A Medical Image Encryption Scheme for Secure Fingerprint-Based Authenticated Transmission
Abstract
:Featured Application
Abstract
1. Introduction
- Ensure the visual security of medical image transmission through a watermarking process.
- Protect the medical image and the physician’s fingerprint through an encryption scheme to make the proposed scheme resistant to white-box attacks.
- Ensure the medical image’s authenticity using the physician’s fingerprint.
- Perform a proposed scheme simulation to evaluate the quality of the reconstructed medical image, the quality of the watermarking, and the accuracy of the reconstructed fingerprint feature in terms of peak signal-to-noise ratio (PSNR), mean structural similarity index measure (MSSIM), distance of histogram intersection, and the relative error.
- Perform a critical security analysis to evaluate the resistance of the proposed scheme to brute-force attacks.
- Compare the proposed scheme’s performance with other visual encryption schemes to validate its effectiveness.
2. Related Work
- a visual protection from black-box attacks;
- encryption protection from white-box attacks;
- an image authentication through a physician’s fingerprint
3. Materials and Methods
3.1. Visually Secure Image Encryption Scheme
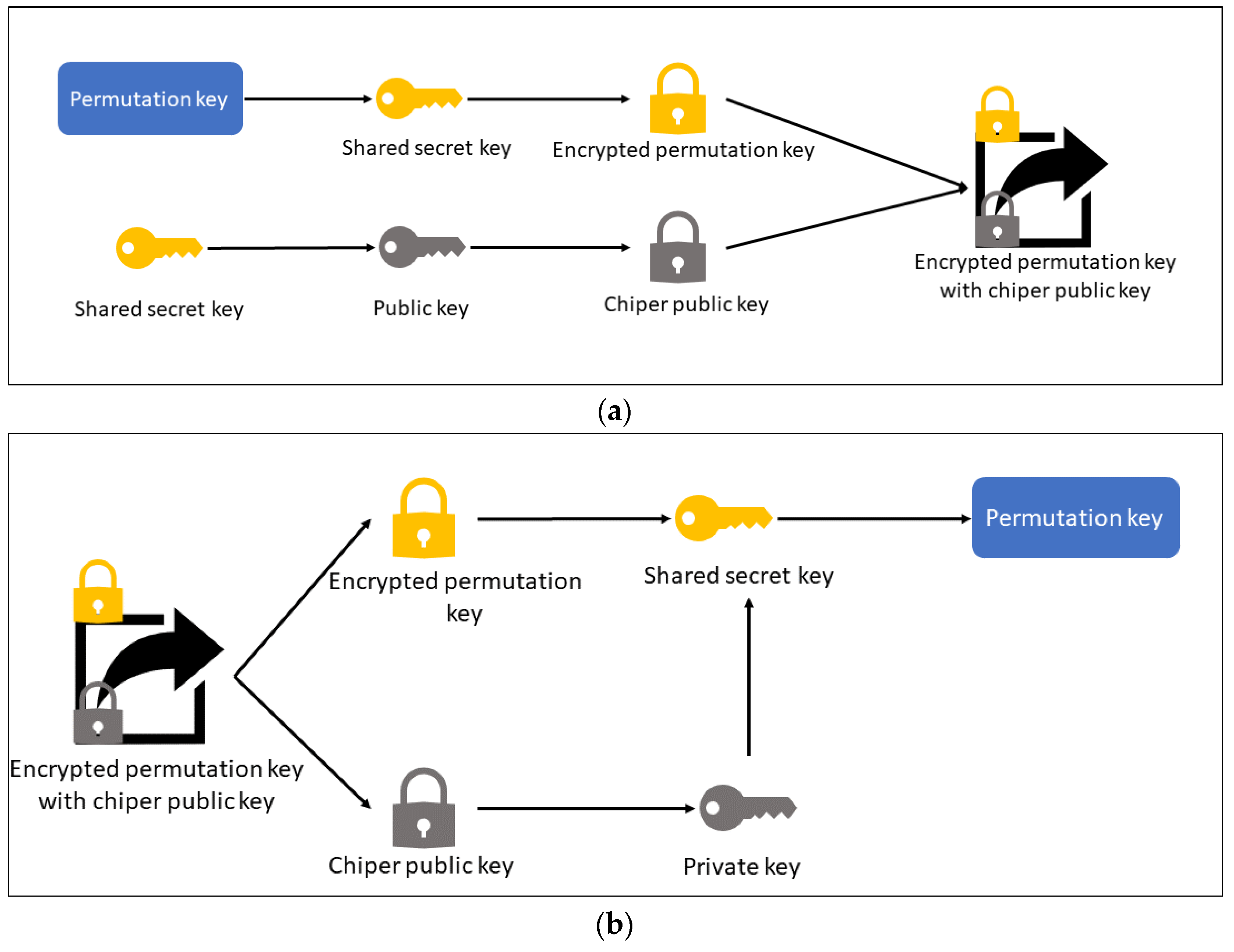
3.2. Key Protection Scheme
3.3. Visually Secure Image Decryption Scheme
4. Simulation Results and Analysis
4.1. Simulation Results
4.2. Histogram Analysis
4.3. Comparation Analysis
4.4. Extracted Fingerprint Feature Analysis
4.5. Key Security Analysis
4.6. Running Efficiency Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arbabshirani, M.R.; Plis, S.; Sui, J.; Calhoun, V.D. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage 2017, 145, 137–165. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mandal, P.K. A Comprehensive Report on Machine Learning-Based Early Detection of Alzheimer’s Disease using Multi-modal Neuroimaging Data. ACM Comput. Surv. 2023, 55, 1–44. [Google Scholar] [CrossRef]
- Gattulli, V.; Impedovo, D.; Pirlo, G.; Semeraro, G. Early Dementia Identification: On the Use of Random Handwriting Strokes. In Lecture Notes in Computer Science; (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2022; Volume 13424 LNCS, pp. 285–300. [Google Scholar]
- Dentamaro, V.; Giglio, P.; Impedovo, D.; Moretti, L.; Pirlo, G. AUCO ResNet: An end-to-end network for COVID-19 pre-screening from cough and breath. Pattern Recognit. 2022, 127, 108656. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Du, Y.; Yang, S.; Zhang, J.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. RetCCL: Clustering-guided contrastive learning for whole-slide image retrieval. Med. Image Anal. 2023, 83, 102645. [Google Scholar] [CrossRef]
- McKinney, S.M.; Sieniek, M.; Godbole, V.; Godwin, J.; Antropova, N.; Ashrafian, H.; Back, T.; Chesus, M.; Corrado, G.S.; Darzi, A.; et al. International evaluation of an AI system for breast cancer screening. Nature 2020, 577, 89–94. [Google Scholar] [CrossRef]
- Balasamy, K.; Suganyadevi, S. A fuzzy based ROI selection for encryption and watermarking in medical image using DWT and SVD. Multimed. Tools Appl. 2021, 80, 7167–7186. [Google Scholar]
- Narayanan, A.; Shmatikov, V. Robust de-anonymization of large sparse datasets. In Proceedings of the 2008 IEEE Symposium on Security and Privacy, Oakland, CA, USA, 18–21 May 2008; pp. 111–125. [Google Scholar]
- Schwarz, C.G.; Kremers, W.K.; Therneau, T.M.; Sharp, R.R.; Gunter, J.L.; Vemuri, P.; Arani, A.; Spychalla, A.J.; Kantarci, K.; Knopman, D.S.; et al. Identification of Anonymous MRI Research Participants with Face-Recognition Software. N. Engl. J. Med. 2019, 381, 1684–1686. [Google Scholar] [CrossRef]
- Kaissis, G.; Ziller, A.; Passerat-Palmbach, J.; Ryffel, T.; Usynin, D.; Trask, A.; Lima, I.; Mancuso, J.; Jungmann, F.; Steinborn, M.-M.; et al. End-to-end privacy preserving deep learning on multi-institutional medical imaging. Nat. Mach. Intell. 2021, 3, 473–484. [Google Scholar] [CrossRef]
- Zear, A.; Singh, A.K.; Kumar, P. A proposed secure multiple watermarking technique based on DWT, DCT and SVD for application in medicine. Multimed. Tools Appl. 2018, 77, 4863–4882. [Google Scholar] [CrossRef]
- Priya, S.; Santhi, B. A Novel Visual Medical Image Encryption for Secure Transmission of Authenticated Watermarked Medical Images. Mob. Netw. Appl. 2021, 26, 2501–2508. [Google Scholar] [CrossRef]
- Wan, W.; Wang, J.; Zhang, Y.; Li, J.; Yu, H.; Sun, J. A comprehensive survey on robust image watermarking. Neurocomputing 2022, 488, 226–247. [Google Scholar] [CrossRef]
- Monrat, A.A.; Schelén, O.; Andersson, K. A survey of blockchain from the perspectives of applications, challenges, and opportunities. IEEE Access 2019, 7, 117134–117151. [Google Scholar] [CrossRef]
- Parida, P.; Pradhan, C.; Gao, X.Z.; Roy, D.S.; Barik, R.K. Image Encryption and Authentication with Elliptic Curve Cryptography and Multidimensional Chaotic Maps. IEEE Access 2021, 9, 76191–76204. [Google Scholar] [CrossRef]
- Yang, X.; Li, T.; Pei, X.; Wen, L.; Wang, C. Medical Data Sharing Scheme Based on Attribute Cryptosystem and Blockchain Technology. IEEE Access 2020, 8, 45468–45476. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, R.; Wang, X.; Gao, K.; Liu, L. A decentralizing attribute-based signature for healthcare blockchain. In Proceedings of the International Conference on Computer Communications and Networks, ICCCN 2018, Hangzhou, China, 30 July–2 August 2018. [Google Scholar]
- Kanwal, S.; Tao, F.; Almogren, A.; Rehman, A.U.; Taj, R.; Radwan, A. A Robust Data Hiding Reversible Technique for Improving the Security in e-Health Care System. CMES Comput. Model. Eng. Sci. 2023, 134, 201–219. [Google Scholar] [CrossRef]
- Salunke, S.; Ahuja, B.; Hashmi, M.F.; Marriboyina, V.; Bokde, N.D. 5D Gauss Map Perspective to Image Encryption with Transfer Learning Validation. Appl. Sci. 2022, 12, 5321. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, X.; Yu, C.; Tang, Z. Reversible Data Hiding in Encrypted Image Based on Multi-MSB Embedding Strategy. Appl. Sci. 2020, 10, 2058. [Google Scholar] [CrossRef]
- Lin, C.-H.; Hu, G.-H.; Chan, C.-Y.; Yan, J.-J. Chaos-Based Synchronized Dynamic Keys and Their Application to Image Encryption with an Improved AES Algorithm. Appl. Sci. 2021, 11, 1329. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, G.; Hou, W.; Huang, P.; Liu, B.; Li, S. A Novel Asymmetric Hyperchaotic Image Encryption Scheme Based on Elliptic Curve Cryptography. Appl. Sci. 2021, 11, 5691. [Google Scholar] [CrossRef]
- Wen, W.; Zhang, Y.; Fang, Y.; Fang, Z. Image salient regions encryption for generating visually meaningful ciphertext image. Neural Comput. Appl. 2018, 29, 653–663. [Google Scholar] [CrossRef]
- Anand, A.; Singh, A.K. An improved DWT-SVD domain watermarking for medical information security. Comput. Commun. 2020, 152, 72–80. [Google Scholar] [CrossRef]
- Cox, I.J.; Doërr, G.; Furon, T. Watermarking is not cryptography. In Lecture Notes in Computer Science; (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2006; Volume 4283 LNCS, pp. 1–15. [Google Scholar]
- Han, B.; Jhaveri, R.; Wang, H.; Qiao, D.; Du, J. Application of Robust Zero-Watermarking Scheme Based on Federated Learning for Securing the Healthcare Data. IEEE J. Biomed. Heal. Inform. 2021, 27, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, J.; Liu, J.; Huang, M.; Chen, Y.W.; Bhatti, U.A. Robust watermarking algorithm for medical images based on log-polar transform. EURASIP J. Wirel. Commun. Netw. 2022, 2022, 1–11. [Google Scholar] [CrossRef]
- Hua, Z.; Zhu, Z.; Yi, S.; Zhang, Z.; Huang, H. Cross-plane colour image encryption using a two-dimensional logistic tent modular map. Inf. Sci. 2021, 546, 1063–1083. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.; Jiang, D. A novel triple-image encryption and hiding algorithm based on chaos, compressive sensing and 3D DCT. Inf. Sci. 2021, 574, 505–527. [Google Scholar] [CrossRef]
- Singh, S.P.; Bhatnagar, G. A Novel Biometric Inspired Robust Security Framework for Medical Images. IEEE Trans. Knowl. Data Eng. 2021, 33, 810–823. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, Z.; Wang, F.; Ni, R.; Wang, J.; Hu, Y. Medical image encryption based on biometric keys and lower-upper decomposition with partial pivoting. Appl. Opt. 2021, 60, 24. [Google Scholar] [CrossRef]
- Tao, S.; Tang, C.; Shen, Y.; Lei, Z. Optical image encryption based on biometric keys and singular value decomposition. Appl. Opt. 2020, 59, 2422. [Google Scholar] [CrossRef]
- Shen, Y.; Tang, C.; Xu, M.; Lei, Z. Optical selective encryption based on the FRFCM algorithm and face biometric for the medical image. Opt. Laser Technol. 2021, 138, 106911. [Google Scholar] [CrossRef]
- Lian, S.; Sun, J.; Wang, Z. A block cipher based on a suitable use of the chaotic standard map. Chaos Solitons Fractals 2005, 26, 117–129. [Google Scholar] [CrossRef]
- Lei, T.; Jia, X.; Zhang, Y.; He, L.; Meng, H.; Nandi, A.K. Significantly Fast and Robust Fuzzy C-Means Clustering Algorithm Based on Morphological Reconstruction and Membership Filtering. IEEE Trans. Fuzzy Syst. 2018, 26, 3027–3041. [Google Scholar] [CrossRef]
- Aparna, P.; Kishore, P.V.V. Biometric-based efficient medical image watermarking in E-healthcare application. IET Image Process. 2019, 13, 421–428. [Google Scholar] [CrossRef]
- Singh, N.; Joshi, S.; Birla, S. Color Image Watermarking with Watermark Authentication against False Positive Detection Using SVD. In Proceedings of the International Conference on Sustainable Computing in Science, Technology and Management (SUSCOM), Jaipur, India, 2 February 2019. [Google Scholar] [CrossRef]
- Parida, P.; Bhoi, N. Wavelet based transition region extraction for image segmentation. Future Comput. Inform. J. 2017, 2, 65–78. [Google Scholar] [CrossRef]
- Albertina, B.; Watson, M.; Holback, C.; Jarosz, R.; Kirk, S.; Lee, Y.; Rieger-Christ, K.; Lemmerman, J. The Cancer Genome Atlas Lung Adenocarcinoma Collection (TCGA-LUAD) (Version 4) [Data Set]. The Cancer Imaging Archive. 2016. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=6881474 (accessed on 28 March 2023).
- Tong, T.; Li, M. Abdominal or Pelvic Enhanced CT Images within 10 Days before Surgery of 230 Patients with Stage II Colorectal Cancer (StageII-Colorectal-CT) [Dataset]. The Cancer Imaging Archive. 2022. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=117113567 (accessed on 28 March 2023).
- Shapey, J.; Kujawa, A.; Dorent, R.; Wang, G.; Dimitriadis, A.; Grishchuk, D.; Paddick, I.; Kitchen, N.; Bradford, R.; Saeed, S.R.; et al. Segmentation of Vestibular Schwannoma from Magnetic Resonance Imaging: An Open Annotated Dataset and Baseline Algorithm. Sci. Data 2021, 8, 286. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). The Clinical Proteomic Tumor Analysis Consortium Cutaneous Melanoma Collection (CPTAC-CM) (Version 10) [Data Set]. The Cancer Imaging Archive; CPTAC: Rockville, MD, USA, 2018; Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=33948224 (accessed on 28 March 2023).
- Saha, A.; Harowicz, M.R.; Grimm, L.J.; Weng, J.; Cain, E.H.; Kim, C.E.; Ghate, S.V.; Walsh, R.; Mazurowski, M.A. Dynamic Contrast-Enhanced Magnetic Resonance Images of Breast Cancer Patients with Tumor Locations [Data Set]. The Cancer Imaging Archive. 2021. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70226903 (accessed on 28 March 2023).
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Maio, D.; Maltoni, D.; Cappelli, R.; Wayman, J.L.; Jain, A.K. FVC2002: Second fingerprint verification competition. In Proceedings of the International Conference on Pattern Recognition, Quebec City, QC, Canada, 11–15 August 2002; Volume 16, pp. 811–814. [Google Scholar]
- Wieclaw, L. A Minutiae-Based Matching Algorithms in Fingerprint Recognition Systems. J. Med. Inform. 2009. Available online: https://www.academia.edu/2508970/A_minutiae_based_matching_algorithms_in_fingerprint_recognition_systems (accessed on 28 March 2023).
- Patel, S.; Vaish, A. Block based visually secure image encryption algorithm using 2D-Compressive Sensing and nonlinearity. Optik 2023, 272, 170341. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Hua, Z.; Zhang, K.; Li, Y.; Zhou, Y. Visually secure image encryption using adaptive-thresholding sparsification and parallel compressive sensing. Signal Process. 2021, 183, 107998. [Google Scholar] [CrossRef]
- Wang, H.; Xiao, D.; Li, M.; Xiang, Y.; Li, X. A visually secure image encryption scheme based on parallel compressive sensing. Signal. Process. 2019, 155, 218–232. [Google Scholar] [CrossRef]
- Ping, P.; Fu, J.; Mao, Y.; Xu, F.; Gao, J. Meaningful Encryption: Generating Visually Meaningful Encrypted Images by Compressive Sensing and Reversible Color Transformation. IEEE Access 2019, 7, 170168–170184. [Google Scholar] [CrossRef]
- Dhall, S.; Gupta, S. Multilayered highly secure authentic watermarking mechanism for medical applications. Multimed. Tools Appl. 2021, 80, 18069–18105. [Google Scholar] [CrossRef]
- Chai, X.; Wu, H.; Gan, Z.; Zhang, Y.; Chen, Y.; Nixon, K.W. An efficient visually meaningful image compression and encryption scheme based on compressive sensing and dynamic LSB embedding. Opt. Lasers Eng. 2020, 124, 105837. [Google Scholar] [CrossRef]







| Medical Image | Reference Image | Reconstructed Medical Image | Visually Meaningful Encrypted Image | ||
|---|---|---|---|---|---|
| PSNR | MSSIM | PSNR | MSSIM | ||
| lungs | Barbara | 52.163 | 0.992 | 32.291 | 0.860 |
| pelvic | Lena | 52.952 | 0.996 | 38.786 | 0.967 |
| head | airplane | 54.401 | 0.997 | 38.586 | 0.972 |
| skin | pepper | 54.861 | 0.998 | 39.718 | 0.971 |
| breast | baboon | 54.590 | 0.996 | 32.998 | 0.920 |
| kidney | girl | 60.526 | 0.999 | 40.316 | 0.975 |
| Reference Image | Embedded Medical Image | Distance of Histogram Intersection |
|---|---|---|
| Barbara | lungs | 0.968 |
| Lena | pelvic | 0.989 |
| airplane | head | 0.992 |
| pepper | skin | 0.992 |
| baboon | breast | 0.987 |
| girl | kidney | 0.980 |
| [48] | [50] | [51] | [52] | [53] | [54] | Proposed Scheme | |
|---|---|---|---|---|---|---|---|
| PSNR | 49.137 | 35.107 | 33.4204 | 32.4235 | 51.6860 | 31.62 | 54.947 |
| MSSIM | 0.92339 | 0.95564 | N/A | 0.8855 | N/A | 0.9887 | 0.9963 |
| [48] | [50] | [51] | [52] | [54] | Proposed Scheme | |
|---|---|---|---|---|---|---|
| Barbara | N/A | N/A | N/A | N/A | N/A | 32.291 |
| Lena | 55.5123 | N/A | N/A | N/A | N/A | 38.786 |
| airplane | 56.5828 | N/A | N/A | N/A | N/A | N/A |
| pepper | 55.5071 | 40.9310 | 32.3513 | 35.1347 | 34.51 | 39.718 |
| baboon | 55.1570 | 40.9187 | 37.1058 | 36.4906 | N/A | 32.998 |
| girl | 57.3175 | N/A | N/A | N/A | N/A | 40.316 |
| Medical Image | Refence Image | Encryption Time (s) | Decryption Time (s) |
|---|---|---|---|
| lungs | Barbara | 13.547 | 10.143 |
| pelvic | Lena | 53.109 | 57.433 |
| head | airplane | 3.419 | 6.188 |
| skin | pepper | 3.316 | 3.665 |
| breast | baboon | 4.884 | 4.156 |
| kidney | girl | 4.056 | 3.345 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, F.; Impedovo, D.; Pirlo, G. A Medical Image Encryption Scheme for Secure Fingerprint-Based Authenticated Transmission. Appl. Sci. 2023, 13, 6099. https://doi.org/10.3390/app13106099
Castro F, Impedovo D, Pirlo G. A Medical Image Encryption Scheme for Secure Fingerprint-Based Authenticated Transmission. Applied Sciences. 2023; 13(10):6099. https://doi.org/10.3390/app13106099
Chicago/Turabian StyleCastro, Francesco, Donato Impedovo, and Giuseppe Pirlo. 2023. "A Medical Image Encryption Scheme for Secure Fingerprint-Based Authenticated Transmission" Applied Sciences 13, no. 10: 6099. https://doi.org/10.3390/app13106099
APA StyleCastro, F., Impedovo, D., & Pirlo, G. (2023). A Medical Image Encryption Scheme for Secure Fingerprint-Based Authenticated Transmission. Applied Sciences, 13(10), 6099. https://doi.org/10.3390/app13106099







