Self-Training with Entropy-Based Mixup for Low-Resource Chest X-ray Classification
Abstract
:1. Introduction
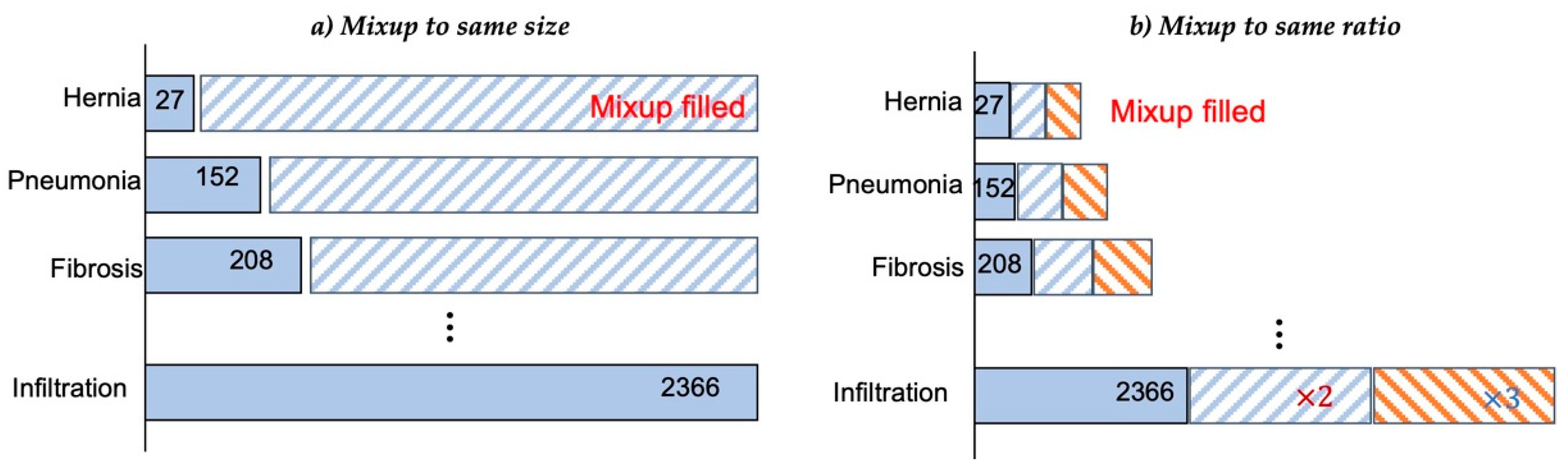
- We propose an entropy-based Mixup method for data augmentation. The entropy is calculated and sorted to select two labeled data for synthesis, and data with high entropy and data with low entropy are selected for mixed synthesis. Two target selection methods to solve the imbalance in the number of samples are compared through experiments: random selection and entropy-based selection.
- We propose a self-training method for utilizing unlabeled data to overcome the low-resource problem. It improves performance by cycling through sample generation, selection, and expansion with sample synthesis. Through self-training, the overfitting problem that occurs during the training process can also be reduced.
- Mixup and self-training are combined to improve the performance of imbalanced multilabel chest X-ray classification. The proposed method applies the Mixup algorithm to a small number of labeled data to alleviate data imbalance and applies self-training with a large number of unlabeled data, replacing the teacher model with the student model repeatedly to perform sampling, which can effectively utilize unlabeled data.
2. Related Work
2.1. Data Augmentation
2.2. Self-Training
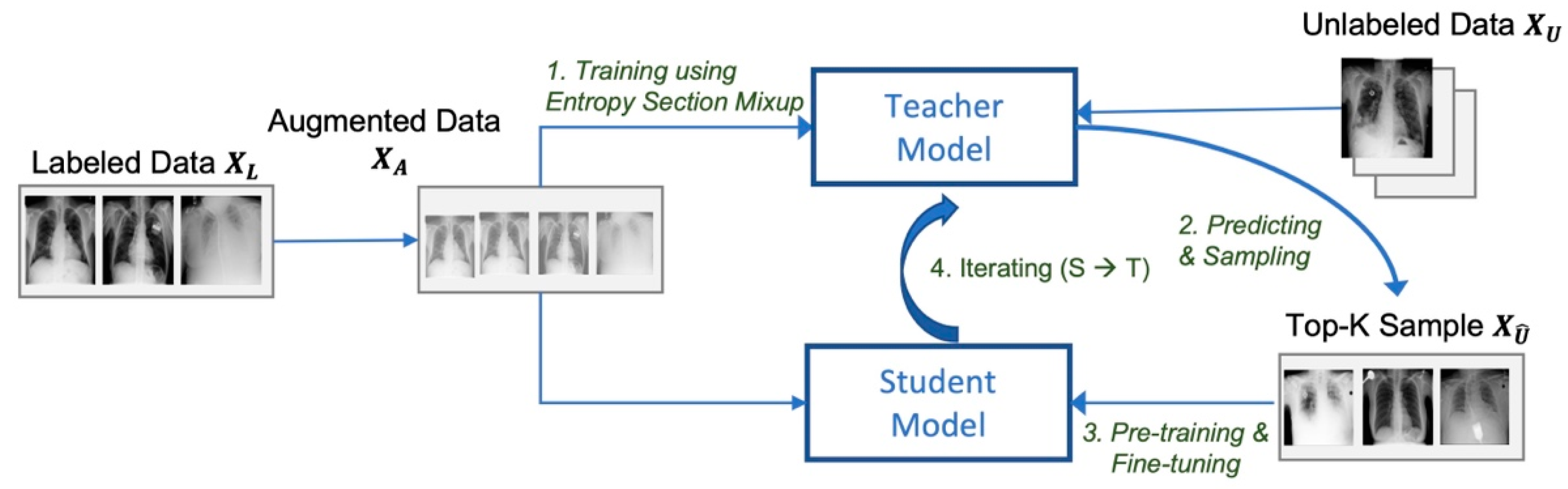
3. Proposed Learning Model
- (1)
- Training the teacher model using data augmented by entropy-based Mixup;
- (2)
- Predicting labels of unlabeled data and top-K sampling of pseudo-label data;
- (3)
- Pretraining and fine-tuning the student model;
- (4)
- Replacing the teacher model with the student model, and going back to step 2.
3.1. Training the Teacher Model Using Entropy-Based Mixup
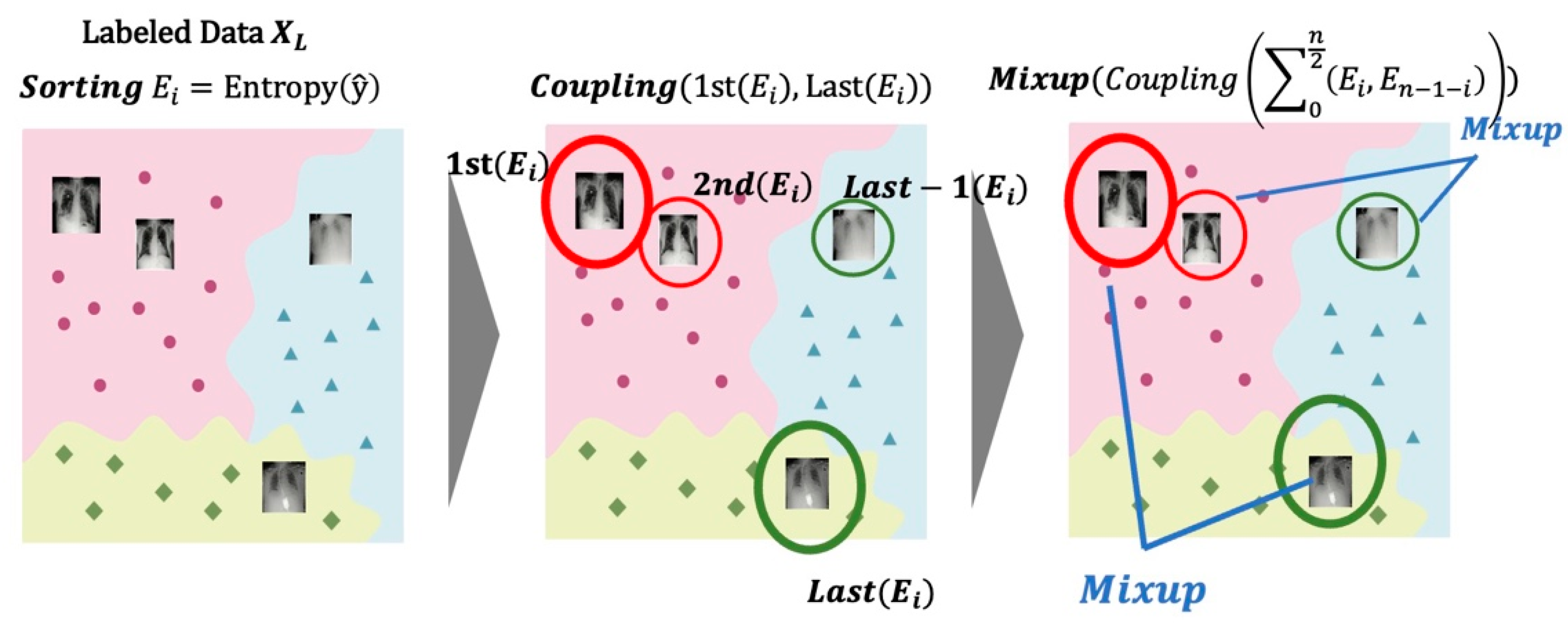
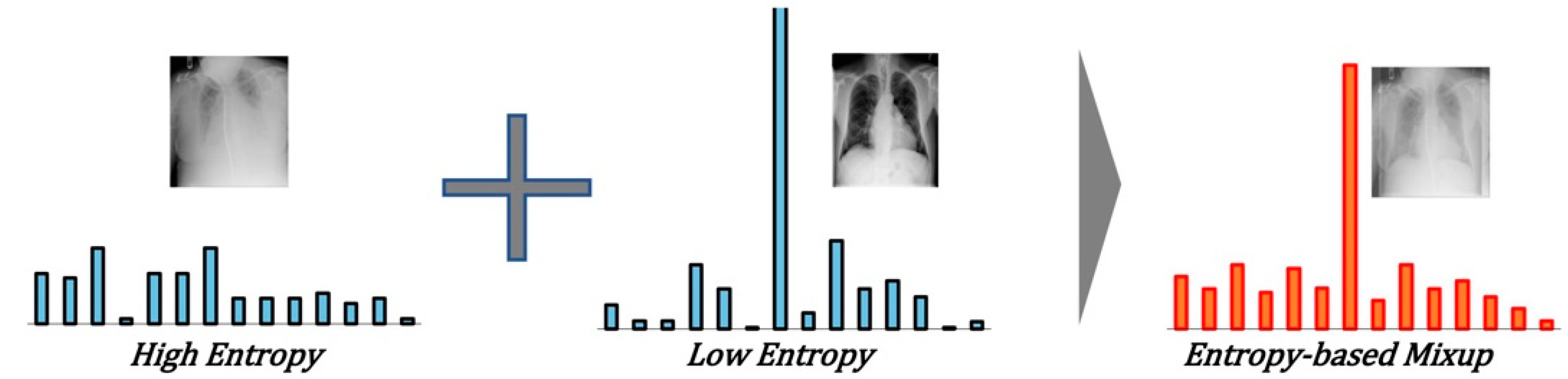
3.2. Self-Training the Student Model and Repetition
4. Experiments
4.1. Datasets and Base Model
4.2. Effects of Entropy-Based Mixup
4.3. Effect of Self-Training
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of Deep Learning Approaches for Multi-Label Chest X-Ray Classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2097–2106. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Zhang, Y.-D.; Zhang, Z.; Zhang, X.; Wang, S.-H. MIDCAN: A multiple input deep convolutional attention network for COVID-19 diagnosis based on chest CT and chest X-ray. Pattern Recognit. Lett. 2021, 150, 8–16. [Google Scholar] [CrossRef]
- Monshi, M.M.A.; Poon, J.; Chung, V. Deep learning in generating radiology reports: A survey. Artif. Intell. Med. 2020, 106, 101878. [Google Scholar] [CrossRef]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Kabra, A.; Chopra, A.; Puri, N.; Badjatiya, P.; Verma, S.; Gupta, P.; Krishnamurthy, B. MixBoost: Synthetic Oversampling using Boosted Mixup for Handling Extreme Imbalance. In Proceedings of the 2020 IEEE International Conference on Data Mining (ICDM), Sorrento, Italy, 17–20 November 2020; pp. 1082–1087. [Google Scholar]
- Thulasidasan, S.; Chennupati, G.; Bilmes, J.A.; Bhattacharya, T.; Michalak, S. On mixup training: Improved calibration and predictive uncertainty for deep neural networks. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Verma, V.; Lamb, A.; Beckham, C.; Najafi, A.; Mitliagkas, I.; Lopez-Paz, D.; Bengio, Y. Manifold Mixup: Better Representations by Interpolating Hidden States. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6438–6447. [Google Scholar]
- Grandvalet, Y.; Bengio, Y. Semi-supervised learning by entropy minimization. Adv. Neural Inf. Process. Syst. 2004, 17. [Google Scholar]
- Xie, Q.; Luong, M.-T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Laine, S.; Aila, T. Temporal ensembling for semi-supervised learning. arXiv 2016, arXiv:1610.02242. [Google Scholar]
- Miyato, T.; Maeda, S.-i.; Koyama, M.; Ishii, S. Virtual adversarial training: A regularization method for supervised and semi-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 41, 1979–1993. [Google Scholar] [CrossRef] [Green Version]
- Van Engelen, J.E.; Hoos, H.H. A survey on semi-supervised learning. Mach. Learn. 2020, 109, 373–440. [Google Scholar] [CrossRef] [Green Version]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef] [Green Version]
- DeVries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6023–6032. [Google Scholar]
- Kim, J.-H.; Choo, W.; Song, H.O. Puzzle Mix: Exploiting Saliency and Local Statistics for Optimal Mixup. In Proceedings of the 37th International Conference on Machine Learning, Virtual Event, 13–18 July 2020; pp. 5275–5285. [Google Scholar]
- Kim, J.-H.; Choo, W.; Jeong, H.; Song, H.O. Co-mixup: Saliency guided joint mixup with supermodular diversity. arXiv 2021, arXiv:2102.03065. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Bellinger, C.; Sharma, S.; Japkowicz, N.; Zaïane, O.R. Framework for extreme imbalance classification: SWIM—Sampling with the majority class. Knowl. Inf. Syst. 2019, 62, 841–866. [Google Scholar] [CrossRef]
- Sohn, K.; Berthelot, D.; Li, C.-L.; Zhang, Z.; Carlini, N.; Cubuk, E.D.; Kurakin, A.; Zhang, H.; Raffel, C. Fixmatch: Simplifying semi-supervised learning with consistency and confidence. arXiv 2020, arXiv:2001.07685. [Google Scholar]
- Lee, D.-H. Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. Workshop Chall. Represent. Learn. ICML 2013, 3, 896. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 702–703. [Google Scholar]
- Berthelot, D.; Carlini, N.; Cubuk, E.D.; Kurakin, A.; Sohn, K.; Zhang, H.; Raffel, C. Remixmatch: Semi-supervised learning with distribution alignment and augmentation anchoring. arXiv 2019, arXiv:1911.09785. [Google Scholar]
- Berthelot, D.; Carlini, N.; Goodfellow, I.; Papernot, N.; Oliver, A.; Raffel, C. Mixmatch: A holistic approach to semi-supervised learning. arXiv 2019, arXiv:1905.02249. [Google Scholar]
- Xie, Q.; Dai, Z.; Hovy, E.; Luong, M.-T.; Le, Q.V. Unsupervised data augmentation for consistency training. arXiv 2019, arXiv:1904.12848. [Google Scholar]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. arXiv 2017, arXiv:1703.01780. [Google Scholar]
- Chen, J.; Yang, Z.; Yang, D. Mixtext: Linguistically-informed interpolation of hidden space for semi-supervised text classification. arXiv 2020, arXiv:2004.12239. [Google Scholar]
- National Institutes of Health-Clinical Center CXR8. Available online: https://nihcc.app.box.com/v/ChestXray-NIHCC (accessed on 1 March 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Müller, R.; Kornblith, S.; Hinton, G.E. When does label smoothing help? Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]

| Without Data Augmentation | Random Mixup (Same Size) | Random: ×2 Mixup (Same Ratio) | Entropy-Based Mixup (Same Ratio) | |
|---|---|---|---|---|
| Atelectasis | 0.6060 | 0.5670 | 0.5644 | 0.6016 |
| Cardiomegaly | 0.5868 | 0.6407 | 0.6492 | 0.6303 |
| Effusion | 0.7295 | 0.6636 | 0.6398 | 0.6701 |
| Infiltration | 0.6242 | 0.6204 | 0.6098 | 0.6190 |
| Mass | 0.5466 | 0.5057 | 0.5437 | 0.5404 |
| Nodule | 0.5658 | 0.5611 | 0.5742 | 0.5677 |
| Pneumonia | 0.5427 | 0.5998 | 0.5659 | 0.5654 |
| Pneumothorax | 0.5951 | 0.4952 | 0.5244 | 0.5912 |
| Consolidation | 0.6489 | 0.6500 | 0.6331 | 0.6470 |
| Edema | 0.6943 | 0.7301 | 0.6948 | 0.6891 |
| Emphysema | 0.5410 | 0.4555 | 0.5559 | 0.5103 |
| Fibrosis | 0.6671 | 0.6935 | 0.6646 | 0.7136 |
| Pleural thickening | 0.6019 | 0.5878 | 0.5203 | 0.6026 |
| Hernia | 0.4424 | 0.5960 | 0.7558 | 0.6670 |
| AUROC average | 0.5994 | 0.5976 | 0.6068 | 0.6154 |
| Teacher Model without Mixup ( ) | Teacher Model with Mixup ( ) | Student Model ( ) | |
|---|---|---|---|
| Atelectasis | 0.6434 | 0.6498 | 0.7123 |
| Cardiomegaly | 0.5465 | 0.5667 | 0.7513 |
| Effusion | 0.7588 | 0.7301 | 0.7731 |
| Infiltration | 0.6423 | 0.6308 | 0.6622 |
| Mass | 0.5704 | 0.5958 | 0.6895 |
| Nodule | 0.5747 | 0.5662 | 0.6829 |
| Pneumonia | 0.6062 | 0.6070 | 0.6293 |
| Pneumothorax | 0.7038 | 0.6532 | 0.7615 |
| Consolidation | 0.7008 | 0.7051 | 0.6935 |
| Edema | 0.7515 | 0.7435 | 0.7951 |
| Emphysema | 0.6403 | 0.6349 | 0.7105 |
| Fibrosis | 0.6494 | 0.7145 | 0.6621 |
| Pleural thickening | 0.6377 | 0.6289 | 0.6895 |
| Hernia | 0.5468 | 0.7043 | 0.6700 |
| AUROC average | 0.6409 | 0.6522 | 0.7059 |
| Improvement | - | +1.8% | +8.2% |
| P | K | AUROC Avg. |
|---|---|---|
| P = 5 | K = 2000 (fix) | 0.6541 |
| P = 10 | 0.7059 | |
| P = 14 | 0.6771 | |
| P = 10(Fix) | K = 1000 | 0.6450 |
| K = 2000 | 0.7059 | |
| K = 4000 | 0.6615 |
| Teacher Model (n = 0) | Student Model (n = 1) | Student Model (n = 2) | Student Model (n = 3) | |
|---|---|---|---|---|
| AUROC average | 0.6522 | 0.7059 | 0.7221 | 0.7409 |
| Improvement | - | +8.2% | +2.3% | +2.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, M.; Kim, J. Self-Training with Entropy-Based Mixup for Low-Resource Chest X-ray Classification. Appl. Sci. 2023, 13, 7198. https://doi.org/10.3390/app13127198
Park M, Kim J. Self-Training with Entropy-Based Mixup for Low-Resource Chest X-ray Classification. Applied Sciences. 2023; 13(12):7198. https://doi.org/10.3390/app13127198
Chicago/Turabian StylePark, Minkyu, and Juntae Kim. 2023. "Self-Training with Entropy-Based Mixup for Low-Resource Chest X-ray Classification" Applied Sciences 13, no. 12: 7198. https://doi.org/10.3390/app13127198





