Abstract
A significant amount of research has investigated automating medical diagnosis using deep learning. However, because medical data are collected through diagnostic tests, deep learning methods used in existing studies have had a disadvantage in that the number of training samples is insufficient and the labeling cost is high. Training approaches considering the common characteristics of medical images are needed. Therefore, in this study, we investigated approaches to overcome the lack of data for representative medical imaging tasks using transfer learning technologies. The tasks were divided into image classification, object detection, and segmentation, commonly needed functions in medical image analyses. We proposed transfer learning approaches suitable for each task that can be applied when there are little medical image data available. These approaches were experimentally validated in the following applications that share similar issues of lacking data: cervical cancer classification (image classification), skin lesion detection and classification (object detection and classification), and pressure ulcer segmentation (segmentation). We also proposed multi-task learning and ensemble learning that can be applied to these applications. Finally, the approaches were compared with state-of-the-art results. In cervical cancer analysis, the performance was improved by 5.4% in sensitivity. Skin lesion classification showed improvement in accuracy of 8.7%, precision of 28.3%, and sensitivity of 39.7%. Finally, pressure ulcer segmentation improved in accuracy by 1.2%, intersection over union by 16.9%, and Dice similarity coefficient by 3.5%.
1. Introduction
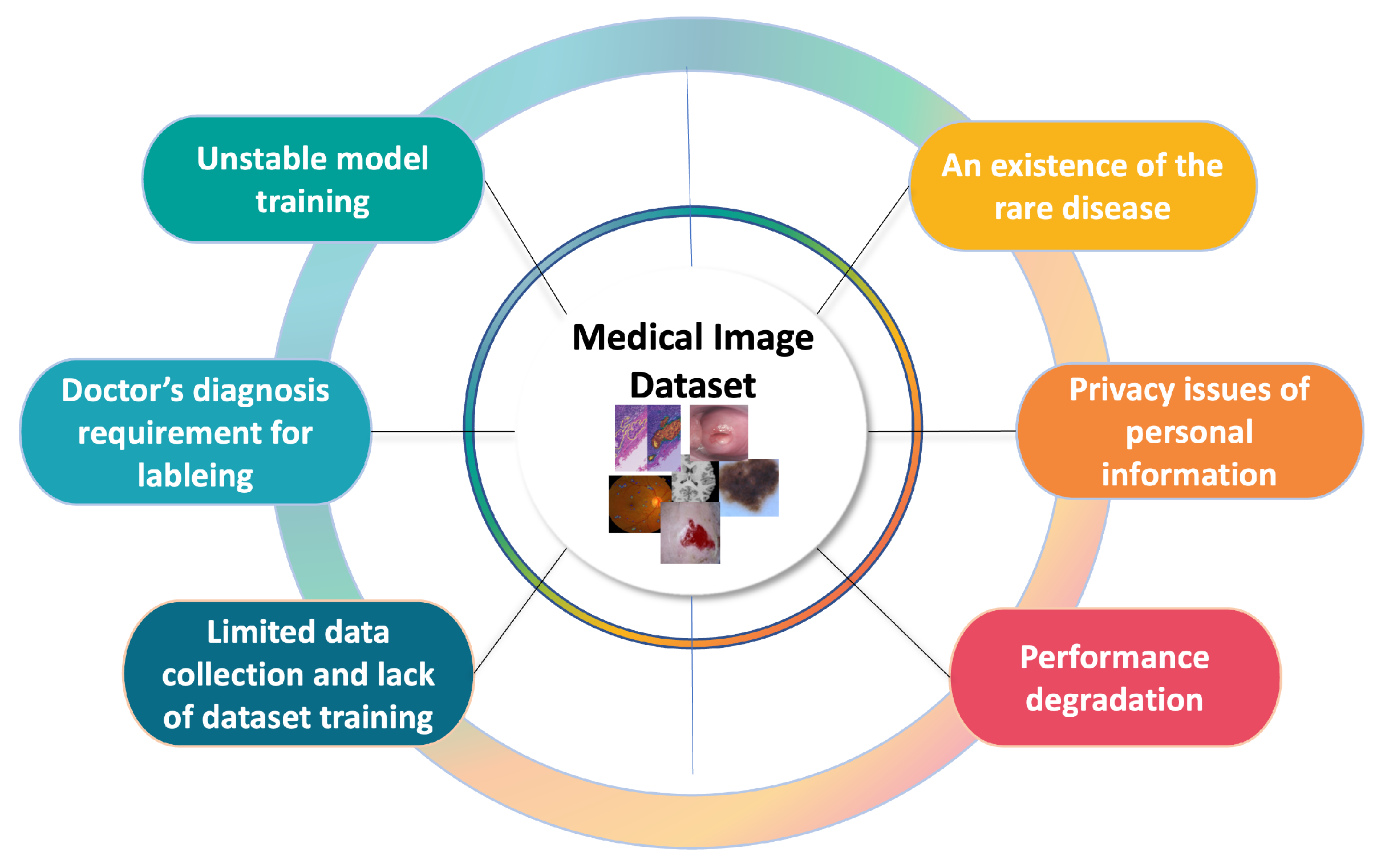
As deep learning research grows, numerous studies are being conducted to automate medical diagnosis using medical image analysis. In particular, the invention of the variant convolutional neural network (CNN) [1,2,3,4,5], which extracts feature maps of images and ensures some degree of shift, scale, and distortion invariance, has accelerated the development of computer vision tasks, including medical image analysis. However, the nature of medical images gives rise to problems including a lack of training data, the high cost of labeling, images left unlabeled by clinicians, and an insufficient number of patients for sampling, as shown in Figure 1. As the problems are connected to the challenges of model training, it is important to focus on how to design or train the model with the limited labeled data (i.e., without collecting more training data). For these reasons, extensive deep learning research has been conducted to solve the problems with automating medical diagnosis. In particular, medical image analysis is primarily divided into three tasks—(1) classification, (2) detection, and (3) segmentation—to solve the chronic problems with medical images associated with each task.
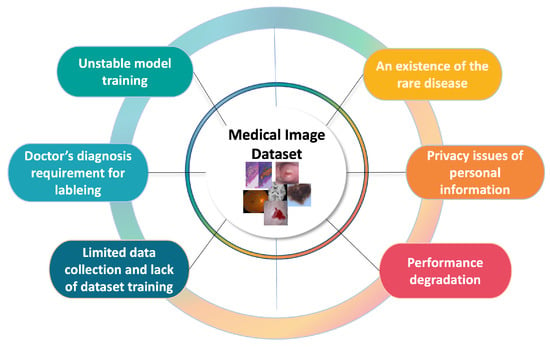
Figure 1.
Diagram of medical image analysis problems.
Image classification, which returns one output value for an input image, is used, for example, for disease classification and cancer diagnosis. In these classification tasks, all the diagnostic tests become samples, and the dataset is significantly smaller than those typically used in computer vision. Furthermore, unlabeled image data cannot be used in supervised learning. Therefore, in the classification task, due to the limited data, a transfer learning method is often used to extract visual features using the weights of a network pre-trained with the ImageNet dataset [6], and the weights are fine-tuned according to the task. One study divided the image into several patches and used it as an input to the model to capture local features from unlabeled data [7]. However, previous works used networks pre-trained with a large dataset, such as the ImageNet dataset [6], and then fine-tuned the networks to improve their performances, but they did not investigate the fact that networks pre-trained with images similar to the tasks might be better than networks pre-trained with the large dataset.
An example of the detection task in medical image analysis is lesion detection. This task consists of locating areas of interest or identifying small lesions in an image, which is a key part of diagnosis. Creating labeled data for this task is the most labor-intensive task for clinicians. Therefore, because such manual labeling is frequently required, semi-supervised learning has been applied, as in [8]. In addition, the authors of [9] combined classification with lesion detection to improve the classification performance. That is, the authors attempted to diagnose a lesion after extracting the region of interest (RoI) from an image. Object detection is often used with classification, wherein the task is used to extract RoIs and classification is then performed using the extracted RoIs due to the lack of a training dataset.
Finally, typical examples of the segmentation task are organ segmentation and lesion segmentation. Examples of organ segmentation include heart analysis and brain analysis requiring quantitative analysis of clinical parameters, such as the specific shape or volume, through the segmentation of organs or the organ’s substructures. The quantitative analysis of organs is performed through magnetic resonance imaging (MRI) or radiography, but there is a lack of data because a limited number of patients undergo the examination. Lesion segmentation is a combination of object detection and segmentation. The most well-known segmentation study used the U-net structure, a model architecture that can make use of the entire image context, and the decoder of U-net uses the encoder information by concatenating the encoder feature [10]. Some studies have combined lesion segmentation with classification. For example, a study on multi-task learning with a two-stream structure performed lesion segmentation and classification simultaneously [11].
In this study, to overcome the lack of data for these tasks, transfer learning approaches are proposed as follows, showing how each approach can be applied to each task: (1) transfer learning using weights learned from similar images; (2) transfer learning through a new type of patch self-supervised learning, a method that can be applied to unlabeled data; and (3) transfer learning using weights learned to distinguish RoIs. The method in (1) assumes that the model performance can be improved by transfer learning using the weights of the model trained with an image similar to the original task because of the lack of a dataset. For (2), a novel type of patch self-supervised learning is proposed to learn information from the image itself to use unlabeled data because of the prohibitive cost of target labeling. Although self-supervised learning is well-known in computer vision, patch self-supervised learning is newly presented to apply to the three target problems (i.e., classification, detection, and segmentation tasks) rather than a single classification problem with medical image analysis in this study. In addition, the cost of lesion and cancer labeling (e.g., abnormal/normal labeling) is very high because skilled clinicians are required. To eliminate this cost, an RoI is identified to be intensively viewed in the image, and because such RoI labeling is a simple task, it is less expensive than the actual target labeling. Therefore, for (3), a transfer learning method using only the image RoI is needed. The weight of the model trained on RoIs is learned again on the target classification. Furthermore, inspired by a clinician’s diagnosis focusing on the RoI of the image, multi-task learning is proposed. Finally, to improve the performance, ensemble learning using appropriate transfer learning approaches for each task is introduced.
We deliberately designed experiments with the limited labeled dataset to solve the above-mentioned problem, and the approaches were experimentally validated with cervical cancer classification, skin lesion detection and classification, and pressure ulcer segmentation, representative applications in medical image analysis. The following were observed from these experiments. The image classification and object classification tasks determine where to look in the image to assess the disease. Therefore, transfer learning using RoIs is effective for these tasks. Moreover, for the task of determining an overall shape, such as segmentation, transferring knowledge with a similar image is effective.
The contributions of this study are as follows:
- Because of the lack of data in medical images, we propose transfer learning approaches: (1) transfer learning with similar images, (2) a new approach using transfer learning with self-supervised learning, and (3) transfer learning with RoIs. We apply these approaches to image classification, object detection, and segmentation tasks, respectively, in representative medical images and demonstrate their performance.
- In addition to these approaches, we propose a multi-task learning and ensemble approach using the optimal transfer learning method appropriate to medical images.
- When these approaches were applied to cervical cancer classification, skin lesion classification, and pressure ulcer segmentation and the results were compared to those of state-of-the-art approaches, for cervical cancer classification, they improved the sensitivity by 5.4%; for skin lesion classification, they improved the accuracy by 8.7%, the precision by 28.3%, and the sensitivity by 39.7%; and for pressure ulcer segmentation, they improved the accuracy by 1.2%, the intersection over union (IoU) by 16.9%, and the Dice similarity coefficient (DSC) by 3.5%.
2. Related Work
In this study, we investigate the methods for solving the problem of the lack of data in medical images based on classification, object detection, and segmentation, which are representative computer vision tasks. A summary of the prior methods is presented in Table 1.
2.1. Classification
The most active area of research is image classification, which is used in disease classification and cancer diagnosis. In particular, various deep learning methods have been proposed for cervical cancer analysis. The study of [12] used the pyramid histogram of local binary patterns (PLBP), pyramid histogram in LAB color space (PLAB), and pyramid histogram of oriented gradients (PHOG) to extract features and classified the extracted features using various machine learning methods, such as random forest, AdaBoost, support vector machine (SVM), and logistic regression. In addition, cervical cancer was classified through an RoI pretreatment method and a pre-trained CaffeNet model. In [13], the authors proposed the LeNet model, which combined additional preprocessing and image size changes using 345 positive and 345 negative images randomly extracted from the NCI data. However, there was a bias because the training set and the validation set contained the same session images. The study of [14] introduced the Faster R-CNN model [15] to detect cervical region and then classified the types of cancer with the extracted region. In [16], the authors used two pre-trained models for cervix detection and cancer classification. The authors trained the models to detect only the cervix area from Intel and MobileODT images using the YOLO (you only look once) algorithm and extracted the RoIs from the U.S. National Cancer Institute (NCI) images through the trained model. Cervical cancer was classified through the CNN-based GoogLeNet model from the extracted RoIs at a high level of performance. Most previous studies used labeled data to extract visual features, which they then employed to classify cancer. Because the amount of labeled data is limited due to the nature of medical images, we tried to improve the model’s performance through RoI extraction. The study of [17] presented a CNN model, but the authors applied patch weight learning and a spatial regulator with Gaussian distribution to detect only cervical region. Ref. [18] proposed an ensembled transfer learning framework for classifying differentiation stage of cervical histopathology images. Ref. [19] proposed variant CNN models with a transfer learning approach to diagnose the stage of cervical cancer by classifying the cervix images using the ODT Kaggle dataset. Also, ref. [20] proposed a cervical cell image generation model based on taming transformers [21] to provide high-quality cervical cancer datasets and used Tokens-to-Token Vision Transformers (T2T-ViTs) [22] combing transfer learning to classify the cervical cancer. As for recent works, refs. [23,24] used variant CNN models and adopted transfer learning to fine-tune models. Ref. [25] introduced a CerviFormer that depends on transformers and used a cross-attention module which enables it to manage very large-scale inputs.
Furthermore, there have been other studies with various applications, including colonoscopy frame classification, polyp detection, pulmonary embolism detection, diabetic macular edema classification, and pneumonia detection. Refs. [26,27] introduced an active incremental fine-tuning method and layer-wise fine-tuning method, respectively, using a pre-trained AlexNet model. The authors demonstrated their method on a small dataset with colonoscopy videos and computed tomography (CT) pulmonary angiography. Ref. [28] experimented with the Inception V3 model pre-trained on the ImageNet dataset on the OCT dataset and chest X-ray images. Refs. [26,27,28] showed that fine-tuned deep models are more robust than models from scratch in various medical imaging tasks. Moreover, ref. [29] introduced a classification task of malignant and benign breast masses and used a deep CNN model with stochastic gradient descent on small mammography images. The authors demonstrated that multi-task transfer learning can be an effective approach for training a deep CNN when training samples are limited. Ref. [30] introduced an ensemble deep transfer learning model for breast cancer classification and used DenseNet201 and ResNet152V2 for transfer learning. As increasing interests of a transformer model, ref. [31] presented a transformer with localization U-Net model for pancreatic cancer classification, and [32] introduced ViT and variant CNN models including pre-trained VGG16 and ResNet50 for breast cancer classification. Also, refs. [33,34] studied transformer-based models for fundus disease and prostate cancer classifications. For further study, ref. [35] introduced variant transformer models, such as scale-aware transformer, and [36] utilized encoded multi-scale features with a transformer to improve the classification performance. In COVID-19 classification, ref. [37] introduced a deep transfer learning model combined with machine learning algorithms for predicting COVID-19 disease with chest CT images. Refs. [38,39] also introduced ViT [40] using a performer and feature aggregation method by transformer. Therefore, because of the introduction of the transformer model, various transformer-based models have been studied to improve classification performance in the medical domain. Ref. [41] proposed a novel image pre-processing method that utilizes histogram equalization and contrast limited adapative histogram equalization to create a three-channel image representation, and the method is evaluated using pre-trained models. Ref. [42] experimented with variant CNN models pre-trained on the ImageNet dataset on a chest X-ray dataset to classify COVID-19.
Some studies have classified the types of lesions. Ref. [43] introduced a regularizer that can be used for the size and number of CNN filters and experimented on the International Skin Imaging Collaboration (ISIC) 2018 dataset to classify a combination of two classes (namely, nevus vs. melanoma) out of seven rather than classifying all seven lesions. Ref. [44] applied a pre-trained CNN model with data augmentation on the ISIC 2018 and 2019 dataset. Ref. [45] presented a ResNet50 model with curriculum learning and momentum contrastive learning and evaluated the method on the ISIC 2019 dataset. Ref. [46] proposed classifying with SVM by re-extracting only the features of the last fully connected layer using various models, such as AlexNet, VGG16, and ResNet18, which were pre-trained with ImageNet. Because the ISIC 2016 and 2017 datasets were used in the experiment, it had a limitation of only two classes. Furthermore, ref. [47] presented a deep belief network using semi-supervised learning that could use both labeled and unlabeled data. The authors proposed an SVM learning method to generate advice weights from misclassified data and applied them to a test set. Although the authors introduced self-supervised learning for classification, no pretext task was given to learn self-information in the image. Rather, an unlabeled image was simply used to adjust the parameter space of the model. In addition, this study had a limitation of only two classes, melanoma and benign. Refs. [48,49] presented a method to transfer knowledge from a pre-trained CNN and used the model to classify seven lesion classes on the HAM10000 dataset. Moreover, ref. [50] suggested a ResNet50 model with task-specific layers for skin lesion classification and demonstrated that learning different tasks together in the same biomedical imaging modalities has the potential to train deep models effectively. As for recent works, ref. [51] introduced Xception and ShuffleNet models using transfer learning and utilized butterfly optimization to improve feature selection. Ref. [52] proposed a Skin-net using residual learning and cross-channel correlation.
2.2. Object Detection
In medical images, object detection is often used in conjunction with classification. Ref. [12] extracted RoIs by calculating the similarity of the color and texture information of the ground truth bounding box using object detection. As detection models have gained increasing interest, variant R-CNN models, including Fast R-CNN [53], Faster R-CNN [15], Mask R-CNN [54], and variant Yolo models [55,56], have been studied. Ref. [14] applied the Faster R-CNN model to detect cervical cancer region and then classified the types of cancer with the detected region. Similarly, ref. [16] used cervical region detection to classify cervical cancer. An image of cervical cancer was detected in the cervix area of an image using a GoogLeNet model to remove unnecessary parts with a magnifying glass device. In preprocessing, the RoI size was reduced, and the data augmentation technique was used to increase the data size. Cancer diagnosis classification was then performed through CNN model training. In the study, the RoI was extracted by applying object detection, but it was also important to see the distribution of blisters in cervical cancer. Ref. [57] presented a Mask R-CNN model to detect pressure ulcer region, and [58] also used Fast R-CNN object detection model to classify COVID-19. Based on the Single Shot MultiBox SDD 300 model, which adjusts the box to the object shape, the lungs in the X-ray image were quickly detected. In addition, negative and positive COVID-19 cases were classified from the lung area detected. In [59], the lung area of chest X-ray images was segmented using U-Net, and it was used to classify pneumonia, COVID-19, and control cases through CNNs. In addition, ref. [60] attempted to detect pneumonia first and then classify COVID-19 and pneumonia using the VGG16 model pre-trained with ImageNet. Therefore, because existing studies show that object detection extracts RoIs approximately as well as segmentation, this study also approaches object detection through segmentation tasks.
2.3. Segmentation
The purpose of image segmentation is to classify all pixels into a specific class [61]. Predicting all pixels is also called pixel-level classification. In the medical field, image segmentation includes organ segmentation and lesion segmentation, which are used in studies to analyze specific shapes or volumes. Recently, various deep learning studies that performed well with little data have been proposed in the medical image segmentation field. For example, ref. [62] introduced a CNN-based end-to-end model with 2700 wound images with a 47.3% IoU. Ref. [63] also created a CNN model, but when the given dataset was small, data augmentation was performed using color change, resulting in a 53% IoU. In 2017, ref. [64] proposed automated segmentation of diabetic foot ulcers and their surrounding skin using a fully convolutional network (FCN). The FCN-16 model using transfer learning performed best, with a 79.4% DSC. However, while the ground truth had a smooth curve, the predicted segmentation had low accuracy due to irregular boundaries. Ref. [65] presented a 3D CNN-based classification framework in which various pathways existed with a pressure ulcer image. The model trained with 193 images resulted in a 92% DSC and 95% area under the curve (AUC). Ref. [66] presented various feature extraction approaches based on color and text and classified four wound tissues, resulting in an average accuracy of 96%. Ref. [67] experimented with variant segmentation models, and the authors studied the UNet with the VGG19 pre-trained on the ImageNet dataset. Ref. [68] proposed an image segmentation framework, WSNet, which leverages wound domain adaptive pre-training on unlabeled wound images and adopted a global–local architecture to learn fine-grained features. Ref. [69] introduced FANet with an edge feature augment module and a spatial relationship feature augment module to utilize the edge information and the spatial relationship between the wound’s and the skin’s regions. Also, refs. [57,70] applied a Mask R-CNN model, but [57] adopted a mesh rasterization method to the model to segment the pressure ulcer. Ref. [71] used a YOLOv5 model to segment and classify pressure ulcers. Ref. [72] experimented with their anatomical structure segmentation method using a CNN with various training techniques, including exponential linear units, batch normalization, and dropout. Moreover, ref. [73] proposed an unsupervised domain adaptation framework based on adversarial networks. The authors divided the method into two parts, a discriminator and a segmentor, and used ResNet18 and FCN, respectively. Ref. [74] tested their method on two tasks, eye vasculature segmentation and neuron membrane detection. The authors proposed two networks, FCN for segmentation and CNN for classification on the segmentor outputs, and connected the networks through a gradient reversal layer for adversarial training. Also, studies using the transformer model have been introduced. Ref. [75] presented a U-shape transformer network for cardiac segmentation and [76] introduced the class-aware adversarial transformer model. Variant UNet models including H-DenseUNet, UNet3+, and nnUNet have been studied for improving the performance of segmentation and then applied to brain tumor, heart, liver, and lung segmentation tasks [77,78,79]. Ref. [80] combined UNet and swin transformer models for brain tumor segmentation. Ref. [81] introduced 3D-UNet and SegResNet, combined with advanced fusion techniques to fuse PET and CT information for tumor segmentation. As for recent works, refs. [82,83] proposed AIM-UNet and CCLnet with cascaded context modules and ladder atrous spatial pyramid pooling, respectively, to segment liver tumors. Ref. [84] experimented with the ResNet model, while [85] utilized the U-Net model on the BraTS 2020 dataset.
Related works in the medical imaging domain are examined and organized into three parts: classification, detection, and segmentation. Based on the examination, this study proposes four transfer learning methods for overcoming the problem of a lack of training samples. It then demonstrates their performance on three representative medical imaging tasks, cervical cancer classification, skin lesion classification, and pressure ulcer segmentation. This study also shows the effective transfer learning method according to the properties of the respective tasks.

Table 1.
Overview of studies using deep learning techniques for medical image analysis. TL* denotes transfer learning.
Table 1.
Overview of studies using deep learning techniques for medical image analysis. TL* denotes transfer learning.
| Reference | Application/Task | Approach | Data | Metric | TL* |
|---|---|---|---|---|---|
| Xu et al., 2017 [12] | Cervical cancer classification | CaffeNet pre-trained with ImageNet, machine learning classifier, three pyramid features: PLAB, PHOG, PLBP | 345 positive/345 negative images by NCI | Accuracy (Acc) | √ |
| Vasudha et al., 2018 [13] | Cervical cancer classification | CNN features, LeNet model | 345 positive/345 negative images randomly selected by NCI | Acc | ✗ |
| Hu et al., 2019 [14] | Cervical cancer classification | Faster R-CNN, CNN pre-trained with the ImageNet dataset and data augmentation | Cervical images by NCI | AUC, Sensitivity, Specificity | √ |
| Alyafeai et al., 2020 [16] | Cervical cancer object detection and classification | YOLO algorithm, GoogLeNet model, data augmentation | Intel & MobileODT dataset, 174 positive/174 negative images by NCI | Acc, specificity, sensitivity | ✗ |
| Xue et al., 2020 [18] | Cervical histopathology classification | Inception-V3, Xception, VGG-16, Resnet-50, and ensembled transfer learning | Histopathology images from China Medical University | Acc, precision, recall, f1-score | √ |
| Zhang et al., 2021 [17] | Cervical cancer classification | CNN with patch weights learning and spatial regulator | 978 records by NCI | Acc, AUC, equal error rate, F1-score, precision, recall | ✗ |
| Dhawan et al., 2021 [19] | Cervical cancer classification | Inception V3, ResNet50, VGG19 | Intel & MobileODT dataset | Precision, recall, f1-score, support, confusion matrix | √ |
| Zhao et al., 2022 [20] | Cervical cancer classification | Taming transformer with pre-trained T2T-ViT | Pap smear dataset consisting of a total of 963 LBC images with four classes, SIPAKMeD consisting of 4049 images of isolated cells with 5 classes, Herlev consisting of 917 isolated single-cell images with seven classes | Acc, F1-score, H-mean, sensitivity, specificity | √ |
| Ghantasala et al., 2023 [23] | Cervical cancer classification | CNN, VGG16 | Melanoma histopathology images | Acc | √ |
| Deo et al., 2023 [25] | Cervical cancer classification | CerviFormer, cross-attention | Pap smear datasets—Sipakmed dataset and Herlev dataset | Acc, precision, recall, f1-score | ✗ |
| Kalbhor and Shinde 2023 [24] | Cervical cancer classification | Pre-trained feature extractor, resnet50, Google Net | Herlev dataset with 917 images | Acc | √ |
| Zhou et al., 2017 [26] | Colonoscopy frame classification, polyp detection, pulmonary embolism detection | Pre-trained AlexNet model with active incremental fine-tuning method | 4000 colonoscopy frames selected from 6 complete colonoscopy videos, 38 short colonoscopy videos from 38 different patients, 121 CT pulmonary angiography datasets with a total number of 326 pulmonary embolisms | AUC, diversity, entropy | √ |
| Tajbakhsh et al., 2016 [27] | Colonoscopy frame classification, polyp detection, pulmonary embolism detection, intima-media boundary segmentation | Pre-trained AlexNet model with layer-wise fine-tuning method | 4000 colonoscopy frames selected from 6 complete colonoscopy videos, 40 short colonoscopy videos from 38 different patients, 121 CT pulmonary angiography datasets with a total number of 326 pulmonary embolism, 92 carotid intima-media thickness videos | AUC, free-response operating characteristic, ROC curve, segmentation error | √ |
| Kermany et al., 2018 [28] | Age-related macular degeneration and diabetic macular edema classification, Pneumonia detection | Inception V3 model pretrained on the ImageNet dataset | 207,130 OCT images. 5232 chest X-ray images from children, including 3883 characterized as depicting pneumonia (2538 bacterial and 1345 viral) and 1349 normal from a total of 5856 patients | Acc, cross-entropy loss, sensitivity, specificity, weighted error ROC curve | √ |
| Xia et al., 2021 [31] | Pancreatic cancer classification | Anatomy-aware transformer with localization u-net | pancreatic ductal adenocarcinom CT scans dataset of 1627 patients | AUC, sensitivity, specificity | ✗ |
| Gheflati B and Rivaz H 2021 [32] | Breast cancer classification | ViT, pretrained VGG16, ResNet50, InceptionV3, and NASNetLarge models based on the breast US dataset | BUSI dataset which has 780 breast US images (133 normal images, 437 malignant masses, and 210 benign tumors), Dataset B which has 163 images (110 benign masses and 53 cancerous masses) | Acc, AUC | √ |
| Rezaeijo et al., 2021 [30] | Breast cancer classification | DenseNet201 and ResNet152V2 | QIN-Breast dataset from TCIA | Acc, precision, recall, f1-score | √ |
| Yang et al., 2021 [33] | Fundus disease classification | Transformer, CNN | OIA dataset | Acc | ✗ |
| Ikromajanov et al., 2022 [34] | Prostate cancer classification | ViT | Kaggle PANDA challenge dataset which has 10,616 images | F1-score, precision, recall | √ |
| Samala et al., 2017 [29] | Classification of malignant and benign breast masses | Deep CNNs with stochastic gradient descent | 1655 digitized-screen film mammography views and 310 digital mammography views with 2454 masses (1057 malignant, 1397 benign) | AUC, ROC curves | √ |
| Masood et al., 2015 [47] | Skin lesion classification | Ensemble, deep belief network, self-advising approach for SVM (RBF kernel, poly kernel) | 290 images with 170 benign, 120 melanoma, Sydney Melanoma Diagnostic Centre, Royal Prince Alfred Hospital | Acc | ✗ |
| Pal et al., 2018 [48] | Skin lesion classification | Various pre-trained CNN models (ResNet50, DenseNet121, MobileNet) | 10,015 images from HAM10000 dataset | Acc | √ |
| Liao and Luo 2017 [50] | Skin lesion classification | ResNet-50 with multi-task learning | DermQuest images with 25 lesion types. 21,657 images that contain both the skin lesion and body location labels | Acc, average precision | √ |
| Carcagnì et al., 2019 [49] | Skin lesion classification | Pre-trained DenseNet121, SVM | 10,015 images from HAM10000 dataset | Precision, recall | √ |
| Mahbod et al., 2019 [46] | Skin lesion classification | Features with a pre-trained model (AlexNet, VGG16, ResNet18), SVM | 2037 images by ISIC 2016, 2017 datasets with 411 malignant melanomas, 254 seborrheic keratoses, 1372 benign nevi | AUC | √ |
| Albahar 2019 [43] | Skin lesion classification | CNN model with an embedded regularizer | 23,906 images by ISIC 2018 with skin melanoma images of benign and malignant lesions | Acc, AUC, sensitivity, specificity | ✗ |
| Nahata and Singh 2020 [44] | Skin cancer classification | Pre-trained CNN and data augmentation | ISIC 2018 and 2019 dataset | Acc, F1-score, precision, recall | √ |
| Sirotkin et al., 2021 [45] | Skin lesion classification | ResNet50 model with curriculum orderings and sequential self-supervised pre-training | ISIC 2019 dataset | AUC, balanced Acc | √ |
| Ahmad et al., 2023 [51] | Skin lesion clasification | Xception, ShuffleNet, data augmentation, butterfly optimization algorithm | ISIC 2018 dataset, HAM10000 dataset | Acc, precision, recall, f1-score, FNR | √ |
| Alsahafi et al., 2023 [52] | Skin lesion classification | Skin-net, cross-channel correlation | ISIC 2019 dataset, ISIC 2020 datset | Acc, precision, sensitivity, specificity, f1-score | ✗ |
| Yu et al., 2021 [35] | Melanoma (Skin) classification | Pre-trained MobileNetv2 to extract patch-wise embeddings and scale-aware transformer with soft labeling | ISIC 2020 dataset | Acc, AUC, F1-score, sensitivity, specificity | √ |
| Wu et al., 2021 [36] | Melanocytic lesions (skin) classification | Encoded multi-scale features with transformer | MPATH-Dx dataset | Acc, AUC, F1-score, sensitivity, specificity | ✗ |
| Wang et al., 2015 [62] | Wound segmentation, infection, detection, healing prediction | Encoder-decoder of ConvNet, SVM, Gaussian process | NYU database | Pixel Acc, mean IoU | ✗ |
| Goyal et al., 2017 [64] | Wound segmentation | FCN for diabetic foot ulcer segmentation, two-tier transfer learning with ImageNet and Pascal VOC dataset | Medetec medical image database, approximately a few hundred wound images | IoU, precision, recall | √ |
| Pholberdee et al., 2018 [63] | Foot ulcer segmentation | CNN, color data augmentation | Lancashire Teaching Hospitals | DSC, matthews correlation coefficient, specificity, sensitivity | ✗ |
| Garcia-Zapirain et al., 2018 [65] | Pressure ulcer segmentation, classification | 3D CNN, RoI, preselected Gaussian kernel 3D hue- saturation- intensity color space images | 193 test color pressure ulcer images, granulation necrotic eschar, and slough tissue images | AUC, DSC, percentage area distance | ✗ |
| Khalil et al., 2019 [66] | Wound segmentation, classification, healing assessment | Various color feature spaces, texture spaces, features extracted through scale-invariant feature transform, non-negative matrix factorization–based feature reduction, various classifiers (naïve Bayes, generalized linear model, random forest) | Medetec wound database with 341 RGB pressure ulcer images, 36 pressure ulcer images from National Pressure Ulcer Advisory Panel website | Acc, precision, sensitivity | ✗ |
| Ohura et al., 2019 [67] | Wound segmentation | SegNet, LinkNet, U-Net, and U-Net with the VGG16 encoder pre-trained on ImageNet | dataset of pressure ulcers, diabetic foot ulcers, and venous leg ulcers data | Acc, AUC, sensitivity, specificity | √ |
| Oota et al., 2023 [68] | Wound segmentation | WSNet with global-local architecture | WoundSeg dataset with 8 types (diabetic, pressure, trauma, venous, surgical, arterial, cellulitis and others) | IoU, DSC | √ |
| Zhang et al., 2023 [69] | Wound segmentation | FANet with a edge feature augment module and a spatial relationship feature augment module, IFANet | MISC dataset with 5523 skin wound images and FUSeg dataset with 1010 images | IoU, sensitivity, specificity | ✗ |
| Zahia et al., 2020 [57] | Pressure ulcer segmentation | Mask R-CNN with mesh rasterization, matching block and measurement block | 210 photographs of pressure injuries including 100 images from Medetec Medical Images | DSC, RMSE, mean absolute error | √ |
| Swerdlow et al., 2023 [70] | Pressure ulcer segmentation | Mask R-CNN | 969 pressure injury images from eKare Inc. | Acc, precision, recall, f1-score, DSC | √ |
| Aldughayfiq et al., 2023 [71] | Pressure ulcer detection and classification | YOLOv5 | Medetec image database and online source images | Mean average precision, recall, f1-score | ✗ |
| Moeskops et al., 2016 [72] | Anatomical structure segmentation | CNN with exponential linear units, batch normalization, and dropout | Brain MRI, breast MRI, cardiac CTA | DSC | √ |
| Dong et al., 2018 [73] | Chest organ segmentation | ResNet18 for discriminator and FCN for segmentor, with unsupervised learning and semi-supervised learning | JSRT dataset for lung and heart segmentation, which contains 257 grayscale chest X-rays. Wingspan dataset which contains 221 grayscale chest X-rays for adult patients with annotated key point for calculation of cardiothoracic ratio | Average percentage error, IoU, mean absolute error, root mean squared error (RMSE) | √ |
| Javanmardi et al., 2018 [74] | Eye vasculature segmentation, neuron membrane detection | FCN for segmentation and CNN for classification on the outputs of the segmentor, connected through a gradient reversal layer. | DRIVE dataset consists of 40 eye fundus images with corresponding pixel labeled ground truth images. STARE dataset with 20 annotated eye fundus images. 100 electron microscopy images of size 1024 × 1024 and 125 electron microscopy images of size 1250 × 1250 | F1-score | √ |
| Li et al., 2018 [77] | Liver tumor segmentation | H-DenseUNet consisting of the 2D U-Net and the 3D Counterpart followed by jointly optimizing the hybrid features in the hybrid feature fusion layer | LiTS dataset containing 131 and 70 contrast-enhanced 3D abdominal CT scans and 3DIRCADb dataset containing 20 venous phase enhanced CT scans | Average symmetric surface distance, Dice, relative volume difference, RMSE, root mean symmetric surface distance, volumetric overlap error | ✗ |
| You et al., 2022 [76] | Liver tumor segmentation | Class-aware adversarial transformer | Synapse1, LiTS, and MP-MRI | Average symmetric surface distance, DSC, jaccard Index, hausdorff distance | √ |
| Özcan et al., 2023 [82] | Liver tumor segmentation | AIM-UNet | HAOS, LiST, 3DIRCADb datsaets | Acc, DSC, IoU | ✗ |
| Rongrong et al., 2023 [83] | Liver tumor segmentation | CCLnet with cascaded context module and Ladder Atrous Spatial Pyramid Pooling, ResNet-34 | LiTS 2017 dataset, 3DIRCADb dataset | Asymmetric metric, DSC, euclidean distance, volumetric overlap | ✗ |
| Huang et al., 2020 [78] | Liver and spleen segmentation | UNet3+ with full-scale skip connections and full-scale deep supervision | ISBI LiTS 2017 dataset containing 131 contrast-enhanced 3D abdominal CT scans. Spleen dataset from the hospital containing 40 and 9 CT volumes | Dice | ✗ |
| Arias-Londono et al., 2020 [59] | Lung segmentation, classification | Deep CNN based on COVID-Net, U-Net, Grad-CAM | 49,983 control X-ray images, 24,114 pneumonia X-ray images, 8573 COVID-19 X-ray images | Acc, F1-score, geometric mean recall, positive predictive value, recall | ✗ |
| Gao et al., 2022 [75] | Cardiac segmentation | U-shape hybrid Transformer Network (a hybrid hierarchical architecture with bidirectional attention) | Large Cardiac MRI dataset, ACDC, M&Ms, M&Ms-2, and UK Biobank | DSC, hausdorff distance | ✗ |
| Hatamizadeh et al., 2022 [80] | Brain tumor segmentation | Swin U-Net transformer | BraTS 2021 datset | DSC, hausdorff distance | ✗ |
| Aggarwal et al., 2023 [84] | Brain tumor detection and segmentation | ResNet | BraTS 2020 dataset with 369 MR images | DSC, IoU, MSE, peak signal noise ratio, sensitivity, specificity | ✗ |
| Montaha et al., 2023 [85] | Brain tumor segmentation | U-Net | BraTS 2020 dataset | DSC | ✗ |
| Fatan et al., 2022 [81] | Head and neck tumor segmentation | 3D-UNet and SegResNet, combined with advanced fusion techniques | PET and CT images from HECTOR Challenge | Dice score | ✗ |
| Isensee et al., 2021 [79] | Variant segmentation | nnUNet with image normalization and voxel spacing preprocessing | Automatic Cardiac Segmentation Challenge, PROMISE12 dataset, LiTS, Multi-Atlas labeling beyond the Cranial Vault challenge | Average foreground dice score | ✗ |
| Saiz et al., 2020 [58] | Lung detection | Single Shot MultiBox Detector, VGG16 pre-trained with ImageNet, Fast R-CNN, contrast-limited adaptive histogram equalization | 887 normal X-ray images and 100 COVID-19 X-ray images | Acc, sensitivity, specificity | √ |
| Brunese et al., 2020 [60] | COVID-19 classification | Variant VGG16 pre-trained with ImageNet, Grad-CAM | 250 images of COVID-19, 2753 X-ray images of pulmonary disease, and 3520 healthy X-ray images | Acc, F-measure, sensitivity, specificity | √ |
| Rezaeijo et al., 2021 [37] | COVID-19 classification | DenseNet201, ResNet50, Xception, VGG16, RandomForest, SVM, Decision tree, Logistic regression | 5480 CT images of confirmed COVID19 and suspected cases | Acc, precision, recall, f1-score | √ |
| Costa et al., 2021 [38] | COVID-19 classification | ViT with Performer | COVIDx dataset | Acc, F1-score, precision, recall, | ✗ |
| Liang 2021 [39] | COVID-19 classification | feature aggregation by transformer, CNN features, data resampling | COV19-CT-DB | macro F1-score | ✗ |
| Sufian et al., 2023 [41] | COVID-19 classification | Histogram equalization and contrast limited adaptive histogram equalization, InceptionV3, MobileNet, ResNet50, VGG16, ViT-B16, ViT-B32 | 2481 CT scans from SARS-CoV-2 CT-Scan dataset | Acc, precision, recall, f1-score | √ |
| Constatinou et al., 2023 [42] | COVID-19 classification | ResNet50, ResNet101, DenseNet121, DenseNet169, InceptionV3 | COVID-QU dataset with 33,920 Chest X-ray images | Acc, precision, recall, f1-score | √ |
3. Method
In this study, the transfer learning approaches were experimentally evaluated with the following representative applications of classification, object detection, and segmentation, as shown in Table 2: (1) cervical cancer classification, (2) skin lesion classification, (3) pressure ulcer segmentation, and (4) chest X-ray object detection and classification. Because chest X-ray object detection extracts the RoI and transforms it into a classification problem, it can be included in the other three classification and segmentation applications. Consequently, the experiments were performed with three applications.

Table 2.
Overview of medical image analysis tasks used for experiment.
Cervical cancer analysis is an image classification problem, and the presence of cancer can be diagnosed by observing the cervical cancer image captured through colposcopy. The NCI dataset, with 978 labeled images and 44,031 unlabeled images, was used in this study for the classification [86]. These images are different from medical images that show the external surface of the skin, such as skin lesions or pressure ulcers, because elements irrelevant to the classification of cervical cancer, such as medical devices, are captured simultaneously with the cervix when it is enlarged during a colposcopy. For a transfer learning approach suitable for cervical cancer analysis, ref. [7] proposed patch self-supervised learning, which can use unlabeled images and learn the self-information by separating unnecessary parts of the image.
Skin lesion classification is performed after extracting the region of the lesion from the skin. This study used the HAM10000 dataset, which consists of 10,015 labeled images with seven types of lesions photographed during dermoscopy [87]. Because the skin and lesion parts are clearly separated in the skin lesion images, a transfer learning approach using only the encoder of the U-Net model is used after training to extract the RoI to improve the classification performance. There has also been a study of multi-task learning with a two-stream structure that performed segmentation and classification simultaneously [88].
Pressure ulcers occur when the skin is under pressure for an extended period. The amount of data available for segmentation is significantly smaller than other datasets because it takes a long time for the disease to appear. Therefore, in this application, the performance can be improved by transfer learning with a wound image similar to the pressure ulcer image.
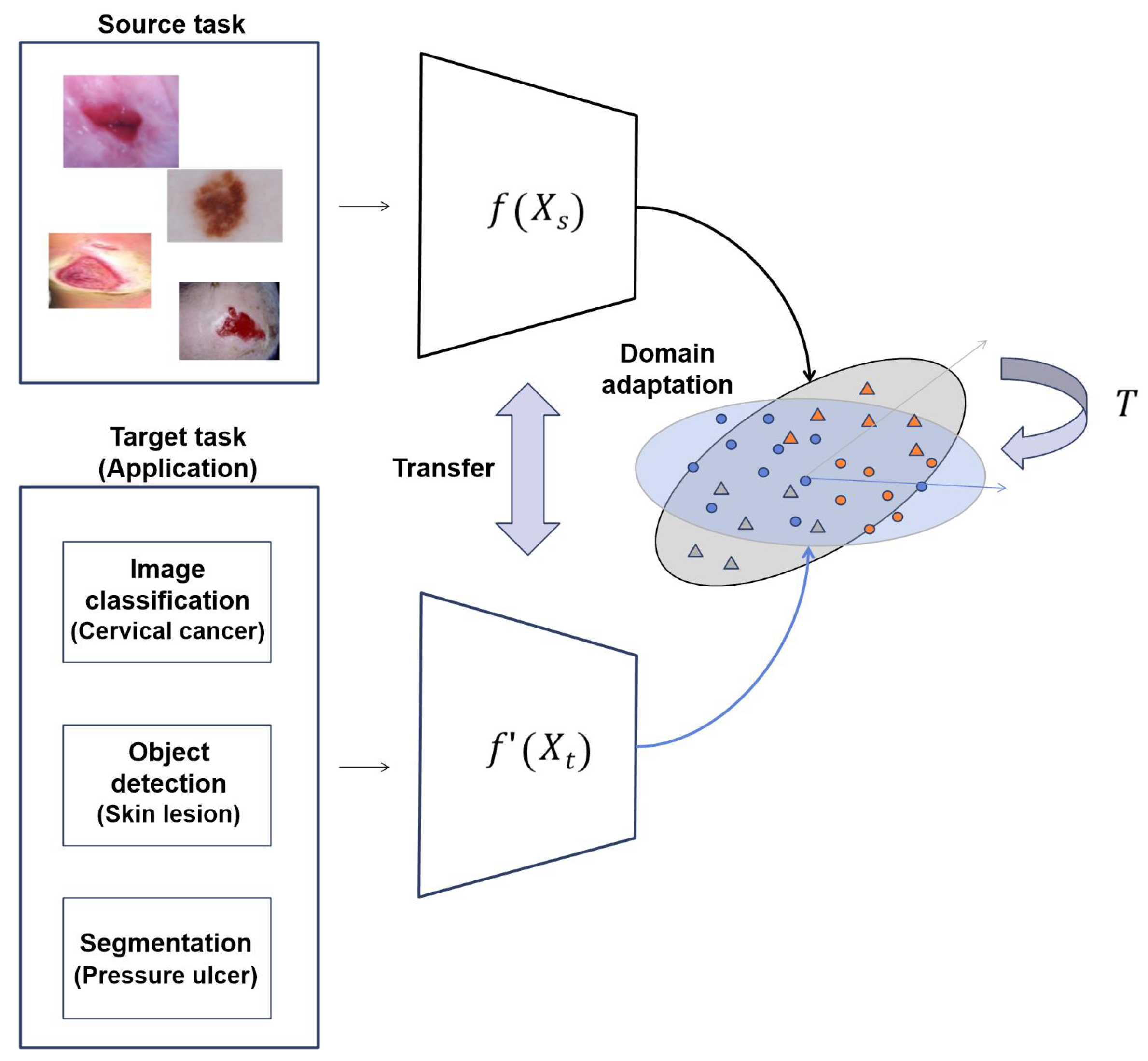
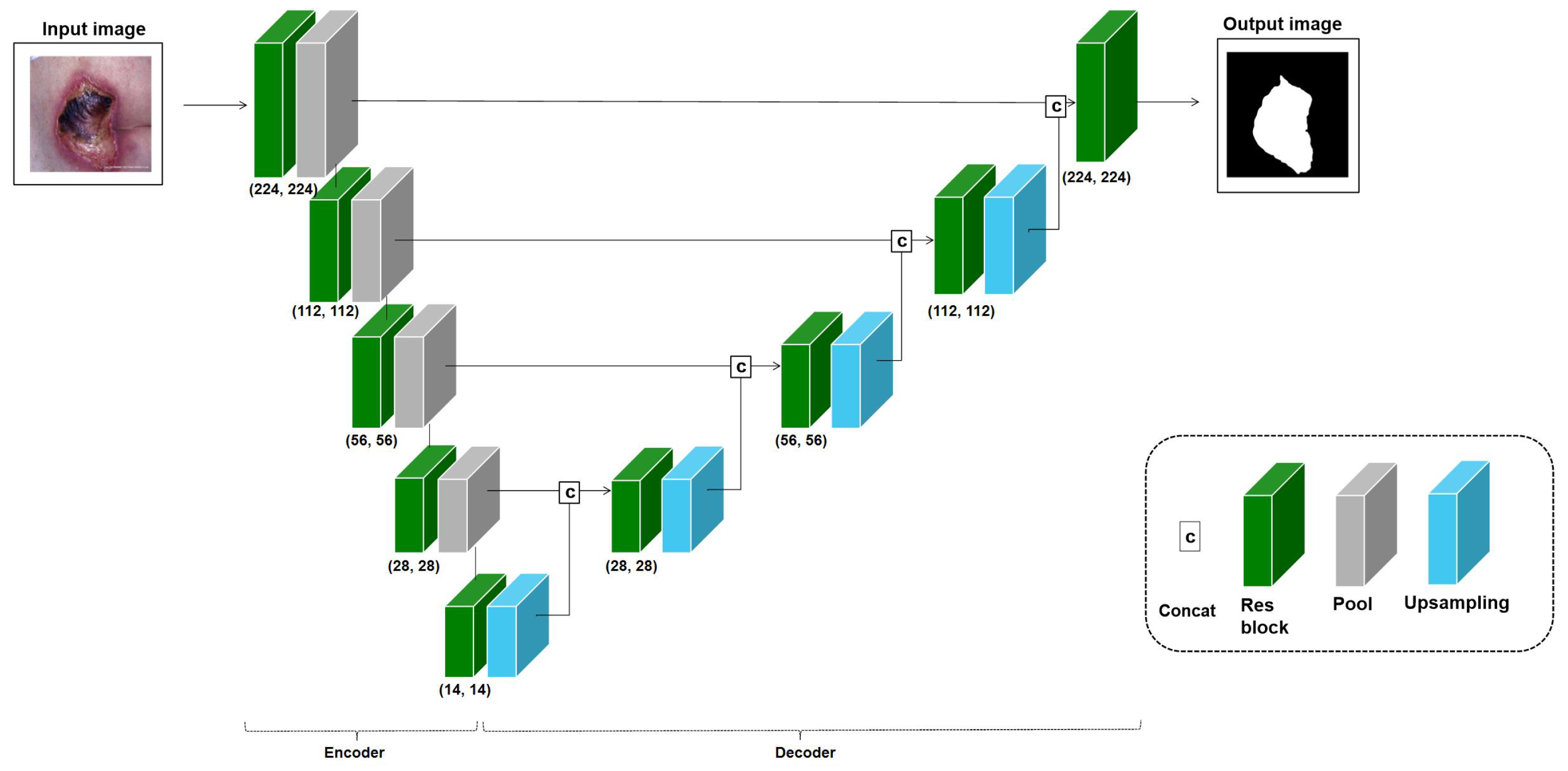
In this study, we propose transfer learning approaches to solve the chronic problems of medical images, as shown in Figure 2. In the given source and target tasks, we train the target task through the transfer learning method of the trained model for the source task. The dataset used in source task is different from the dataset used in the target task. In Equation (1), and , where h, w, and c are the height, width, and channel of input images, are input images of the source task and target task, respectively. , where k is the number of classes, is the output of the source task and is the output of the target task. is an encoder of images for the source task, and is an encoder for the target task, but the initial weight of is the model weight trained with the proposed transfer learning method .
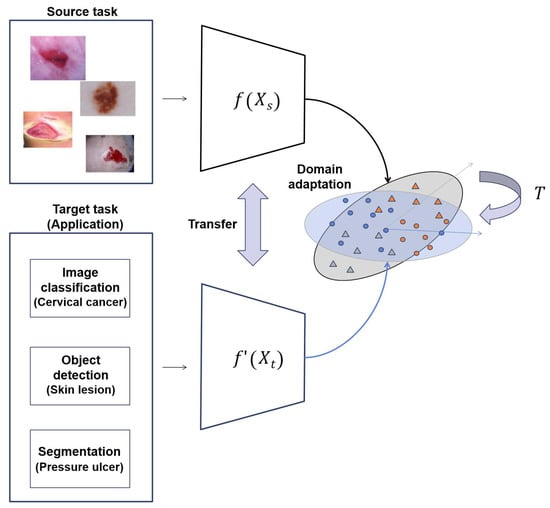
Figure 2.
Overall structure of proposed method.
For the transfer learning method, we propose the four following methods: (1) transfer learning with similar images, (2) transfer learning with self-supervised learning, (3) transfer learning with RoIs, and (4) transfer learning with multi-task learning. We experimentally verify that the proposed transfer learning can be applied to the representative applications. Furthermore, optimal ensemble learning appropriate to each application is presented. The performance is improved by constructing an ensemble model appropriate for the applications.
3.1. : Transfer Learning with Similar Images
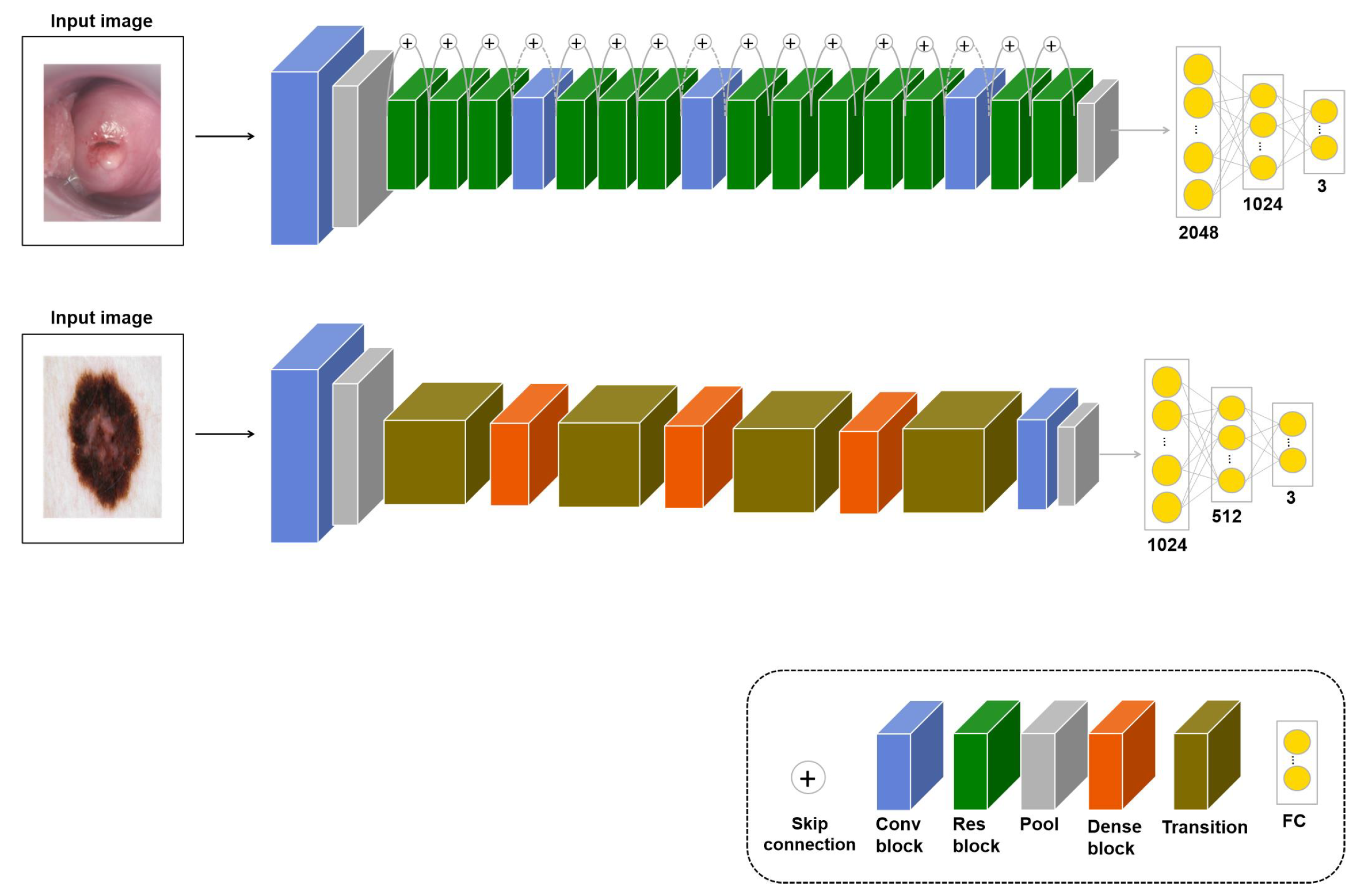
In a deep learning model, the generalization performance increases with the number of training samples. However, when there is a lack of data, there are not enough samples to train the model. Therefore, various augmentation techniques have been studied to increase the training samples, but there is a limitation to viewing the same image multiple times because it augments the original dataset. Therefore, in this study, if the transferred model with an image similar to the original dataset was fine-tuned again to fit the application, it was assumed that the performance would improve. For the learning, the input is images similar to the original dataset, and in the fine-tuning stage, the application dataset is used as the input. This approach, denoted as , measures the performance for the three applications. Models of this approach according to task types are shown in Figure 3.
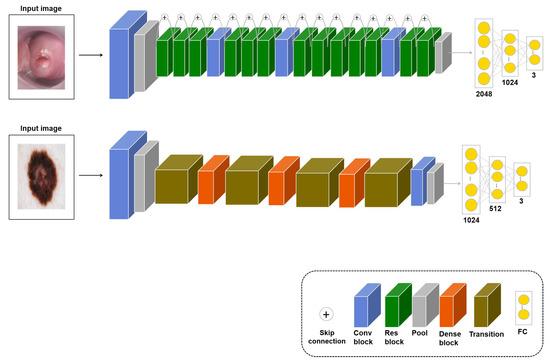
Figure 3.
models for cervical cancer classification (upper-row model, based on ResNet50) and skin lesion classification (bottom-row model, based on DenseNet121). The pressure ulcer segmentation model for is shown in Figure 5. For skin lesion classification, a transition layer is added to reduce the number of channels and the size of the feature map.
3.2. : Transfer Learning with Self-Supervised Learning
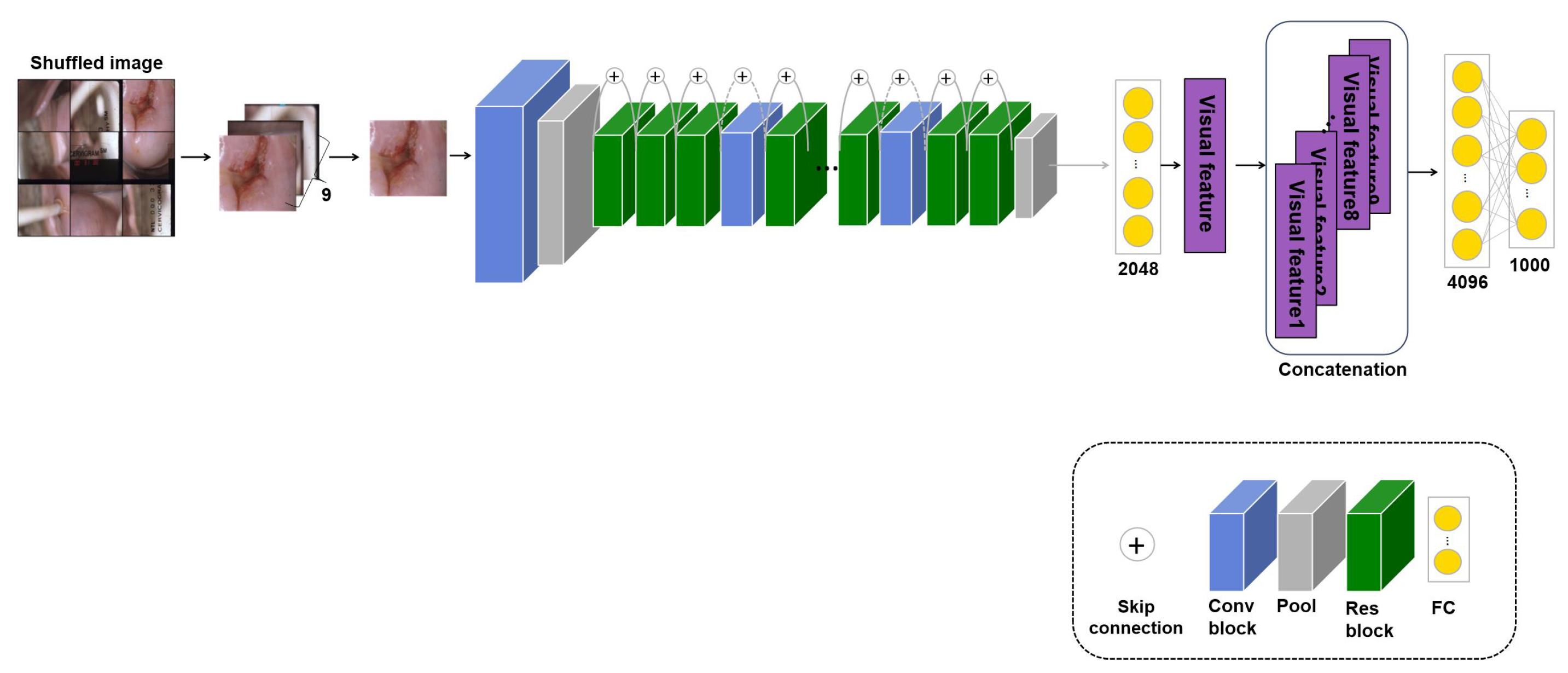
Medical images have a high labeling cost, so there are many cases where data are available but the ground truth is not given. Therefore, an approach that can use unlabeled images is proposed. Methods using self-supervised learning have often been proposed; of these, patch self-supervised learning for classification learns self-information in unlabeled images without image labels [7,89]. Therefore, the proposed approach uses a transferred model with its own information in an unlabeled image to improve the performance. However, although this study has been evaluated on all three tasks, not merely the classification task, such experiments have not been conducted for medical image analysis before this study. For self-supervised learning, the image is divided into nine patches, which are shuffled to learn the original arrangement from the random patches. There are 9! ways to arrange nine patches, but because some are similar arrangements, such as [1,2,3,4,5,6,7,8,9] and [1,2,3,4,5,6,7,9,8], only 1000 cases are considered. Thus, for the self-supervised learning, the input is unlabeled images divided into nine patches and the predicted output is a case to arrange the patches. Also, a cross-entropy loss is used to calculate the error between the predicted output and the true output. The loss is as follows:
where is the amount of data and and are the true and the predicted output, respectively. Furthermore, fewer cases are considered for skin lesion and pressure ulcer images because parts of the skin other than the lesion look similar. There are various other approaches to self-supervised learning, such as contrastive learning, which creates and compares positive and negative samples. However, although such methods can be applied to general datasets, such as dog or cat images, they are not appropriate for medical images, which require specific domain knowledge. Therefore, this study proposes patch self-supervised learning, which learns the original image from randomly shuffled patch images, as shown in Figure 4. For the self-supervised learning, the input is unlabeled images divided into nine patches and the fine-tuning stage uses the application dataset with labels for the input. The model’s weight trained with patch self-supervised learning is transferred and empirically fine-tuned to the application task. Experiments denoted as using a transferred model with such self-supervised learning were performed on the three applications, and the performance was measured.
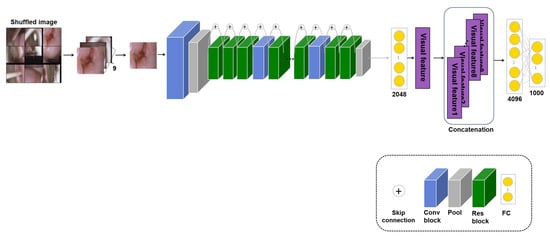
Figure 4.
Model: Patch self-supervised learning model based on ResNet50 for cervical cancer classification.
3.3. : Transfer Learning with RoI
Medical images have crucial areas, or RoIs, that should be focused on based on the task purpose, such as lesion classification and segmentation. Ref. [9] proposed improving the classification performance using the extracted RoI. Inspired by this study, the transfer learning approach with RoIs was used and evaluated on the three applications in this study. This RoI approach is frequently used for classification tasks, but in this study, the object detection and segmentation tasks, including classification, were also performed, so an approach using segmentation appropriate to each application’s medical images was introduced. The RoI training model is based on a U-Net structure that can improve segmentation performance, but the encoder of the U-Net structure uses the baseline model of the application. Hence, if the baseline model for cervical cancer classification is ResNet50, the U-Net for the RoI learning approach consists of a ResNet50 encoder and a decoder using an up-sampling layer, as shown in Figure 5. In the RoI learning, the application dataset is used as the input, and the output is a segmented object area. Also, in the fine-tuning stage, the application dataset is used as the input, and the output is a class type. We trained the model with end-to-end learning to output the RoI first, then used the encoder’s weight for transfer learning and experimentally fine-tuned the model to evaluate the task performance. This transferred U-Net encoder was used to improve the classification performance of the application. The approach using RoI is denoted as , and the performance was measured for the three applications. The total number of parameters of the segmentation model is 28,976,386.
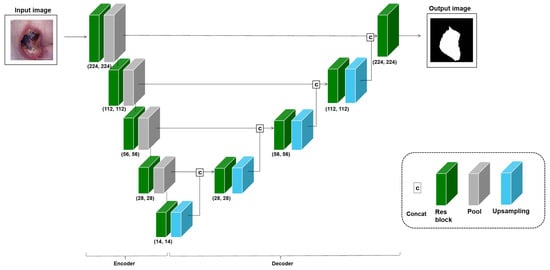
Figure 5.
Model: U-Net based on a ResNet18 variant for pressure ulcer segmentation. For training, the encoder and decoder depend on the baseline model’s number of convolution blocks for each application.
3.4. : Transfer Learning with Multi-Task Learning
When doctors diagnosis a lesion or classify cancer, they diagnose it by looking at the RoI identified, the type of lesion, and the occurrence of cancer at the same time. Therefore, inspired by doctors’ diagnoses, this study assumed that performance can be improved by performing RoI segmentation and classification simultaneously. Thus, multi-task learning was conducted to simultaneously perform segmentation and classification with the same image using the baseline model of each application. For the classification branch, global averaged pooling and fully connected layers were added from the final visual feature of the baseline model. For example, as seen in Figure 5, pooling and fully connected layers were added from the final feature extracted from the U-Net encoder for the classification branch. For the segmentation branch, a decoder using the up-sampling layer was added to the baseline model of the application and transformed into a U-Net model, as shown in Figure 5. The approach is referred to as , and the performance was measured for the three applications. In this approach, the loss weight is given using the following equation as a two-stream loss calculation inspired by [90]. is the output of a neural network with weights on input . Equation (3) is the multi-task likelihood function with segmentation output and classification output . We designed the multi-task model with two branches of classification and segmentation and used the encoder’s weight for transfer learning. In addition, the encoder was designed with a single-task output through the fully connected layer and used to evaluate the task performance.
Equation (3) leads to Equation (4) being minimized with temperatures and (the input is scaled by each sigma according to the output type):
In Equation (5), we adapt segmentation and classification likelihood to squash a scaled version of the model output through a sigmoid function and softmax function with positive scalars and . The sigmoid function is used for the segmentation task because the task is to classify pixels into 0 or 1, and the softmax function is used to classify various types for the classification task (e.g., 7 types for skin lesion classification). The scalars relate to the model’s uncertainty, as measured in the entropy. Equation (5) can be written as Equations (6) and (7):
The final joint loss is given in Equation (8):
where and . We minimize Equation (8) with respect to W, , and .
In this study, after measuring the performance of the four approaches for each application, we propose an effective transfer learning approach and a high-performance ensemble approach.
4. Experiments
We experimented with the proposed approaches for the following three applications: (1) cervical cancer classification, (2) skin lesion classification, and (3) pressure ulcer segmentation. The sizes of the images used for training were different in this experiment. However, a consistent image size was needed for each application. Therefore, all image sizes were empirically set to 512 × 512 pixels for cervical cancer classification, 448 × 448 for skin lesion classification, and 224 × 224 for pressure ulcer segmentation. In addition, in the pressure ulcer images, the pixel brightness was adjusted in pre-processing to make the ulcer boundary clearly visible through the anti-aliasing technique. We also used data augmentation, such as rotation and flip, to increase the number of training samples for the applications. In addition, 70% and 20% of the data were randomly selected for training and validation, respectively, while the remaining 10% was selected for testing. We kept the ratio between classes roughly the same for all three parts.
The experiment was performed on the baseline model for each application with similar images, and the model was fine-tuned for the target dataset. Therefore, in this experiment, the performance for the three applications was measured using the weight of the transferred model to be trained with similar images.
The experiment used the novel patch self-supervised learning that learned self-information in the image, and it was assumed that the performance would increase if the transferred model with unsupervised learning was fine-tuned to fit the application. Therefore, in this experiment, transfer learning with self-supervised learning to predict the arrangement of nine shuffled patches of an image was used for the three applications. Cervical cancer classification had unlabeled images, so the approach was performed with these images, whereas skin lesion classification and pressure ulcer segmentation were trained with images with the labels removed for consistency.
The experiment assumed that performance would increase if the transferred model used to segment only the important area (the RoI) in the image was fine-tuned to the application. Therefore, in this experiment, the baseline model was transformed into a U-Net model by attaching the decoder, and the model was trained to segment the RoI. After training, the U-Net encoder was fine-tuned, and the performance was measured. However, because pressure ulcer segmentation was a targeted task, RoI training was replaced by classifying the pressure ulcers in the dataset, including wounds and ulcer images. The encoder baseline model U-Net for pressure ulcer segmentation was extracted and used to train the classification. The transferred model was transformed into a U-Net model to segment the pressure ulcers.
When making a diagnosis, physicians examine important parts of the image simultaneously to diagnose a lesion or cancer. Therefore, multi-task learning was carried out inspired by the idea of RoI detection and classification being performed simultaneously [88]. In each application experiment, the model consisted of two branches: segmentation and classification. The classification branch was the baseline model of the application, and the segmentation branch was the U-Net model, which segmented the RoI by attaching an up-sampling layer from the extracted visual features of the baseline model. Furthermore, the weight of the loss of the two branches was adjusted using Equation (8). In addition, the training of the baseline model of each application was based on the following three models (application tasks): ResNet50 (cervical cancer classification), DenseNet121 (skin lesion classification), and ResNet18 (pressure ulcer segmentation). The baseline models selected empirically were trained on the ImageNet dataset and then fine-tuned with the application datasets.
4.1. Cervical Cancer Classification
4.1.1. Datasets
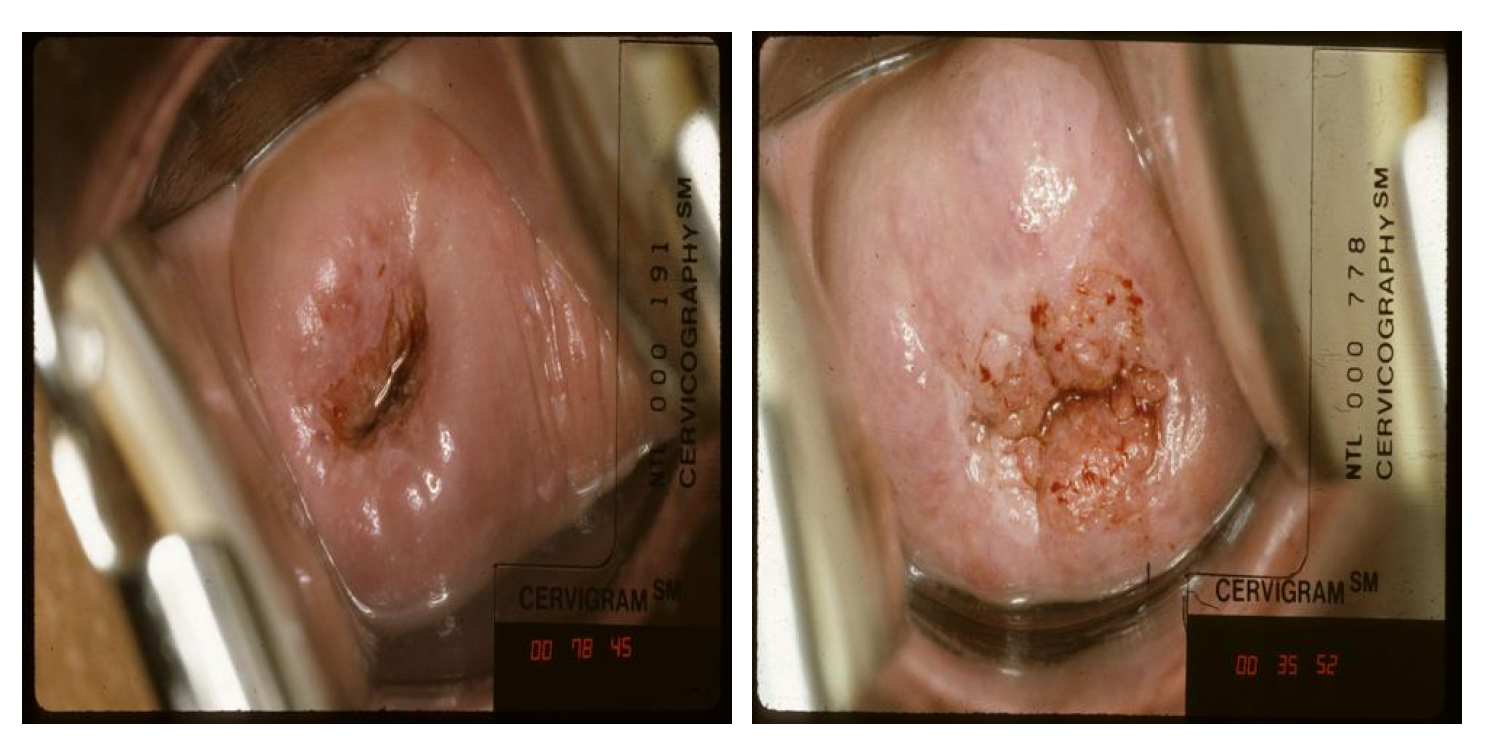
NCI dataset: In this study, the NCI dataset collected from the U.S. NCI was used for cervical cancer classification. The dataset was collected from approximately 10,000 women over 18 years old by the project Costa Rica: Proyecto epidemiologico guanacaste [91]. Each patient visited the hospital several times during the project and had several checkups per visit [86]. The total number of images is 45,009, consisting of 978 labeled images within one year of the inspection date and 44,031 unlabeled images. In addition, cervical intraepithelial neoplasia (CIN) grades (CIN 0, 1, 2, 3, and 4) are used to label each patient’s cervical image with the ground truth. In Figure 6, CIN 2 or higher is classified as abnormal, and CIN0 and CIN1 are classified as normal. Therefore, in the experiment, the data were converted into a binary classification problem by dividing it into positive results (CIN 2+) and negative results (CIN0/1). The NCI dataset was used for the training and test sets.
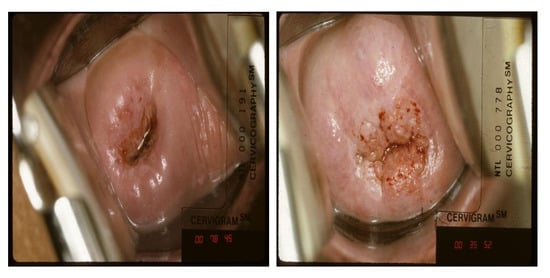
Figure 6.
Sample of negative class (left) and positive class (right) from the NCI dataset.
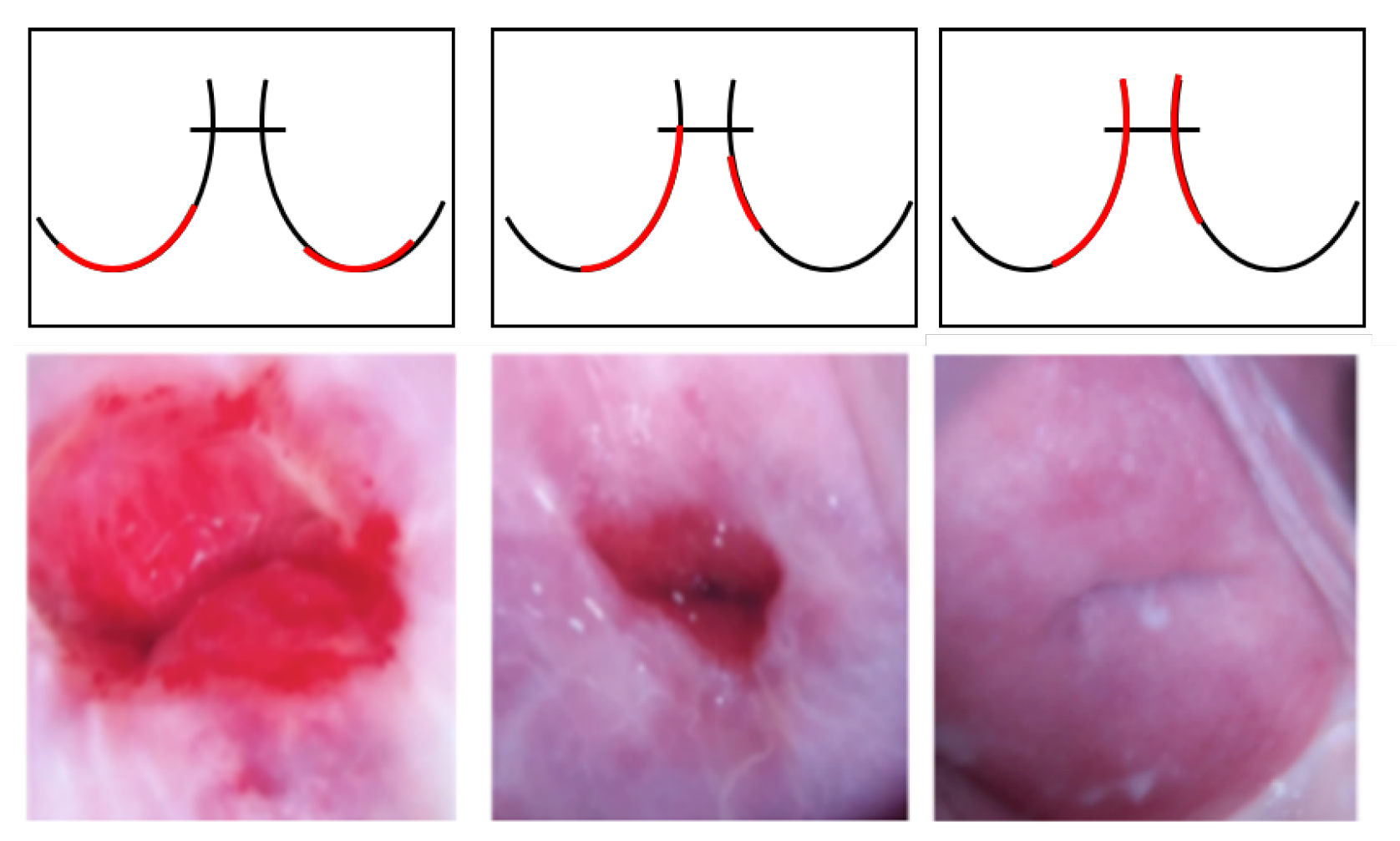
Intel and MobileODT Cervical Cancer Screening dataset: In addition to the NCI data, transfer learning was performed using similar cervical images for the experiments. The Intel and MobileODT Cervical Cancer Screening dataset is used to classify the transformation zone of the cervix [92]. Identifying the transformation zone can significantly improve the quality and efficiency of cervical cancer screening because, even if it is negative, additional screening may be required for the transformation zone. The transformation zone is classified into three types in Figure 7: Type 1, completely ectocervical, fully visible, and small or large; Type 2, endocervical component, fully visible, and may have an ectocervical component, which may be small or large; and Type 3, has an endocervical component, not fully visible, and may have an ectocervical component, which may be small or large. The dataset was used for the cervical cancer classification experiment because it could improve performance by first identifying the transformation zone and then classifying cancer with similar image training.
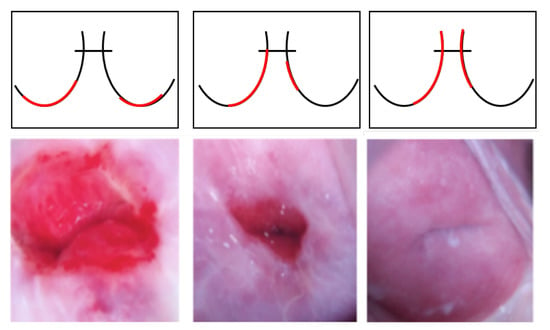
Figure 7.
Samples of the transformation zone in the Intel and MobileODT dataset according to type (from left to right, Type 1, Type 2, and Type 3). The red line shows different transformation zone locations based on the different types.
4.1.2. Metrics
The following three evaluation metrics were used to measure the cervical cancer classification performance:
4.1.3. Results
The training for cervical cancer classification used the Intel and MobileODT dataset. Because it was important to see the visible transformation zone to identify cervical cancer, we pre-trained it to classify the transformation zone. As shown in Table 3, cervical cancer classification using transfer learning with a similar image showed better performance than the baseline. In cervical cancer classification, there are many parts outside the transformation zone (the area where cervical cells are most likely to become cancerous) that are unnecessary for diagnosing cancer.

Table 3.
Transfer learning results for cervical cancer classification.
The model learns the global context of the image to separate the unnecessary parts and then learn the local features needed for image classification. After training with the patch self-supervised learning method was completed, cervical cancer was classified using the transferred model. The row in Table 3 shows that cervical cancer classification using transfer learning with the self-supervised learning model had higher accuracy and sensitivity than the baseline.
learning extracts the cervical cancer RoI. As shown in the cervix image in Figure 3, when the cervix is viewed from the bottom, it looks like a circular shape with a center, which is a hole in the lower part of the cervix. When there is a blob-like region around the center, the transformation zone covers most of the abnormal cervical cancer tissue. Therefore, because it is important to see the circular part when determining cervical cancer, the baseline model-based U-Net was trained to segment the circular RoI. Then, the U-Net encoder was extracted, and cervical cancer classification was performed using the encoder. As shown in Table 3, the most improved performance was obtained when the model with RoI transfer learning was used.
Furthermore, in the same way that clinicians detect and diagnose transformation zones simultaneously, multi-task learning that performed segmentation and classification only in the circular area demonstrated better accuracy and specificity than the baseline model.
Finally, the transfer learning approaches with greater accuracy than the baseline model were ensembled and evaluated on the test set. The ensemble had the highest sensitivity. Because the and had lower specificity than the baseline model, the ensemble model had low specificity. However, the sensitivity of the ensemble model was the highest of all the approaches. As sensitivity is an important metric in medical diagnosis, this is a significant result. The results showed that classifying an image that had unnecessary parts (e.g., a medical device or photograph date) hampered training features, so transfer learning with RoIs was an effective approach.
4.2. Skin Lesion Classification
4.2.1. Datasets
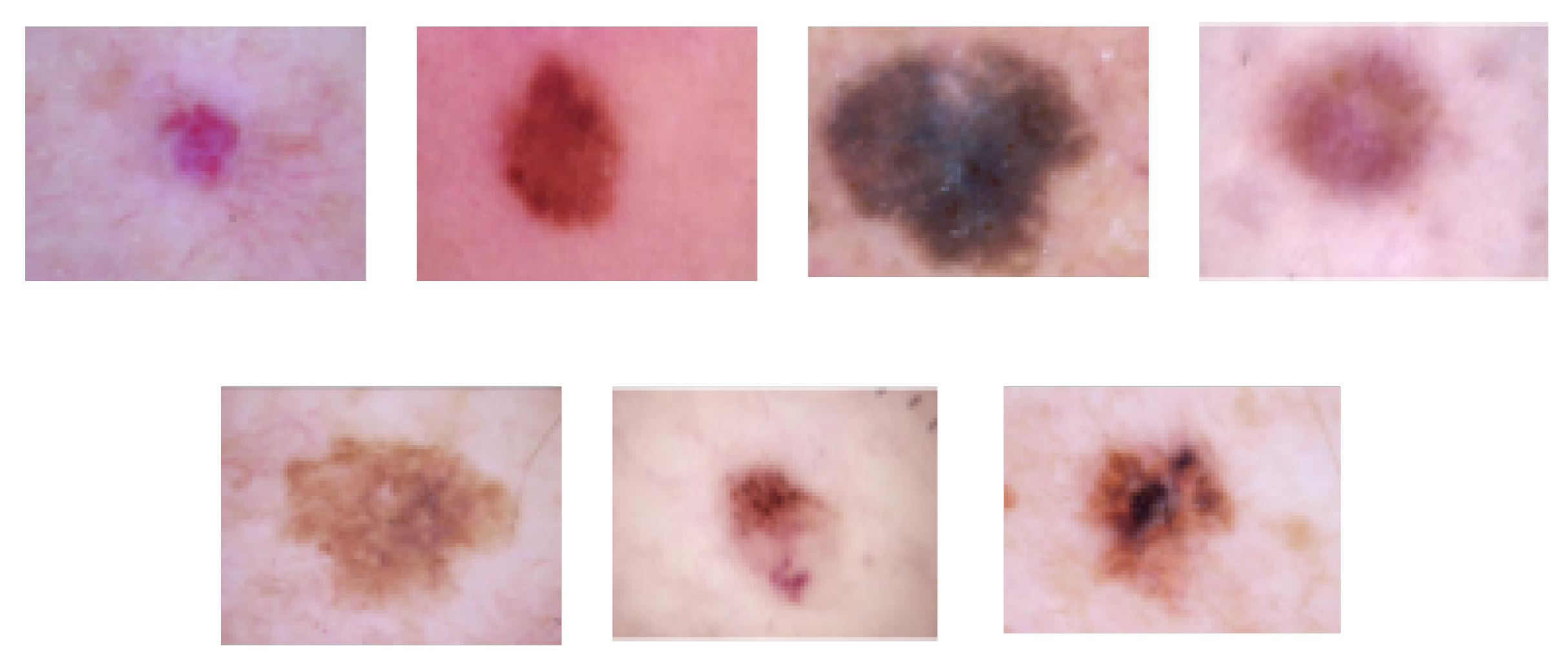
HAM10000 dataset: The HAM10000 dataset consists of 10,015 dermoscopy images and is classified into the following seven lesion classes in Figure 8 [87]: (1) actinic keratoses and intraepithelial carcinoma/Bowen’s disease (akiec), (2) basal cell carcinoma (bcc), (3) benign keratosis-like lesions (solar lentigines/seborrheic keratoses and lichen-planus-like keratoses, bkl), (4) dermatofibroma (df), (5) melanoma (mel), (6) melanocytic nevi (nv), and (7) vascular lesions, including angiomas, angiokeratomas, pyogenic granulomas, and hemorrhage (vasc). The HAM10000 dataset was used for training and testing for skin lesion classification.
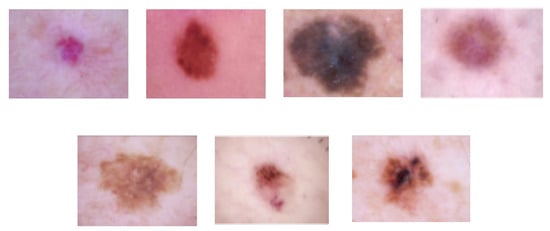
Figure 8.
Skin lesion images of each class (from left to right, vasc, nv, mel, df, bkl, bcc, and akiec) in the HAM10000 dataset.
International Skin Imaging Collaboration (ISIC) dataset: The transfer learning of skin lesion classification used the dataset collected by the ISIC, which is sponsored by the International Society for Digital Imaging of the Skin to improve the diagnostic performance of melanoma, commonly known as skin cancer, and provides the largest publicly available skin lesion image dataset [93]. In this study, the ISIC 2017 dataset was used for training, and the data were classified into the following three types: (1) melanoma, (2) nevus, and (3) seborrheic keratosis. The total number of images in the dataset is 2750, as shown in Table 4.

Table 4.
Number of images in the ISIC dataset according to skin lesion type.
4.2.2. Metrics
The accuracy and sensitivity metrics defined in Section 4.1.2 were also used to measure the skin lesion classification performance. The following precision evaluation metric was also used:
4.2.3. Results
The experiment for skin lesion classification used the ISIC 2017 dataset. Table 5 shows the performance when the approach was applied to skin lesion classification using a transferred model with similar images. Because the ISIC 2017 dataset has fewer classes than the HAM10000 dataset, training for seven types on the HAM10000 datasets using the transferred weight from showed slightly worse accuracy and precision but better sensitivity.

Table 5.
Transfer learning results for skin lesion classification.
Because labeling lesion diagnosis is expensive, there are many unlabeled images. Therefore, transfer learning with patch self-supervised learning was used as an approach for using images without labels. Unlike the cervical images, the skin lesion images were taken with a dermatoscope, and most skin patches look similar to each other except for the area with the lesion. When the approach, which predicts the arrangement of patches, was applied, the accuracy was lower than the baseline accuracy, but the precision was 2.2% better, as shown in Table 5.
Because the images of the HAM10000 dataset were photographed with a dermatoscope, the areas where skin lesions exist were clearly separated, as shown in Figure 8. However, the model needed to learn to separate the lesion area from normal skin. Therefore, it was trained to segment lesions according to the lesion shape. After a decoder was attached to the baseline model DenseNet121 using an up-sampling layer and the model was transformed into a U-Net model, the U-Net model was trained to segment the RoI in accordance with the lesion shape. Then, the U-Net encoder was extracted, and the skin lesions were classified using the encoder. As shown in Table 5, the accuracy and precision were improved by 1.8% and 5.8%, respectively, compared to the baseline.
When learning was used to train both skin lesion segmentation and classification simultaneously, as inspired by clinicians’ diagnoses, the performance was worse than when training a single task. Because skin images taken with a magnifying glass were clearly distinguished by the average pixel value of the skin lesions and other pixels, multi-task learning appeared to cause confusion. In addition, according to [94], multi-task learning has a destructive interference relationship in which the performance of one task is improved while the performance of another task is degraded. The author stated that the division of information between tasks is a fine line to walk.
However, when ensemble learning was used with different initial weights using , the accuracy was higher than the baseline accuracy, showing the highest accuracy, precision, and sensitivity. Therefore, for the dataset photographed with a dermatoscope or magnifying glass or containing images that clearly distinguish between the normal area and the RoI (that is, between healthy skin and the lesion), as shown in Figure 8, transfer learning using the trained weights to segment the RoI based on the shape performed best.
4.3. Pressure Ulcer Segmentation
4.3.1. Datasets
Dongguk University Ilsan Hospital pressure ulcer images (DGU): Pressure ulcers are caused by constant pressure on the skin, so it is important to observe the size and depth of the ulcer. For pressure ulcer segmentation, the experiment was conducted using pressure ulcer images collected at Dongguk University Ilsan Hospital [95]. The dataset includes 101 images taken with a digital camera, and the data were labeled by a clinical specialist.
Medetec Wound Database (MWD): The MWD is an open dataset that includes various chronic wound types such as malignant wounds and surgical wound infections [96]. The dataset contains 492 images, and it was used for the transfer learning of for pressure ulcer segmentation.
Advancing the Zenith of Healthcare wound dataset (AZH): For additional training data, this study used the AZH dataset [97], which includes 1109 images of foot ulcers collected over two years from 889 patients during several clinical visits. Annotation of the images was performed by a wound treatment specialist at a wound clinic. Because the AZH images look similar to pressure ulcer images, as shown in Figure 9, the AZH dataset was used for the experiment.
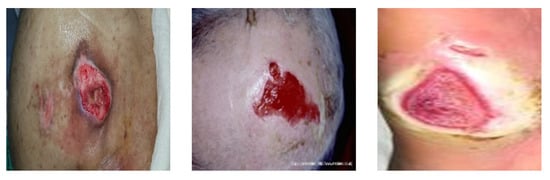
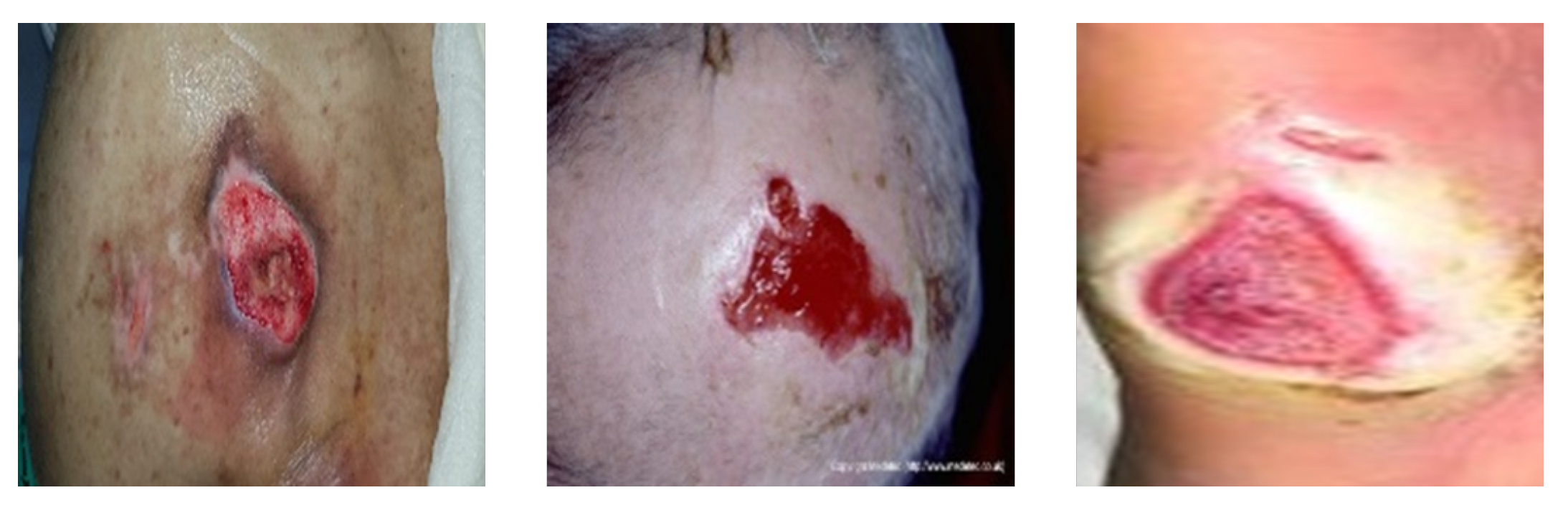
Figure 9.
Pressure ulcer image examples from the DGU, MWD, and AZH datasets, respectively.
4.3.2. Metrics
Because pressure ulcer segmentation is a segmentation task, the following three evaluation metrics were used to measure the performance: pixel-level accuracy, IoU, and DSC.
4.3.3. Results
The training for pressure ulcer segmentation used the MWD and AZH datasets. The row in Table 6 indicates that pressure ulcer segmentation using transfer learning with a similar image performed better overall than the baseline. Because wound images are very similar to ulcer images, they provide increased training data. Therefore, during transfer learning with a similar wound image, the DSC performance was very high.

Table 6.
Transfer learning results for pressure ulcer segmentation.
The experiment for pressure ulcer segmentation performed self-supervised learning using the MWD and AZH datasets with the labels removed. In pressure ulcer and wound images, as well as skin lesion images, it was difficult to predict the arrangement of normal skin parts. When pressure ulcer segmentation was trained using the weights transferred with , the accuracy was degraded.
Because the purpose of pressure ulcer segmentation is to segment RoI, the experiment was slightly modified to distinguish pressure ulcers from the wound and ulcer images. The baseline model for pressure ulcer segmentation was U-Net. The U-Net encoder was trained to classify wounds and ulcers by combining the encoder with U-Net. As seen in Table 6, the performance after transfer learning was worse because the weight trained to distinguish ulcers and wounds was not helpful in training segmentation, as it learned only local features instead of the overall shape.
for pressure ulcers was also evaluated with pressure ulcer classification and segmentation, similar to skin lesion multi-task learning, but the DSC performance was inferior to the single-task baseline model training. Therefore, multi-task learning may not be suitable for segmenting and classifying lesions in skin images, such as skin lesion images or pressure ulcer images.
When ensemble learning was performed with different initial weights using , which had a higher performance than the baseline, it had the highest accuracy, IoU, and DSC. Therefore, it was most effective to use transfer learning with similar images in the application of learning a global shape, such as pressure ulcer segmentation.
4.4. Comparison with State-of-the-Art Approaches
In this study, we compared state-of-the-art approaches using transfer learning for each application as follows: (1) cervical cancer classification, Alyafeai et al. [16], Zhang et al. [17], Xu et al. [12], and Hu et al. [14]; (2) skin lesion classification, Pal et al. [48], Carcagnì et al. [49], and Sirotkin et al. [45]; and (3) pressure ulcer segmentation, Wang et al. [62], Khalil et al. [66], Zahia et al. [57], Goyal et al. [64], and Ohura et al. [67]. For comparison with the state-of-the-art approaches, we followed the common metrics used in prior publications.
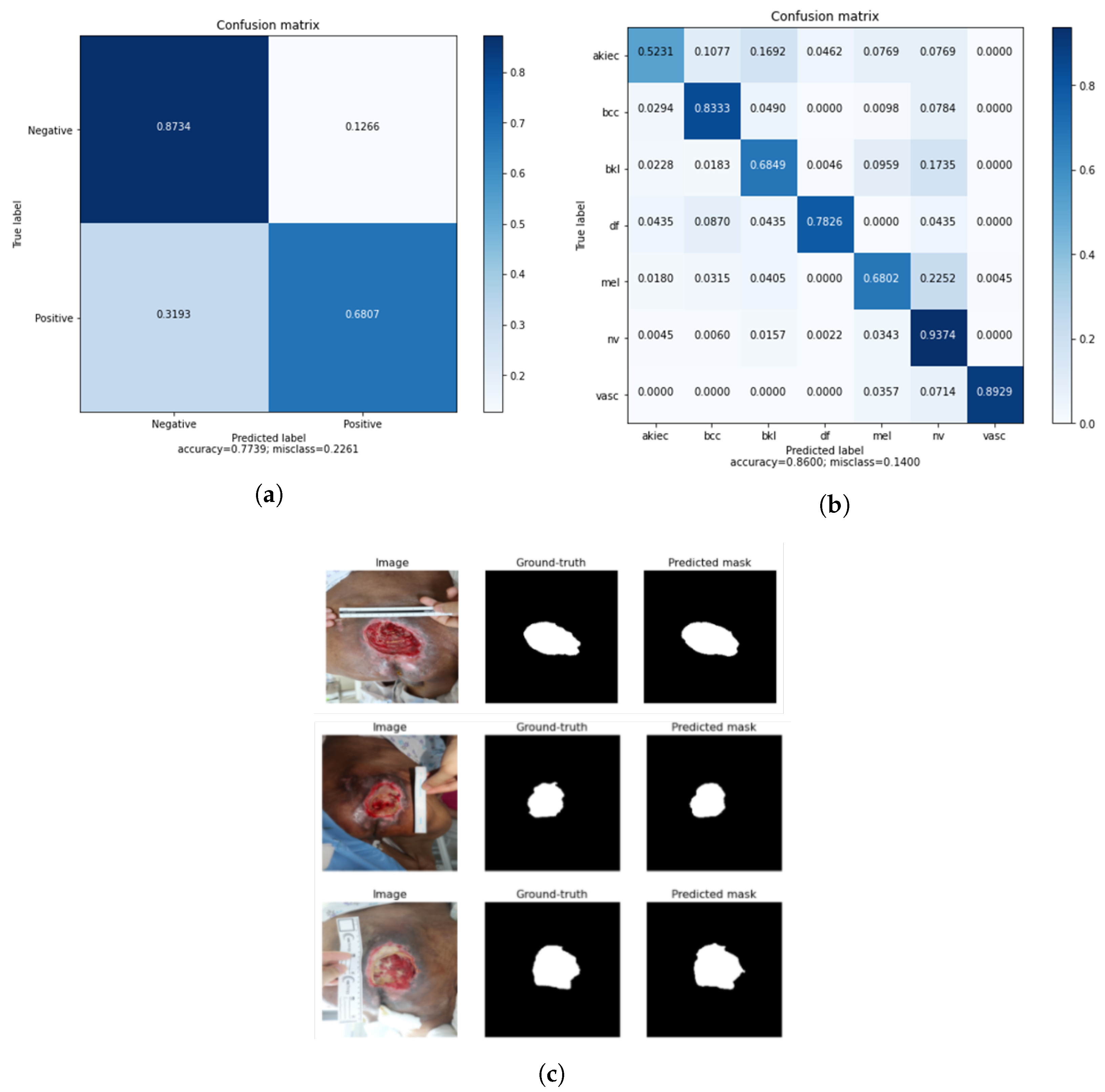
As shown in Table 7, each application experiment’s ensemble performance showed improvement over the state-of-the-art results. In the classification of cervical cancer, the ensemble model was selected from the approach with the highest accuracy, and the performance was improved by 3.5% and 18.2% in accuracy and sensitivity, respectively, compared to [12] using transfer learning. Also, the performance was improved by 19.76% and 9.35% in accuracy and sensitivity, compared to [14]. The accuracy and sensitivity of the ensemble were 4.6% and 24.1% higher, respectively, than those in [16], and the accuracy of [17] was 3.44%, which was slightly higher compared to the ensemble model. However, sensitivity is important in medical diagnosis, so the sensitivity being higher than that in the other studies was significant. In skin lesion classification, compared to the existing state-of-the-art approaches using transfer learning [48,49], the accuracy improved by 8.7%, precision by 28.3%, and sensitivity by 39.7%. Also, when compared to [45] using transfer learning with online deep clustering and momentum contrastive learning, the accuracy improved by 10.6%. Because the HAM10000 dataset has seven skin lesion types and it is important to consider the overall performance balance, the ensemble model of skin classification consisted of the approach with the highest accuracy, . The accuracy and sensitivity performance were also significant, and overall, the ensemble model performed better than the state-of-the-art approaches. Finally, for pressure ulcer segmentation, the accuracy and IoU were 4% and 52.6%, respectively, better than those in [62], and the accuracy was 3% higher than that in [66]. When compared to [57], the IoU and DSC of the ensemble model were 16.9% and 6.4% higher those in [57]. In addition, the DSC was 3.5% better than that in [64] using transfer learning, where only the DSC metric could be compared. The accuracy and DSC were 1.1% and 8.4% better than those in [67] using transfer learning. As shown in Figure 10a, the cervical cancer classification ensemble performance had the highest true positive and true negative values. In addition, the skin cancer classification ensemble performance obtained a balanced true positive value for each class, as shown in Figure 10b. Furthermore, the pressure ulcer mask predicted by the ensemble was similar to the ground truth, as shown in Figure 10c.

Table 7.
Overall results compared to state-of-the-art approach results for the three applications: (a) cervical cancer classification (image classification), (b) skin lesion classification (object detection and classification), (c) pressure ulcer segmentation (segmentation). TL denotes transfer learning, and * denotes reproduced results following their parameter settings for our dataset. Ensemble** performances marked in bold are compared to state-of-the-art approaches.
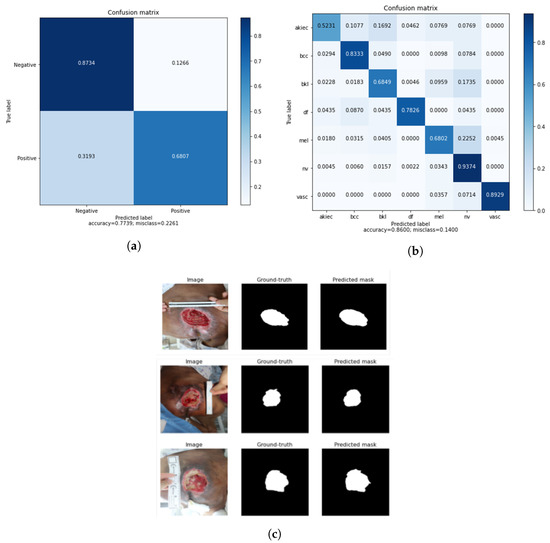
Figure 10.
(a) Confusion matrix of the cervical cancer classification ensemble model; (b) confusion matrix of the skin lesion classification ensemble model; and (c) predicted segmentation output of the pressure ulcer segmentation ensemble model.
Therefore, the proposed approach to overcome a lack of data in medical image analysis was effective in applications representative of medical imaging tasks. Consequently, the image and object classification task focused on the RoI to classify diseases or lesions effectively using transfer learning with RoIs. Furthermore, the segmentation task of learning a global shape was accomplished by transfer learning with similar images.
5. Discussion
This study proposed the following four transfer learning approaches to overcome the problem of the lack of medical data: (1) transfer learning with similar images, (2) transfer learning with self-supervised learning, (3) transfer learning with RoI, and (4) transfer learning with multi-task learning. Inspired by physicians’ medical diagnosis methods, we introduced transfer learning approaches for representative analysis problems. The methods were developed based on mapping to computer vision problems, which have been extensively studied. To the best of our knowledge, we are the first to investigate efficient transfer learning techniques for solving representative problems in medical image analysis. However, there are several problems in the methods, as follows. First, the transfer learning methods are used in experiments on applications, including cervical cancer classification, skin lesion detection and classification, and pressure ulcer segmentation. However, medical image analysis encompasses variant domains such as tumors and breast cancer, depending on the organ of the body. Thus, experiments of transfer learning on variant datasets are conducted to cover the hypotheses we introduced. Second, transfer learning needs further investigation when there is a possibility of negative transfer. In particular, when compared to the transfer learning approaches in Table 7, transfer learning with similar images performs well, while others show different results. This is because learning only works if the initial and target tasks are similar enough for the first round of training to be relevant. For these reasons, to make further improvements and to be used in practice, it is necessary to study more transfer learning approaches with various datasets.
6. Conclusions and Future work
In this study, we investigated approaches to overcome the lack of data in medical image analysis, and we proposed transfer learning approaches. Multi-task learning inspired by clinicians’ diagnoses and ensemble learning were also proposed. We applied the proposed approaches to representative medical image analysis problems: cervical cancer classification, skin lesion detection and classification, and pressure ulcer segmentation, and we showed improvement in their performance. For lesion classification, the region of the image to be focused on was found, so the transfer learning approach using RoI was effective. However, it was more efficient to extract the RoI by considering the characteristics of the area to be focused on (e.g., in cervical cancer classification, because the transformation zone was important, the circular RoI was important). In addition, transferring the weights trained with similar images was effective for learning a global shape in pressure ulcer segmentation. Therefore, in this study, a transfer learning approach that can overcome the lack of medical images has been proposed for applications that are representative of medical imaging tasks. For future work, we will research an approach that can automatically detect and learn the RoIs to improve the performance, and we will aim to further improve the transfer learning to cover various medical domains.
Author Contributions
Conceptualization, J.C. and J.K.; Methodology, J.C. and J.K.; Software, J.C.; Validation, J.C.; Formal Analysis, J.C.; Investigation, J.C.; Resources, J.C.; Data Curation, J.C.; Writing—Original Draft Preparation, J.C.; Writing—Review and Editing, J.C. and J.K.; Visualization, J.C.; Supervision, J.C. and J.K.; Project Administration, J.K.; Funding Acquisition, J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the MSIT(Ministry of Science and ICT), Korea, under the ITRC(Information Technology Research Center) support program(IITP-2023-2020-0-01789), and the Artificial Intelligence Convergence Innovation Human Resources Development(IITP-2023-RS-2023-00254592) supervised by the IITP(Institute for Information & Communications Technology Planning & Evaluation).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Publicly available datasets were used in this study. The datasets can be found here: (1) Intel and MobileODT Cervical Cancer Screening dataset (https://www.kaggle.com/c/intel-mobileodt-cervical-cancer-screening, accessed on 5 January 2023), (2) HAM10000 dataset (https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/DBW86T, accessed on 5 January 2023), (3) International Skin Imaging Collaboration dataset (https://api.isic-archive.com/api/docs/swagger/, accessed on 5 January 2023), (4) Medetec Wound Database (http://www.medetec.co.uk/files/medetec-image-databases.html, accessed on 5 January 2023), and (5) Advancing the Zenith of Healthcare wound dataset (https://github.com/uwm-bigdata/wound-segmentation, accessed on 5 January 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yann, L.; Patrick, H.; Bottou, L.; Yoshua, B. Object recognition with gradient-based learning. In Shape, Contour and Grouping in Computer Vision. Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1999; Volume 1681, pp. 319–345. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 11966–11976. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Chae, J.; Zimmermann, R.; Kim, D.; Kim, J. Attentive Transfer Learning via Self-Supervised Learning for Cervical Dysplasia Diagnosis. J. Inf. Process. Syst. 2021, 17, 453–461. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, H. Self-transfer learning for fully weakly supervised object localization. arXiv 2016, arXiv:1602.01625. [Google Scholar]
- Chae, J.; Kim, H.; Yang, H. A Dual Attention Network for Skin Lesion Classification. Korea Software Congress 2020, 47, 460–462. [Google Scholar]
- Hesamian-Hesam, M.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, H.; Li, Y.; Liu, Q.; Xu, X.; Wang, S.; Yap, P.T.; Shen, D. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med. Image Anal. 2021, 70, 101918. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, H.; Xin, C.; Kim, E.; Long, L.R.; Xue, Z.; Antani, S.; Huang, X. Multi-feature based benchmark for cervical dysplasia classification evaluation. Pattern Recognit. 2017, 63, 468–475. [Google Scholar] [CrossRef]
- Vasudha, A.M.; Juneja, M. Cervix cancer classification using colposcopy images by deep learning method. Int. J. Eng. Technol. Sci. Res. (IJETSR) 2018, 5, 426–432. [Google Scholar]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. JNCI J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Alyafeai, Z.; Ghouti, L. A fully automated deep learning pipeline for cervical cancer classification. Expert Syst. Appl. 2020, 141, 112951. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, Y.; Liu, Z.; Zimmermann, R. A Spatial Regulated Patch-Wise Approach for Cervical Dysplasia Diagnosis. Proc. AAAI Conf. Artif. Intell. 2021, 35, 733–740. [Google Scholar] [CrossRef]
- Xue, D.; Zhou, X.; Li, C.; Yao, Y.; Rahaman, M.M.; Zhang, J.; Chen, H.; Zhang, J.; Qi, S.; Sun, H. An Application of Transfer Learning and Ensemble Learning Techniques for Cervical Histopathology Image Classification. IEEE Access 2020, 8, 104603–104618. [Google Scholar] [CrossRef]
- Dhawan, S.; Singh, K.; Arora, M. Cervix Image Classification for Prognosis of Cervical Cancer using Deep Neural Network with Transfer Learning. EAI Endorsed Trans. Pervasive Health Technol. 2021, 7, e5. [Google Scholar] [CrossRef]
- Zhao, C.; Shuai, R.; Ma, L.; Liu, W.; Wu, M. Improving cervical cancer classification with imbalanced datasets combining taming transformers with t2t-vit. Multimed. Tools Appl. 2022, 81, 24265–24300. [Google Scholar] [CrossRef]
- Esser, P.; Rombach, R.; Ommer, B. Taming Transformers for High-Resolution Image Synthesis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 13–19 June 2020; pp. 12868–12878. [Google Scholar]
- Yuan, L.; Chen, Y.; Wang, T.; Yu, W.; Shi, Y.; Jiang, Z.; Tay, F.H.; Feng, J.; Yan, S. Tokens-to-Token ViT: Training Vision Transformers From Scratch on ImageNet. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, BC, Canada, 11–17 October 2021; pp. 558–567. [Google Scholar] [CrossRef]
- Ghantasala, G.S.P.; Hung, B.T.; Chakrabarti, P. An Approach For Cervical and Breast Cancer Classification Using Deep Learning: A Comprehensive Survey. In Proceedings of the 2023 International Conference on Computer Communication and Informatics (ICCCI), Budapest, Hungary, 27–29 September 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Kalbhor, M.M.; Shinde, S.V. Cervical cancer diagnosis using convolution neural network: Feature learning and transfer learning approaches. In Proceedings of the Soft Comput, Chongqing, China, 5–7 January 2023. [Google Scholar] [CrossRef]
- Deo, B.S.; Pal, M.; Panigarhi, P.K.; Pradhan, A. CerviFormer: A Pap-smear based cervical cancer classification method using cross attention and latent transformer. arXiv 2023, arXiv:2303.10222. [Google Scholar]
- Zhou, Z.; Shin, J.; Zhang, L.; Gurudu, S.; Gotway, M.; Liang, J. Fine-tuning convolutional neural networks for biomedical image analysis: Actively and incrementally. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4761–4772. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Cha, K.H.; Richter, C.D. Multi-task transfer learning deep convolutional neural network: Application to computer-aided diagnosis of breast cancer on mammograms. Phys. Med. Biol. 2017, 62, 8894. [Google Scholar] [CrossRef]
- Rezaeijo, S.M.; Ghorvei, M.; Mofid, B. Predicting Breast Cancer Response to Neoadjuvant Chemotherapy Using Ensemble Deep Transfer Learning Based on CT Images. J. X-ray Sci. Technol. 2021, 29, 835–850. [Google Scholar]
- Xia, Y.; Yao, J.; Lu, L.; Huang, L.; Xie, G.; Xiao, J.; Yuille, A.; Cao, K.; Zhang, L. Effective Pancreatic Cancer Screening on Non-contrast CT Scans via Anatomy-Aware Transformers. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer: Cham, Switzerland, 2021; pp. 259–269. [Google Scholar]
- Gheflati, B.; Rivaz, H. Vision transformer for classification of breast ultrasound images. arXiv 2021, arXiv:2110.14731. [Google Scholar]
- Yang, H.; Chen, J.; Xu, M. Fundus disease image classification based on improved transformer. In Proceedings of the International Conference on Neuromorphic Computing (ICNC), Wuhan, China, 11–14 October 2021; pp. 207–214. [Google Scholar]
- Ikromjanov, K.; Bhattacharjee, S.; Hwang, Y.B.; Sumon, R.I.; Kim, H.C.; Choi, H.K. Whole Slide Image Analysis and Detection of Prostate Cancer using Vision Transformers. In Proceedings of the 2022 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Korea, Republic of Korea, 21–24 February 2022; pp. 399–402. [Google Scholar] [CrossRef]
- Yu, Z.; Mar, V.; Eriksson, A.; Chandra, S.; Bonnington, P.; Zhang, L.; Ge, Z. End-to-End Ugly Duckling Sign Detection for Melanoma Identification with Transformers. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer: Cham, Switzerland, 2021; pp. 176–184. [Google Scholar]
- Wu, W.; Mehta, S.; Nofallah, S.; Knezevich, S.; May, C.; Chang, O.; Elmore, J.; Shapiro, L. Scale-aware transformers for diagnosing melanocytic lesions. IEEE Access 2021, 9, 163526–163541. [Google Scholar] [CrossRef]
- Rezaeijo, S.M.; Ghorvei, M.; Abedi-Firouzjah, R.; Mojtahedi, H.; Zarch, H.E. Detecting COVID-19 in chest images based on deep transfer learning and machine learning algorithms. Egypt. J. Radiol. Nucl. Med. 2021, 52, 145. [Google Scholar] [CrossRef]
- Costa, G.; Paiva, A.; Júnior, G.B.; Ferreira, M. COVID-19 automatic diagnosis with CT images using the novel Transformer architecture. In Proceedings of the Anais do XXI Simpósio Brasileiro de Computação Aplicada à Saúde, SBC, Online, 15–18 June 2021; pp. 293–301. [Google Scholar] [CrossRef]
- Liang, S. A hybrid deep learning framework for covid-19 detection via 3d chest ct images. arXiv 2021, arXiv:2107.03904. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations(ICLR), Virtual, 3–7 May 2021. [Google Scholar]
- Sufian, M.M.; Moung, E.G.; Hijazi, M.H.A.; Yahya, F.; Dargham, J.A.; Farzamnia, A.; Sia, F.; Mohd Naim, N.F. COVID-19 Classification through Deep Learning Models with Three-Channel Grayscale CT Images. Big Data Cogn. Comput. 2023, 7, 36. [Google Scholar] [CrossRef]
- Constantinou, M.; Exarchos, T.; Vrahatis, A.G.; Vlamos, P. COVID-19 Classification on Chest X-ray Images Using Deep Learning Methods. Int. J. Environ. Res. Public Health 2023, 20, 2035. [Google Scholar] [CrossRef] [PubMed]
- Albahar, M. Skin lesion classification using cnn with novel regularizer. IEEE Access 2019, 7, 38306–38313. [Google Scholar] [CrossRef]
- Nahata, H.; Singh, S. Deep Learning Solutions for Skin Cancer Detection and Diagnosis. In Machine Learning with Health Care Perspective: Machine Learning and Healthcare; Springer International Publishing: Berlin/Heidelberg, Germany, 2020; Volume 13, pp. 159–182. [Google Scholar] [CrossRef]
- Sirotkin, K.; Escudero-Vinolo, M.; Carballeira, P.; SanMiguelv, J. Improved skin lesion recognition by a Self-Supervised Curricular Deep Learning approach. arXiv 2021, arXiv:2112.12086. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Wang, C.; Ecker, R.; Ellinge, I. Skin lesion classification using hybrid deep neural networks. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1229–1233. [Google Scholar] [CrossRef]
- Masood, A.; Al-Jumaily, A.; Anam, K. Self-supervised learning model for skin cancer diagnosis. In Proceedings of the International IEEE/EMBS Conference on Neural Engineering (NER), Montpellier, France, 22–24 April 2015; pp. 1012–1015. [Google Scholar] [CrossRef]
- Pal, A.; Ray, S.; Garain, U. Skin disease identification from dermoscopy images using deep convolutional neural network. arXiv 2018, arXiv:1807.09163. [Google Scholar]
- Carcagnì, P.; Leo, M.; Cuna, A.; Mazzeo, P.L.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of Skin Lesions by Combining Multilevel Learnings in a DenseNet Architecture. In Image Analysis and Processing—ICIAP 2019; Springer: Cham, Switzerland, 2019; pp. 335–344. [Google Scholar]
- Liao, H.; Luo, J. A deep multi-task learning approach to skin lesion classification. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence Workshops, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Ahmad, N.; Shah, J.H.; Khan, M.A.; Baili, J.; Ansari, G.J.; Tariq, U.; Kim, Y.J.; Cha, J.H. A novel framework of multiclass skin lesion recognition from dermoscopic images using deep learning and explainable AI. Front. Oncol. 2023, 13, 1151257. [Google Scholar] [CrossRef]
- Alsahafi, Y.S.; Kassem, M.A.; Hosny, K.M. Skin-Net: A novel deep residual network for skin lesions classification using multilevel feature extraction and cross-channel correlation with detection of outlier. J. Big Data 2023, 10, 105. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Washington, DC, USA, 7–13 December 2015; pp. 1440–1448. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.; Liao, H. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Zahia, S.; Garcia-Zapirain, B.; Elmaghraby, A. Integrating 3D Model Representation for an Accurate Non-Invasive Assessment of Pressure Injuries with Deep Learning. Sensors 2020, 20, 2933. [Google Scholar] [CrossRef]
- Saiz, F.; Barandiaran, I. COVID-19 detection in chest X-ray images using a deep learning approach. Int. J. Interact. Multimed. Artif. Intell. 2020, 1, 11–14. [Google Scholar] [CrossRef]
- Arias-Londoño, J.; Gómez-García, J.; Moro-Velázquez, L.; Godino-Llorente, J. Artificial intelligence applied to chest X-ray images for the automatic detection of COVID-19: A thoughtful evaluation approach. IEEE Access 2020, 8, 226811–226827. [Google Scholar] [CrossRef]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Wang, C.; Yan, X.; Smith, M.; Kochhar, K.; Rubin, M.; Warren, S.M.; Wrobel, J.; Lee, H. A unified framework for automatic wound segmentation and analysis with deep convolutional neural networks. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2415–2418. [Google Scholar] [CrossRef]
- Pholberdee, N.; Pathompatai, C.; Taeprasartsit, P. Study of chronic wound image segmentation: Impact of tissue type and color data augmentation. In Proceedings of the 15th International Joint Conference on Computer Science and Software Engineering (JCSSE), Nakhonpathom, Thailand, 11–13 July 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Goyal, M.; Yap, M.H.; Reeves, N.D.; Rajbhandari, S.; Spragg, J. Fully convolutional networks for diabetic foot ulcer segmentation. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 618–623. [Google Scholar] [CrossRef]
- Garcia-Zapirain, B.; Elmogy, M.; El-Baz, S.; Elmaghraby, A. Classification of pressure ulcer tissues with 3D convolutional neural network. Med. Biol. Eng. Comput. 2018, 56, 2245–2258. [Google Scholar] [CrossRef]
- Khalil, A.; Elmogy, M.; Ghazal, M.; Burns, C.; El-Baz, A. Chronic wound healing assessment system based on different features modalities and non-negative matrix factorization (NMF) feature reduction. IEEE Access 2019, 7, 80110–80121. [Google Scholar] [CrossRef]
- Ohura, N.; Mitsuno, R.; Sakisaka, M.; Terabe, Y.; Morishige, Y.; Uchiyama, A.; Okoshi, T.; Shinji, I.; Takushima, A. Convolutional neural networks for wound detection: The role of artificial intelligence in wound care. J. Wound Care 2019, 28, S13–S24. [Google Scholar]
- Oota, S.R.; Rowtula, V.; Mohammed, S.; Liu, M.; Gupta, M. WSNet: Towards an Effective Method for Wound Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 2–7 January 2023; pp. 3234–3243. [Google Scholar]
- Zhang, P.; Chen, X.; Yin, Z.; Zhou, X.; Jiang, Q.; Zhu, W.; Xiang, D.; Tang, Y.; Shi, F. Interactive Skin Wound Segmentation Based on Feature Augment Networks. IEEE J. Biomed. Health Inform. 2023, 27, 3467–3477. [Google Scholar] [CrossRef]
- Swerdlow, M.; Guler, O.; Yaakov, R.; Armstrong1, D.G. Simultaneous Segmentation and Classification of Pressure Injury Image Data Using Mask-R-CNN. Comput. Math. Methods Med. 2023, 2023, 3858997. [Google Scholar] [CrossRef]
- Aldughayfiq, B.; Ashfaq, F.; Jhanjhi, N.Z.; Humayun, M. YOLO-Based Deep Learning Model for Pressure Ulcer Detection and Classification. Healthcare 2023, 11, 1222. [Google Scholar] [CrossRef]
- Moeskops, P.; Wolterink, J.M.; van der Velden, B.H.M.; Gilhuijs, K.G.A.; Leiner, T.; Viergever, M.A.; Išgum, I. Deep Learning for Multi-task Medical Image Segmentation in Multiple Modalities. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer: Cham, Switzerland, 2016; pp. 478–486. [Google Scholar]
- Dong, N.; Kampffmeyer, M.; Liang, X.; Wang, Z.; Dai, W.; Xing, E. Unsupervised Domain Adaptation for Automatic Estimation of Cardiothoracic Ratio. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer: Cham, Switzerland, 2018; pp. 544–552. [Google Scholar]
- Javanmardi, M.; Tasdizen, T. Domain adaptation for biomedical image segmentation using adversarial training. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI), Washington, DC, USA, 4–7 April 2018; Volume 2018, pp. 554–558. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, M.; Liu, D.; Metaxas, D. A multi-scale transformer for medical image segmentation: Architectures, model efficiency, and benchmarks. arXiv 2022, arXiv:2203.00131. [Google Scholar]
- You, C.; Zhao, R.; Liu, F.; Chinchali, S.P.; Topcu, U.; Staib, L.H.; Duncan, J.S. Class-Aware Generative Adversarial Transformers for Medical Image Segmentation. arXiv 2022, arXiv:2201.10737. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Qiaowei, Z.; Iwamoto, Y.; Han, X.H.; Chen, Y.W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Crimi, A., Bakas, S., Eds.; Springer: Cham, Switzerland, 2022; pp. 272–284. [Google Scholar]
- Fatan, M.; Hosseinzadeh, M.; Askari, D.; Sheikhi, H.; Rezaeijo, S.M.; Salmanpour, M.R. Fusion-Based Head and Neck Tumor Segmentation and Survival Prediction Using Robust Deep Learning Techniques and Advanced Hybrid Machine Learning Systems. In Head and Neck Tumor Segmentation and Outcome Prediction; Andrearczyk, V., Oreiller, V., Hatt, M., Depeursinge, A., Eds.; Springer: Cham, Switzerland, 2022; pp. 211–223. [Google Scholar]
- Özcan, F.; Uçan, O.N.; Karaçam, S.; Tunçman, D. Fully Automatic Liver and Tumor Segmentation from CT Image Using an AIM-Unet. Bioengineering 2023, 10, 215. [Google Scholar] [CrossRef]
- Bi, R.; Guo, L.; Yang, B.; Wang, J.; Shi, C. 2.5D cascaded context-based network for liver and tumor segmentation from CT images. Electron. Res. Arch. 2023, 31, 4324–4345. [Google Scholar] [CrossRef]
- Aggarwal, M.; Tiwari, A.K.; Sarathi, M.P.; Bijalwan, A. An early detection and segmentation of Brain Tumor using Deep Neural Network. BMC Med. Inform. Decis. Mak. 2023, 23, 78. [Google Scholar] [CrossRef]
- Montaha, S.; Azam, S.; Rafid, A.K.M.R.H.; Hasan, M.Z.; Karim, A. Brain Tumor Segmentation from 3D MRI Scans Using U-Net. SN Comput. Sci. 2023, 4, 386. [Google Scholar] [CrossRef]
- National Library of Medicine (U.S.)—The Cleveland Clinic (n.d.). An Innovative Treatment for Cervical Precancer (UH3). 2017. ClinicalTrials.gov Identifier: NCT03084081. Available online: https://classic.clinicaltrials.gov/ct2/show/study/NCT03084081 (accessed on 5 January 2023).
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zeng, Z.; Yeo, S.; Tan, C.; Tey, H.L.; Su, Y. A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv 2017, arXiv:1703.01025. [Google Scholar]
- Chae, J.; Zhang, Y.; Zimmermann, R.; Kim, D.; Kim, J. An Attention-Based Deep Learning Model with Interpretable Patch-Weight Sharing for Diagnosing Cervical Dysplasia. In Intelligent Systems and Applications; Springer: Cham, Switzerland, 2022; pp. 634–642. [Google Scholar]
- Kendall, A.; Gal, Y.; Cipolla, R. Multi-task learning using uncertainty to weigh losses for scene geometry and semantics. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition(CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7482–7491. [Google Scholar] [CrossRef]
- Herrero, R.; Schiffman, M.; Bratti, C.; Hildesheim, A.; Balmaceda, I.; Sherman, M.; Greenberg, M.; Cárdenas, F.; Gómez, V.; Helgesen, K.; et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: The Guanacaste Project. Rev. Panam. Salud Publica 1997, 1, 362–375. [Google Scholar] [CrossRef]
- Intel & MobileODT Cervical Cancer Screening Competition. 2017. Available online: https://www.kaggle.com/c/intel-mobileodt-cervical-cancer-screening (accessed on 5 April 2023).
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 168–172. [Google Scholar] [CrossRef]
- Crawshaw, M. Multi-task learning with deep neural networks: A Survey. arXiv 2020, arXiv:009.09796. [Google Scholar]
- Chae, J.; Hong, K.; Kim, J. A pressure ulcer care system for remote medical assistance: Residual U-Net with an attention model based for wound area segmentation. arXiv 2021, arXiv:2101.09433. [Google Scholar]
- Homas, S. Medetec Wound Database. Available online: http://www.medetec.co.uk/files/medetec-image-databases.html (accessed on 5 April 2023).
- Wang, C.; Anisuzzaman, D.M.; Williamson, V.; Dhar, M.; Rostami, B.; Niezgoda, J.; Gopalakrishnan, S.; Yu, Z. Fully automatic wound segmentation with deep convolutional neural networks. Sci. Rep. 2020, 10, 21897. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).