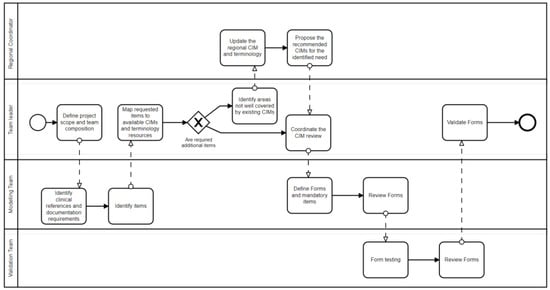
Figure 3.
Representation of the clinical information modeling Process implemented in the MedicalForms system.
3.4. System Description
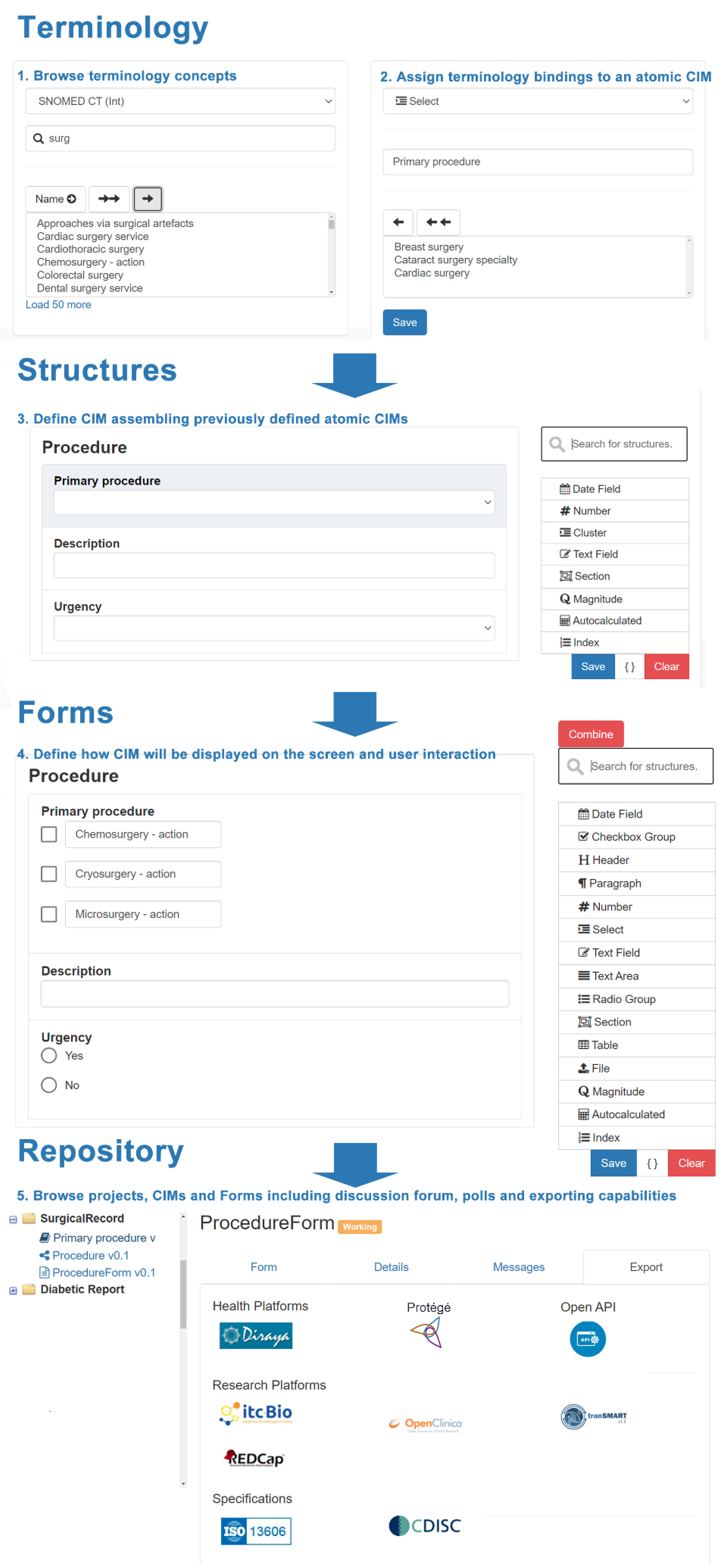
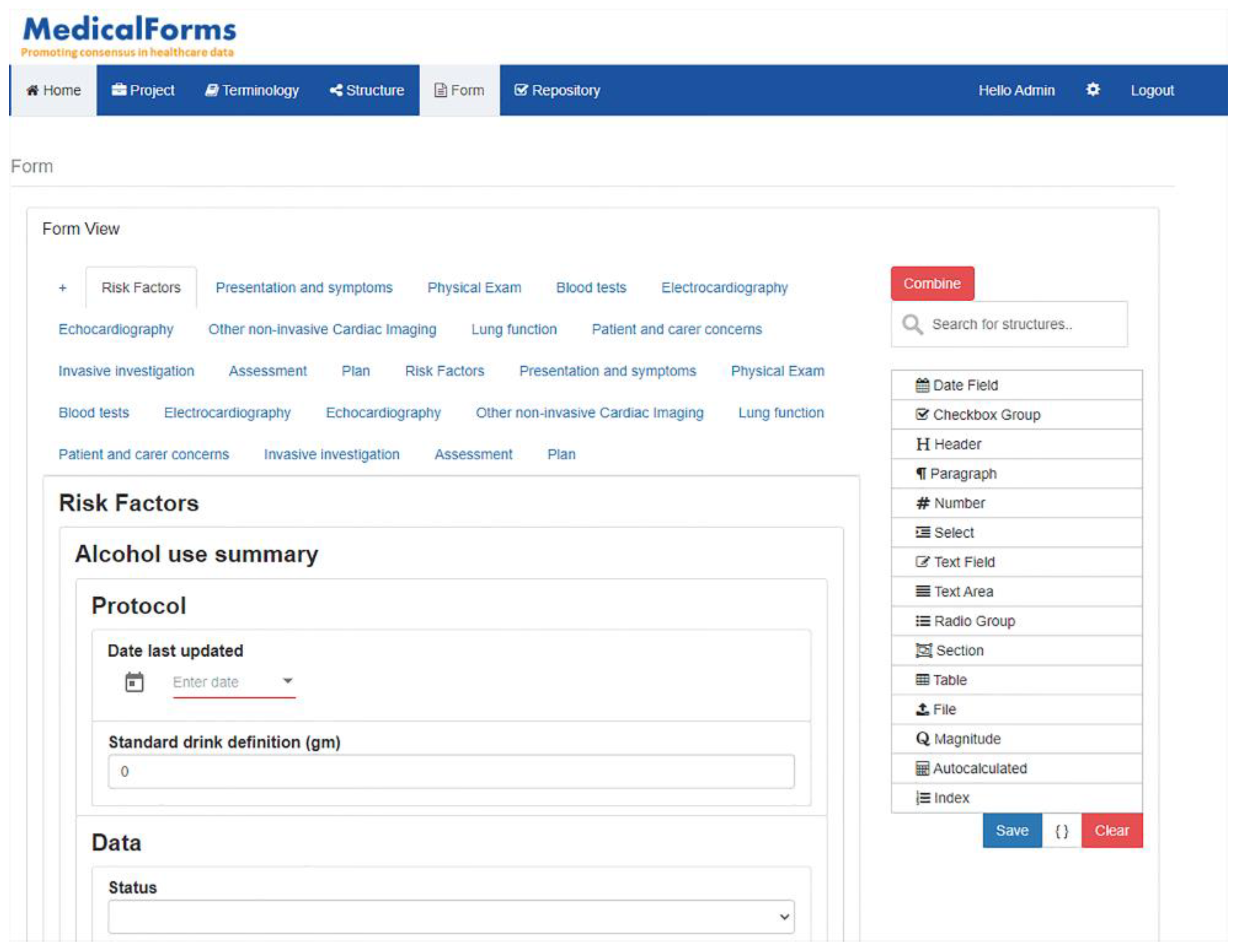
The system was specifically designed to be understood by clinicians and end users without knowledge about technical specifications. They can work on one or multiple projects, including the following areas:
- Terminology: Allows to define atomic semantic structures corresponding to basic data types specifying bindings to terminology concepts. This section allows basic semantic structures to be generated that will be equivalent for the most commonly used data types included in ISO 21090 for dates, number, text, sections, cluster, physical magnitude and autocalculated results. Although it was expected to include additional data types as long as they were required, this subset was enough to address multiple use cases described in future sections.
- Structures: Allows to assemble and organize previously defined atomic structures establishing hierarchical and complex structures. This section includes an additional type of item that allows for defining indexes that provide associated value to a specific answer in a dropdown menu. They are relevant for medical scores where multiple options are mapped to a value rather than a terminology concept.
- Forms: Allows to define how structures are presented on screen detailing user interaction and supporting specialization through combination and post-coordination capabilities. This section adds new elements focused on specifying presentation capabilities for those structures previously defined, including table, radiobutton, checkbox, file and differs between text area and text field.
The system includes a repository where users can navigate between the defined semantic structures and forms. Figure 4 details how the system allows the creation and reuse of semantic structures and forms, and the multiple options for exportation.
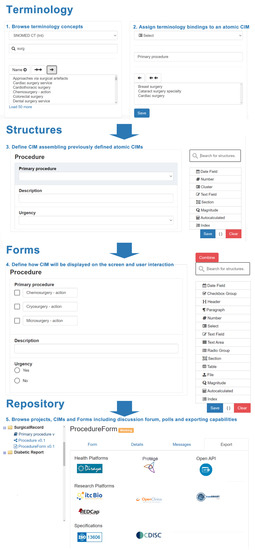
Figure 4.
Details for the clinical information modeling process to define an implementable form.
3.5. Semantic Infrastructure
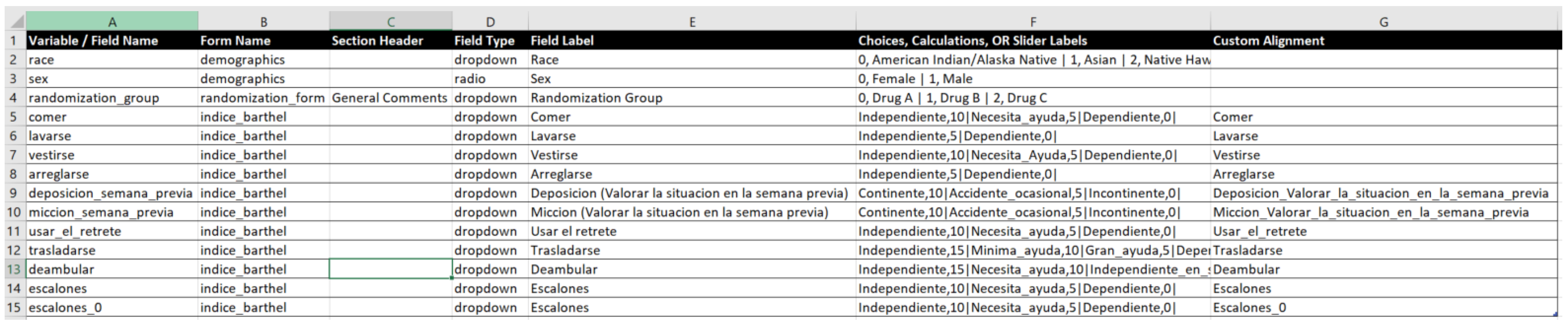
In order to support the definition of terminology subsets, structures and forms, the MedicalForms system defined a set of basic elements. The following table (Table 1) describes the most relevant classes of the MedicalForms JSON information structure. This structure has been defined according to the needs for specifying multiple elements able to be included in MedicalForms according to the type of interaction with the end user (type class), how many items can be interrelated (typeStructure class) and its representation on the screen (typeShow class). Appendix B. Semantic Infrastructure description describes the multiple elements included as part of the MedicalForms system specifying its associated attributes and how they are mapped to ISO 21090 Datatypes, ISO 13606 and openClinica CTMS.

Table 1.
Description of the most relevant classes of the MedicalForms JSON information structure.
3.6. Semantics Capabilities for Information Consistency
The system allows the establishment of a highly controlled CIMP where the project leaders and editorial team are allowed to generate any semantic structure or form based on the existing terminologies already incorporated in the system. Once the need for a concept that is not included in the existing terminologies is identified, the coordinator is required to be contacted for additions. This will ensure that all data items collected will be consistent with the terminologies approved and well known in an organization.
The system includes simplified functionality for post-coordinating multiple terminology concepts. This allows us to define semantic structures based on pre-coordinated terminology concepts that are post-coordinated when they are implemented as screen forms. This is a very relevant point as semantic structures based on pre-coordinated concepts are able to support a larger number of clinical scenarios with easier management of semantics to support aggregation and compatibility for data exploitation and interoperability. On the other hand, clinicians prefer to use post-coordinated terms when they are working on a specialized domain since it reduces the number of items that need to be recorded in the form. Current initiatives such as CIMI WG are working with a similar approach defining iso-semantic transformation from the precoordinated CIM to each specific scenario implemented.
A library was developed containing generic CIMs complemented with educational material about how to manage with consistency negation, thresholds, indexes and calculation questionnaires, as well as options for others in multiple option questions.
3.7. Promoting the Participation of Clinicians in CIMP
The MedicalForms system was designed with special focus on supporting the participation of clinicians as part of the CIMP. Rather than applying technical concepts from EHR specifications, we chose the words “terminologies”, “structures” and “forms” to explain the modeling process emphasizing the benefits from reusing the library of available structures able to be specialized when they are adapted in forms. User interface was simplified to allow clinicians to drag and drop any structure within a CIM in order to support freedom in the definition. Lastly, functionalities designed to support consensus building were included. There is a forum associated with each CIM and a survey tool that helps to obtain feedback from multiple clinical experts working in a project.
3.8. Validating the Management of Semantics for International and Large Healthcare Provider Use Cases
The semantic validation was performed based on the Heart Failure Summary developed within SemanticHealthNet and represented as openEHR archetypes and templates. As part of this project, 18 openehr archetypes were applied to develop a template focused on representing a clinically-orientated summary of a first visit to a consultant-led heart failure clinic (e.g., as a clinic visit record or letter to the GP) but meeting the needs of research/audit and care pathways where possible. Figure 5 shows how the Heart Failure Summary has been implemented, including multiple tabs for each of the main sections of the form. In addition, there were two structures generated and one form according to according to the defined Andalusian Stroke Integrated Care process to validate the system capabilities to address large healthcare provider needs
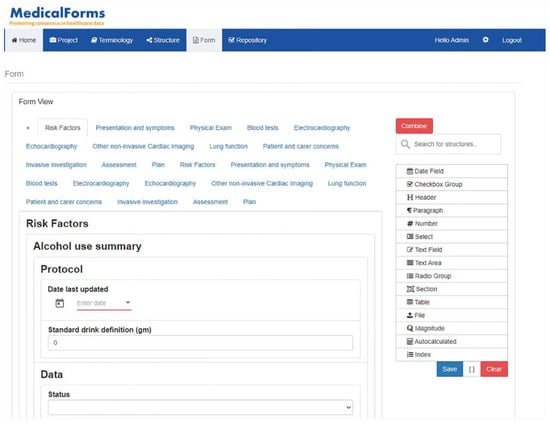
Figure 5.
Representation of the Heart Failure Clinic’s First Visit Summary Form with multiple tabs for each section (v.1).
3.9. End User Perception
Next, Table 2 presents the perception from a group of nine members of the Clinical Documentation Committee (group 1) and a group of six principal investigators (group 2) collected through the Technology Acceptance Questionnaire. Global results show that both groups agree with the usefulness (4.2), attitude toward using (4.2) and intention to use (4.1) the tool. Although the overall result in the easy to use dimension was (3.6), there were differences found in the perception of how easy to use the tool was. On the one hand, group 1 only received 45 min of training about the system and resulted in a neutral perception (3.1). On the other hand, group 2 received 45 min of training and later agreed (4.3) that the system was easy to use.

Table 2.
Evaluation of end user perception through Technology Acceptance Model. Each domain provides the mean ± the standard deviation.
3.10. Implementation
The system was applied to the development of 10 research registries, 1 decision support system and 3 mobile apps where our team participated. The included 60 users acted in the following roles: 1 regional coordinator, 6 project leaders, 9 editorial team and 44 validators. Table 3 details the different domains, medical units, participants and resultant forms and semantic structures for each of the projects carried out with the MedicalForms system.

Table 3.
Details of the projects where the MedicalForms system was applied.
4. Discussion
The MedicalForms system successfully managed and ensured semantic consistency in defined EHR specifications and forms for clinical practice and research. It was able to establish a coordinated management of information between EHR, CTMS, analytics platform and mobile apps. In addition, the system demonstrated the potential for reusing standardized approaches across multiple clinical domains. There were no particularities found in any of the clinical domains where the system was applied.
Clinician involvement: This research expects to contribute to guide future evolution of existing CIMTs towards obtaining a greater involvement of clinicians. Our survey based on the TAM questionnaire shows a good level of acceptance. The proposed simplified language that does not require learning new concepts reduces the learning curve associated with other approaches. Our approach confirmed results from a previous international survey [14] pointing out that applying screens and forms as part of the CIP is beneficial for clinicians in understanding system requirements.
Obtaining consensus in large healthcare providers is a time-consuming task. The proposed functionalities for supporting coordination between multiple clinical experts are expected to contribute towards systemizing this process resulting in a reduction of time in the long term.
Combined information modeling process with form design and data analytics: It is possible to find CIMTs that define how the information is to be structured in the screen form (for example, those based on the openEHR template), but these are not capable of producing implementable forms because this specification does not determine the options for representing a cluster such as dropdown, checkbox or radiobutton. Our research aims to provide a complementary approach that is not only focused on the definition of interoperability specifications for EHR communication, but on producing implementable forms for patient care and research with integrated data analytics.
Screen presentation harmonization: The necessity for finding a balance between standardization of common data elements and allowing for innovation, in order to preserve the capability for different interfaces to be created by vendors, has previously been reported [14]. It has been identified that several systems allow the definition of screen forms by importing a configuration file. It is of no surprise that more systems adopt this approach and possible advances in the harmonization of definitions could be implemented.
Semantic infrastructure definition: Our initiative is based on the CIMs since it can be identified as the more generic mechanism to adopt the two-level modeling approach. Based on CIM, we were able to produce forms and semantic structures compatible with existing tools such as openClinica, RedCap, Diraya and TranSMART with a reduced level of complexity compared to existing EHR specifications. In addition, we took care to satisfy requirements included as part of the ISO 13606, openEHR and CDISC reference models. Last, we analyzed existing HL7 specifications (v2, CD and FHIR) and planned a journey to support future compliance as long as we are involved in projects related to these specifications.
Semantic Web technologies: Associated with the capabilities for exporting CIMs in OWL format, it is expected that the MedicalForms system contributes towards the adoption of ontology-based tools. These technologies apply defined relationships between concepts from existing medical ontologies to provide advanced capabilities for clinical decision support, healthcare data analysis, retrieving synthesis information from the entered data and visualization capabilities.
Issues exporting semantic structures as implementable forms: The MedicalForm system was designed with the goal of supporting the definition of CIMs according to multiple EHR interoperability specifications. As a consequence, the system was required to be flexible enough to define semantic structures with no limitation on the number of depth levels that sections can have in compliance with the ISO 13606 and openEHR specifications. This causes certain limitations when we want to export implementable forms in systems such as OpenClinica or Diraya due to the maximum depth of sections in these systems being just one level. Although systems with increased flexibility of depth can be found, they always have a maximum level of depth. This is reasonable since the screen presentation capabilities will not be able to display an unlimited number of subsections. Two approaches were tested to overcome this problem: providing guidance to modelers about the limitation of depth of subsections and providing automatic mapping that reduces the number of sublevels included in the structure.
Issues designing implementable forms for data analytics: Given that the system has been designed to support the establishment of how data will be managed in data collection tools and analytics tools, it was recommended to define from the beginning those items that will be applied in the data analysis. This is especially relevant when data analysis is performed from variables that are derived from transformations and calculations from the original collected data. There were identified benefits from specifying these transformations at the moment of designing data collection since they were allowed to define automatic transformations for data analysis of the collected data.
Limitations and future work: This research was focused initially on the definition of a semantic consistent transformation between multiple EHR specifications with a generic reference model such as openEHR, ISO 13606 and CDISC. Additional work will be required to define a library of resources correctly mapped with other specifications whose reference model is dependent on the information carried out such as HL7 FHIR and HL7 CDA. It is expected that it will be carried out as long as future use cases based on these specifications will emerge. They will be able to benefit from the ongoing cross-mapping work carried out by openEHR and HL7.
5. Conclusions
The MedicalForms system proposes a consistent software-based methodology to support clinical information modeling providing support for terminology subset, information models and forms definition and validation. The system proposes innovative approaches for incorporating clinicians as part of the CIMP supporting the automatic mapping of defined semantic structures to some of the most relevant specifications (openEHR, ISO 13606 and CDISC) and widely deployed tools (OpenClinica, RedCap and Diraya).
System capabilities for semantic management have been validated through the definition of semantic structures and forms in complex international and large healthcare provider scenarios such as those addressed by the European Heart Failure Summary, the defined SemanticHealthNet project and the Andalusian Stroke integrated care process. The evaluation of the MedicalForms system against the requirements defined as part of the Delphi study about tool requirements [23] shows that this tool is above average in areas related to supporting clinician involvement, collaboration, terminology server communication and searching capabilities and metadata of the CIMs.
It has been implemented in a reference hospital that provides hospital and community care services to 480,000 people. The system provides support for developing more than 100 structures and 50 forms in 15 projects related to healthcare and research data collection. As a result, we expect that our system-based methodology could contribute to improve the semantic consistency in information between multiple research projects and healthcare scenarios in large healthcare providers and international scenarios. It is expected that our scalable approach could gradually incorporate new specifications and systems in the future. In addition, our initiative could be identified as a relevant example for other tools focused on clinical information modeling.
6. Patents
The Andalusian Technology Transfer Office registered MedicalForms: Herramienta de Modelado de Información Clínica (1909023179153) through the Save Creative digital copyright registration service.
Author Contributions
Conceptualization, methodology and writing—original draft preparation, A.M.-C. and J.M.-C.; software, evaluation and writing—review and editing, S.S.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partially funded by the Andalusian Ministry of Health and Families, grant numbers PIN-0315-2016, PIN-0441-2017 and PIN-0185-2018.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Virgen Macarena University Hospital and Virgen del Rocio University Hospital (PIN-0315-2016 on 30 December 2016).
Informed Consent Statement
Patient consent was waived because this study was focused on optimizing the definition and implementation of health information systems rather than evaluating individual patient information.
Data Availability Statement
Data supporting reported results can be found on the project website https://www.hospitalmacarena.es/entrada-blog/medicalforms/ (accessed on 2 November 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
This annex shows the results of evaluating the MedicalForms system against the requirements defined as part of the Delphi study about tool requirements.

Table A1.
Self-evaluation of the tool against the requirements for CIMT.
Table A1.
Self-evaluation of the tool against the requirements for CIMT.
| ID | Description | MedicalForms |
|---|---|---|
| 1 | Can your tool represent data types according to a specified data type standard? | Yes |
| 2 | Is your tool able to define and manage the following datatypes: Boolean, Integer, Double, date, date-time, URI, Multimedia, Concept Descriptor, Physical Quantity, String with Language | Yes |
| 3 | Is your tool able to define a CIM according to a formal syntax that conforms to an open (published) specification? | Yes |
| 4 | Users can easily determine which CIM specification and which version of that specification is supported by the tool | Yes |
| 5 | The tool developer has demonstrated a process of verifying that the CIMs produced or modified using the tool do conform to each CIM specification that is supported (e.g., through import of models into other conformant tools, or by demonstrated parsing tests against the published specification) | Not available yet |
| 6 | Does your tool allow to export/import according to a specified international standard for CIM representation? | Yes |
| 7 | If the tool supports more than one CIM specification, users can select which one to use when designing a new model | Yes |
| 8 | If the tool supports more than one CIM specification, it is clear to a user which specification a CIM conforms to when opening (viewing or editing) an existing model | Yes |
| 9 | Does your tool allow importing/exporting CIM in XML format, according to a publicly accessible XML schema? | Yes |
| 10 | Does your tool allow importing/exporting CIM in ADL format, to a specified version? | Not available yet |
| 11 | Is your tool able to validate that a defined or imported CIM is conformant to the selected specification? | Yes |
| 12 | Is your tool able to show any validation errors a specific CIM has according to the selected specification? | Yes |
| 13 | With a valid CIM, does your modeling tool emit an XML Schema (or similar, please state) against which instances of it may be validated? | Not available yet |
| 14 | Does your modeling tool emit library code by which valid instances in XML Schema (or other, as above) may be parsed into a common object-oriented programming environment (if so, please state which)? | Not available yet |
| 15 | Does your tool support defining for which purpose a CIM is recommended to be applied? | Yes |
| 16 | Does your tool support defining for which usage a CIM is recommended to be applied? | Yes |
| 17 | Does your tool support defining for which clinical domain or clinical user a CIM is recommended to be applied? | Yes |
| 18 | Does your tool allow the creation of new versions of a previously defined CIM? | Yes |
| 19 | Does your tool allow displaying previous versions of a CIM, detailing the changes made in the current version? | Not available yet |
| 20 | Does your tool contain or link to a repository of all previous versions of any particular CIM? | Yes |
| 21 | Does your tool contain or reference, or can it generate a track of changes between all previous versions of a particular CIM? | Not available yet |
| 22 | Does your tool allow to select an existing CIM and define further constrains making possible to specialize its definition for local scenario ensuring compatibility with the generic definition? | Yes |
| 23 | If your tool has been used to create a CIM that is a specialization (or localization) of another, does the specialized CIM include a reference to the more general one it has specialized? | Not available yet |
| 24 | Does your tool allow to identify all those specialized versions of CIM defined for local scenario from a CIM defined for generic scenario | Not available yet |
| 25 | Is your tool able to support the registration of multiple users so that the actions of different users on the same CIM can be attributed to each user? | Yes |
| 26 | Does your tool allow creating profiles for modeling experts such as author, editor or reviewer and their organizations? | Yes |
| 27 | Does your tool include a simplified view for clinical experts to define or review clinical concepts that should be included as CIM nodes? | Yes |
| 28 | Does your tool include a simplified view for clinical experts to define or review clinical concepts that can be bound, or are bound, to CIM nodes? | Yes |
| 29 | Does your tool provide a representation of CIM nodes and value sets in the form of a Prototype screen form? | Yes |
| 30 | Does your tool provide a representation of CIM nodes and value sets in the form of a MindMap? | Yes |
| 31 | Does your tool allow searching CIMs based on the CIM name? | Yes |
| 32 | Does your tool allow searching CIMs based concept codes and attributes associated with CIM nodes? | Yes |
| 33 | Does your tool allow searching CIMs based on domain? | Yes |
| 34 | Does your tool allow searching CIMs based on value sets and terms bound to nodes? | Yes |
| 35 | Each node of a CIM could be mapped to a term within a published international terminology (automatically, or by end users manually or a combination of these) | Yes |
| 36 | Each value list created or reviewed by a user can be drawn from or mapped to terms from a published international terminology | Yes |
| 37 | Does your tool allow user to define value sets that will be bounded to CIM nodes | Yes |
| 38 | Does your tool allow mapping nodes to one or multiple terminology concepts? | Yes |
| 39 | Terminology bindings will be defined according to the chosen specification? | Yes |
| 40 | Does your tool allow a user to enter language translations of terms and concepts used within a CIM definition? | Yes |
| 41 | If the tool supports the use of more than one published international terminology, a user may map each node name to more than one terminology (including multiple language translations of each term, if relevant) | Yes |
| 42 | If the tool supports the use of more than one published international terminology, a user may map each term in a value list to more than one terminology (including multiple language translations of each term, if relevant) | Yes |
| 43 | Does your tool allow mapping nodes to one or multiple ontology concepts? | Yes |
| 44 | An author constructing a terminology value list can use the tool to identify one or more suitable terms from a published international terminology and incorporate such terms and any relevant child concepts | Not available yet |
| 45 | Does your tool allow searching in large taxonomies that will be bound to CIM nodes | Yes |
| 46 | A mechanism exists through use of the tool for a CIM author or reviewer to determine the semantic relationships between node names within a CIM by reference to their concept relationships within a published international terminology | Not available yet |
| 47 | A mechanism exists through use of the tool for a CIM author or reviewer to determine the semantic relationships between a node name and its value list if this is a terminology value set | Yes |
| 48 | Does your tool support connecting with remote (online) Terminology servers that conform to published standards and specification (e.g., CTS2) | Not available yet |
| 49 | Does your tool support connecting with Terminology servers based on specifications to provide functionalities for terminology service administration. Some functionalities could include the ability to load terminologies, export terminologies, activate terminologies, and retire terminologies. | Yes |
| 50 | Does your tool support connecting with Terminology servers based on specifications to provide functionalities for search and query concepts within terminology server based on some search criteria. This includes restrictions to specific associations or other attributes of the terminology, including navigation of associations for result sets. | Yes |
Appendix B
The Table A2 describes the multiple elements included as part of the MedicalForm system specifying its associated attributes and how they are mapped to ISO 21090 Datatypes, ISO 13606 and openClinica CTMS. In addition, Table A2 provides an example of JSON structure applied for management of terminology subsets, semantic structures and forms.

Table A2.
MedicalForm element attributes and mapping to standards and openClinica CMTS.
Table A2.
MedicalForm element attributes and mapping to standards and openClinica CMTS.
| MedicalForm Class | Description | Attributes | Multiple Items | Is Able to Contain and Organize Elements | Aligned with |
|---|---|---|---|---|---|
| Section | Supports information organization on the screen | Header Code required | No | Yes | ISO13606 Section openClinica mapping could produce Section, header or subheader |
| Tab | Supports information organization with screen visibility capabilities | Header Code required | No | Yes | ISO13606 Section openClinica Section |
| Table | Supports information organization with screen visibility capabilities | Header Code required | No | Yes | ISO13606 Section openClinica grid |
| Header | Describes information on the screen for data recording | Header Code required | No | No | ISO21090 TS openClinica Header or subheader |
| Number | Records a number | header: code maxValue minValue: 0, value decimalNumber defaultValue helptext conditionalView required | No | No | Depending on number of decimal specified it is mapped to either ISO21090 INT or REAL datatypes |
| Magnitude | Records a physical magnitude | header code unit maxValue minValue”: 0, value defaultValue maxLength helptext conditionalView required | No | No | ISO21090 PQ datatypes OpenClinica Text specifying unit |
| Text | Records either a simple text | header code value defaultValue helptext | No | No | ISO21090 ST datatypes OpenClinica text |
| TextArea | Records either a simple text | header code value defaultValue maxLength rows helptext | No | No | ISO21090 ST datatypes OpenClinica textarea |
| Select | Display a set of options presented in dropdown menu | header code item.display item.value item.code helptext conditionalView | Yes | No+ | ISO 13606 Cluster composed by ITEMS based on ISO21090 CD openClinica single-select |
| Checkbox | Display a set of options presented in dropdown menu | header code item.display item.value item.code helptext conditionalView | Yes | No+ | ISO 13606 Cluster composed by ITEMS based on ISO21090 CD openClinica checkbox |
| RadioGroup | Display a set of options presented in dropdown menu | header code item.display item.value item.code helptext conditionalView | Yes | No+ | ISO 13606 Cluster composed by ITEMS based on ISO21090 CD openClinica radio |
| Autocalculated | Defines a value calculated from previous entries | header formula unit decimalNumber maxValue minValue value conditionalView | No | No | Depending on multiple options it can be mapped either ISO21090 INT, REAL or PQ datatypes OpenClinica calculation |
| File | Includes the file upload button in a form | header value hidden | No | No | OpenClinica file |

Table A3.
Example of MedicalForm JSON for terminology subsets, semantic structures and forms.
Table A3.
Example of MedicalForm JSON for terminology subsets, semantic structures and forms.
| Terminology Subset | Semantic Structure | Form |
|---|---|---|
A terminology subset called “Primary procedure” containing 2 items:
| A structure that contains a title “Plastic surgery care plan”, and cluster called “Primary procedure” containing 2 items:
| A form with a tab structure that contains subset called “Surgery Record”, that contains a “header”: “Plastic surgery care plan”, and a mandatory radiobutton called “Primary procedure” containing 2 options:
|
| [ { “_id”: “5ad7501dd06474080de79760”, “type”: “select”, “typeStructure”: “complex”, “typeShow”: “item”, “value”: ““, “class”: “form_model”, “header”: “Primary procedure”, “code”: “399455000”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “form”: “5ad7501dd06474080de7975f”, “__v”: 0, “container”: [], “options”: [ { “_id”: “5ad5d408d06474080de7793e”, “display”: “Chemosurgery—action”, “code”: “129403009”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 }, { “_id”: “5ad5d408d06474080de77a91”, “display”: “Cryosurgery—action”, “code”: “129393004”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 } ] } ] | [ { “_id”: “61f1c8a2b930b800130bea63”, “type”: “section”, “typeStructure”: “text”, “typeShow”: “container”, “class”: “header”, “header”: “Plastic surgery care plan”, “value”: ““, “code”: “773436007”, “form”: “61f1c8a2b930b800130bea61”, “__v”: 0, “container”: [ { “options”: [ { “_id”: “5ad5d408d06474080de7793e”, “display”: “Chemosurgery—action”, “code”: “129403009”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 }, { “_id”: “5ad5d408d06474080de77a91”, “display”: “Cryosurgery—action”, “code”: “129393004”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 } ], “container”: [], “_id”: “61f1c8a2b930b800130bea62”, “__v”: 0, “form”: “61f1c8a2b930b800130bea61”, “version”: “2017”, “terminology”: “SNOMED CT (Int)”, “code”: “399455000”, “header”: “Primary procedure”, “class”: “form_model”, “value”: ““, “typeShow”: “item”, “typeStructure”: “complex”, “type”: “select” } ], “options”: [] } ] | [ { “type”: “tab”, “typeStructure”: “text”, “typeShow”: “container”, “class”: “header”, “header”: “Surgery Record”, “value”: ““, “hidden”: true, “container”: [ { “_id”: “61f1c8a2b930b800130bea63”, “type”: “section”, “typeStructure”: “text”, “typeShow”: “container”, “class”: “header”, “header”: “Plastic surgery care plan”, “value”: ““, “code”: “773436007”, “form”: “61f1c8a2b930b800130bea61”, “__v”: 0, “container”: [ { “type”: “radiobutton”, “typeStructure”: “complex”, “typeShow”: “item”, “class”: “form_model”, “header”: “Primary procedure”, “options”: [ { “_id”: “5ad5d408d06474080de7793e”, “display”: “Chemosurgery—action”, “code”: “129403009”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 }, { “_id”: “5ad5d408d06474080de77a91”, “display”: “Cryosurgery—action”, “code”: “129393004”, “terminology”: “SNOMED CT (Int)”, “version”: “2017”, “__v”: 0 } ], “edit”: false, “typeInformationModel”: “select” } ], “options”: [] } ], “edit”: false } ] |
Appendix C
Next presents how the Barthel clinical information model has been exported in a format compatible with Diraya EHR system, openClinica, and RedCap. It is relevant that the CroniCare system supports the definitions of the OpenClinica platform and is able to generate mobile app forms based on the same configuration files.

Figure A1.
OpenClinica.
Figure A1.
OpenClinica.
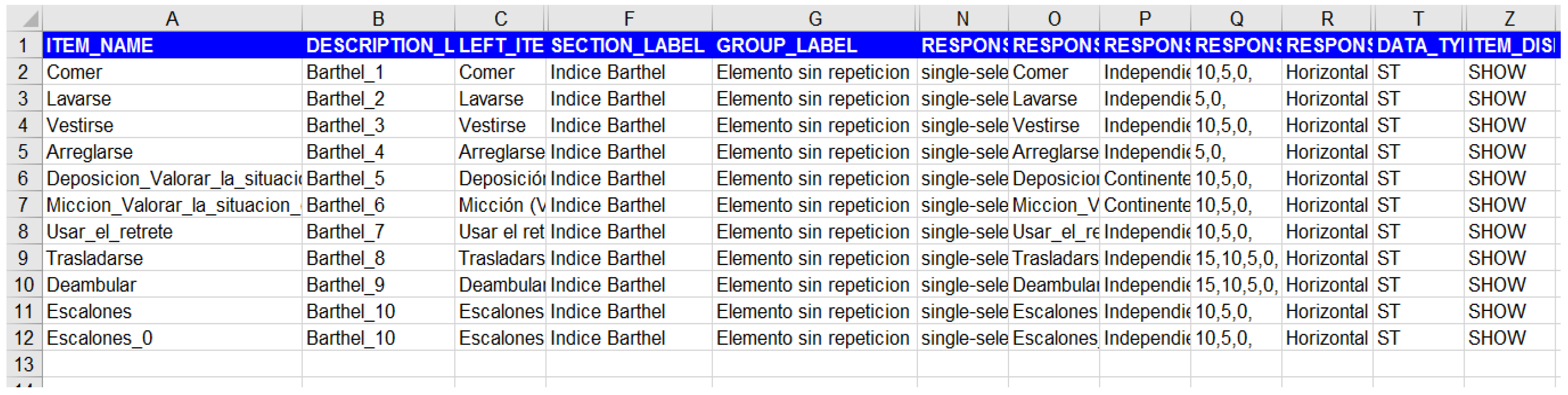
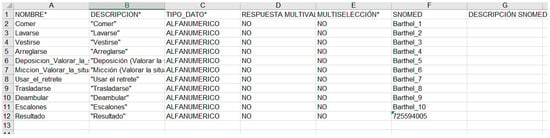
Figure A2.
Diraya—Andalusian Health Service Electronic Health Record system.
Figure A2.
Diraya—Andalusian Health Service Electronic Health Record system.
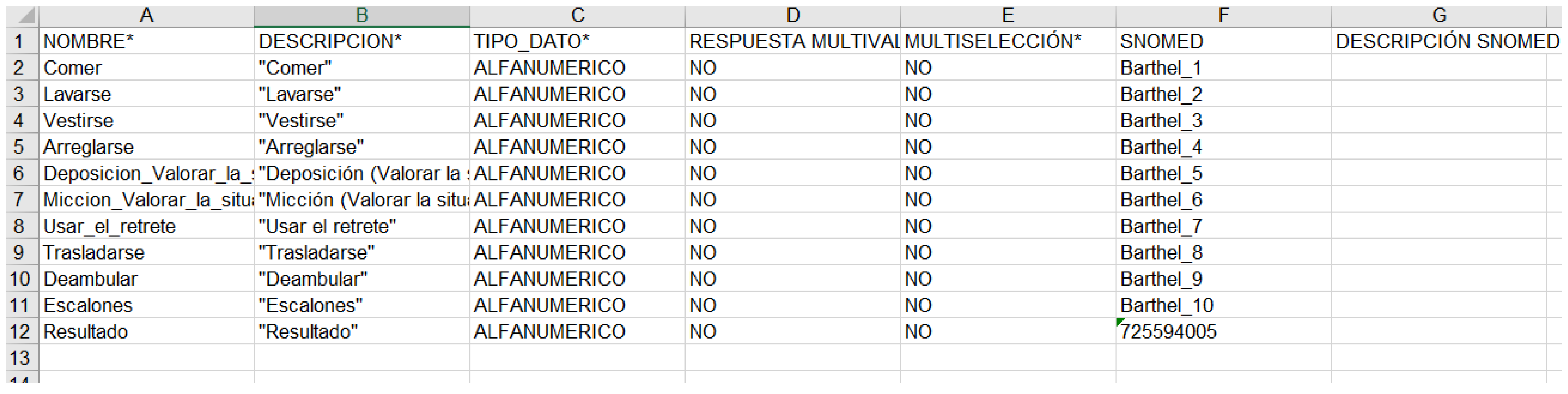
Figure A3.
RedCap.
Figure A3.
RedCap.
Appendix D
Additional materials can be found on the project website:
- URL: https://www.hospitalmacarena.es/entrada-blog/medicalforms/ (accessed on 2 November 2021).
From this website you will be able to access and test the MedicalForm system by using the following user:
- user: guest@medicalforms.org
- password: guest
References
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Gayar, O.F.; Ambati, L.S.; Nawar, N. Wearables, artificial intelligence, and the future of healthcare. In AI and Big Data’s Potential for Disruptive Innovation; IGI Global: Hershey, PA, USA, 2020; pp. 104–129. [Google Scholar]
- Moreno-Conde, A.; Moner, D.; Cruz, W.D.D.; Santos, M.R.; Maldonado, J.A.; Robles, M.; Kalra, D. Clinical information modeling processes for semantic interoperability of electronic health records: Systematic review and inductive analysis. J. Am. Med. Inform. Assoc. 2015, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Conde, A.; Thienpont, G.; Lamote, I.; Coorevits, P.; Parra, C.; Kalra, D. European Interoperability Assets Register and Quality Framework Implementation. In Exploring Complexity in Health: An Interdisciplinary Systems Approach; IOS Press: Amsterdam, The Netherlands, 2016; pp. 690–694. [Google Scholar]
- ISO/DTS 13972; Health Informatics—Clinical Information Models—Characteristics, Structures and Requirements. International Standardization Organization: Geneva, Switzerland, 2022.
- Tague Nancy, R. Plan–Do–Study–Act cycle. In The quality Toolbox, 2nd ed.; ASQ Quality Press: Milwaukee, WI, USA, 2005; pp. 390–392. ISBN 978-0873896399. [Google Scholar]
- Kalra, D.; Beale, T.; Heard, S. The openEHR foundation. Stud. Health Technol. Inform. 2005, 115, 153–173. [Google Scholar] [PubMed]
- Website of the HL7 Clinical Information Modelling Initiative Website. Available online: https://hl7.org/Special/Committees/cimi/index.cfm (accessed on 30 October 2021).
- ISO/HL7 27932; Data Exchange Standards—HL7 Clinical Document Architecture. International Standardization Organization: Geneva, Switzerland, 2009.
- ISO 13606-1:2019; Health Informatics—Electronic Health Record Communication. International Standardization Organization: Geneva, Switzerland, 2009.
- Huser, V.; Sastry, C.; Breymaier, M.; Idriss, A.; Cimino, J.J. Standardizing data exchange for clinical research protocols and case report forms: An assessment of the suitability of the Clinical Data Interchange Standards Consortium (CDISC) Operational Data Model (ODM). J. Biomed. Inform. 2015, 57, 88–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical Data Acquisition Standards Harmonization Model; Version 1.2; Clinical Data Interchange Standards Consortium: Austin, TX, USA, 2021.
- Moreno-Conde, A.; Austin, T.; Moreno-Conde, J.; Parra-Calderón, C.L.; Kalra, D. Evaluation of clinical information modeling tools. J. Am. Med. Inform. Assoc. 2016, 23, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno Conde, A. Quality Framework for Semantic Interoperability in Health Informatics: Definition and Implementation. Ph.D. Thesis, University College London, London, UK, 2016. Chapter 4.3 International Study of Experts on Best Modelling Practices. Available online: https://discovery.ucl.ac.uk/id/eprint/1529311/ (accessed on 30 October 2021).
- Villalba-Mora, E.; Casas, I.; Lupiañez-Villanueva, F.; Maghiros, I. Adoption of health information technologies by physicians for clinical practice: The Andalusian case. Int. J. Med.Inform. 2015, 84, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Diraya EHR System Software Repository. Available online: https://www.juntadeandalucia.es/repositorio/usuario/listado/fichacompleta.jsf?idProyecto=626 (accessed on 30 October 2021).
- Cavelaars, M.; Rousseau, J.; Parlayan, C.; de Ridder, S.; Verburg, A.; Ross, R.; Rienk Visser, G.; Rotte, A.; Azevedo, R.; Boiten, J.-W.; et al. OpenClinica. J. Clin. Bioinform. 2015, 5, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheufele, E.; Aronzon, D.; Coopersmith, R.; McDuffie, M.T.; Kapoor, M.; Uhrich, C.A.; Palchuk, M.B. tranSMART: An open source knowledge management and high content data analytics platform. AMIA Summits Transl. Sci. Proc. 2014, 2014, 96. [Google Scholar] [PubMed]
- Murphy, S.N.; Weber, G.; Mendis, M.; Gainer, V.; Chueh, H.C.; Churchill, S.; Kohane, I. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J. Am. Med. Inform.Assoc. 2010, 17, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno Conde, A.; Moreno Conde, J.; Salas, S.; Gonzalez, V.; Valido, A.; Guardia, P.; Luque, L.; Reales, A.; Machuca, A.; Montblanc, E.; et al. CroniCare: Platform for the Design and Implementation of Follow-Up, Control and Self-Management Interventions for Chronic and Multimorbidity Patients Based on Mobile Technologies. In Proceedings of the MedInfo 2021, Virtual, 2–4 October 2021. [Google Scholar]
- CroniCare System Website. Available online: https://www.hospitalmacarena.es/entrada-blog/cronicareproject/ (accessed on 30 October 2021).
- Moreno-Conde, A.; Jódar-Sánchez, F.; Kalra, D. Requirements for clinical information modelling tools. Int. J. Med. Inform. 2015, 84, 524–536. [Google Scholar] [CrossRef] [PubMed]
- The Heart Failure Summary Template. Available at the Projects Section of the OpenEHR Clinical Knowledge Manager. Available online: https://ckm.openehr.org/ckm/ (accessed on 30 October 2021).
- Jiménez Hernández, M.D.; Lama Herrera, C.M.; Morales Serna, J.C.; Ras Luna, J.; Sanz Amores, R. Stroke: Integrated Care Process. 2015 Andalusian Ministry of Health. Available online: https://juntadeandalucia.es/export/drupaljda/salud_5af1957075765_pai_ictus_abril_2015.pdf (accessed on 30 October 2021).
- Venkatesh, V.; Davis, F.D. A theoretical extension of the technology acceptance model: Four longitudinal field studies. Manag. Sci. 2000, 46, 186–204. [Google Scholar] [CrossRef] [Green Version]
- MedicalForms System Website. Available online: https://www.hospitalmacarena.es/entrada-blog/medicalforms/ (accessed on 30 October 2021).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).