Investigation of Fruit Growth Patterns, Olive Fly Bactrocera oleae (Rossi) Infestation, and Genetic Diversity in Italian Olive Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Fruit Olive Fly Infestation
2.3. Statistical Analysis of Morphological Data
2.4. DNA Isolation
2.5. SSR Amplification and Electrophoresis
2.6. SSR Data Analysis
2.7. Co-Inertia Analysis (COIA)
3. Results
3.1. Drupe Growth
3.2. Olive Fruit Fly Infestation of the Drupes
3.3. Genetic Diversity
3.4. Co-Inertia between the Two Principal Component Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daane, K.M.; Johnson, M.W. Olive fruit fly: Managing an ancient pest in modern times. Annu. Rev. Entomol. 2010, 55, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Koprivnjak, O.; Dminić, I.; Kosić, U.; Majetić, V.; Godena, S.; Valenčič, V. Dynamics of oil quality parameters changes related to olive fruit fly attack. Eur. J. Lipid Sci. Technol. 2010, 112, 1033–1040. [Google Scholar] [CrossRef]
- Gómez-Caravaca, A.M.; Cerretani, L.; Bendini, A.; Segura-Carretero, A.; Fernández-Gutiérrez, A.; Del Carlo, M.; Compagnone, D.; Cichelli, A. Effects of fly attack (Bactrocera oleae) on the phenolic profile and selected chemical parameters of olive oil. J. Agric. Food Chem. 2008, 56, 4577–4583. [Google Scholar] [CrossRef]
- Girolami, V. Studies on the biology and population ecology of Dacus oleae (Gmelin). 1. Influence of environmental abiotic factors on the adult and on the immature stages. Redia 1979, 62, 147–191. [Google Scholar]
- Kounatidis, I.; Papadopoulos, N.; Mavragani Tsipidou, P.; Cohen, Y.; Tertivanidis, K.; Nomikou, M.; Nestel, D. Effect of elevation on spatio-temporal patterns of olive fly (Bactrocera oleae) populations in northern Greece. J. Appl. Entomol. 2008, 132, 722–733. [Google Scholar] [CrossRef]
- Malheiro, R.; Casal, S.; Baptista, P.; Pereira, J.A. A review of Bactrocera oleae (Rossi) impact in olive products: From the tree to the table. Trends Food Sci. Technol. 2015, 44, 226–242. [Google Scholar] [CrossRef]
- Pavlidi, N.; Gioti, A.; Wybouw, N.; Dermauw, W.; Ben-Yosef, M.; Yuval, B.; Jurkevich, E.; Kampouraki, A.; Van Leeuwen, T.; Vontas, J. Transcriptomic responses of the olive fruit fly Bactrocera oleae and its symbiont Candidatus Erwinia dacicola to olive feeding. Sci. Rep. 2017, 7, 42633. [Google Scholar] [CrossRef]
- Ben-Yosef, M.; Pasternak, Z.; Jurkevitch, E.; Yuval, B. Symbiotic bacteria enable olive fly larvae to overcome host defences. R. Soc. Open Sci. 2015, 2, 150170. [Google Scholar] [CrossRef]
- Notario, A.; Sánchez, R.; Luaces, P.; Sanz, C.; Pérez, A.G. The Infestation of Olive Fruits by Bactrocera oleae (Rossi) Modifies the Expression of Key Genes in the Biosynthesis of Volatile and Phenolic Compounds and Alters the Composition of Virgin Olive Oil. Molecules 2022, 27, 1650. [Google Scholar] [CrossRef]
- Spadafora, A.; Mazzuca, S.; Chiappetta, F.F.; Parise, A.; Perri, E.; Innocenti, A.M. Oleuropein-specific-β-glucosidase activity marks the early response of olive fruits (Olea europaea) to mimed insect attack. Agric. Sci. China 2008, 7, 703–712. [Google Scholar] [CrossRef]
- Aluja, M.; Mangan, R.L. Fruit fly (Diptera: Tephritidae) host status determination: Critical conceptual, methodological, and regulatory considerations. Annu. Rev. Entomol. 2008, 53, 473–502. [Google Scholar] [CrossRef] [PubMed]
- Scarpati, M.L.; Scalzo, R.L.; Vita, G. Olea europaea volatiles attractive and repellent to the olive fruit fly (Dacus oleae, Gmelin). J. Chem. Ecol. 1993, 19, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Malheiro, R.; Casal, S.; Baptista, P.; Pereira, J.A. Physico-chemical characteristics of olive leaves and fruits and their relation with Bactrocera oleae (Rossi) cultivar oviposition preference. Sci. Hortic. 2015, 194, 208–214. [Google Scholar] [CrossRef]
- Neuenschwander, P.; Michelakis, S.; Holloway, P.; Berchtol, W. Factors affecting the susceptibility of fruits of different olive varieties to attack by Dacus oleae (Gmel.) (Dipt., Tephritidae). Z. Für Angew. Entomol. 1985, 100, 174–188. [Google Scholar] [CrossRef]
- Katsoyannos, B.I.; Kouloussis, N.A. Captures of the olive fruit fly Bactrocera oleae on spheres of different colours. Entomol. Exp. Appl. 2001, 100, 165–172. [Google Scholar] [CrossRef]
- Gonçalves, M.F.; Malheiro, R.; Casal, S.; Torres, L.; Pereira, J.A. Influence of fruit traits on oviposition preference of the olive fly, Bactrocera oleae (Rossi) (Diptera: Tephritidae), on three Portuguese olive varieties (Cobrançosa, Madural and Verdeal Transmontana). Sci. Hortic. 2012, 145, 127–135. [Google Scholar] [CrossRef]
- Rizzo, R.; Caleca, V.; Lombardo, A. Relation of fruit color, elongation, hardness, and volume to the infestation of olive cultivars by the olive fruit fly, B actrocera oleae. Entomol. Exp. Et Appl. 2012, 145, 15–22. [Google Scholar] [CrossRef]
- Fletcher, B. The biology of dacine fruit flies. Annu. Rev. Entomol. 1987, 32, 115–144. [Google Scholar] [CrossRef]
- Rattanapun, W.; Amornsak, W.; Clarke, A.R. Bactrocera dorsalis preference for and performance on two mango varieties at three stages of ripeness. Entomol. Exp. Appl. 2009, 131, 243–253. [Google Scholar] [CrossRef]
- Rallo, L.; Barranco, D.; Díez, C.M.; Rallo, P.; Suárez, M.P.; Trapero, C.; Pliego-Alfaro, F. Strategies for olive (Olea europaea L.) breeding: Cultivated genetic resources and crossbreeding. Adv. Plant Breed. Strateg. Fruits 2018, 3, 535–600. [Google Scholar]
- Grasso, F.; Coppola, M.; Carbone, F.; Baldoni, L.; Alagna, F.; Perrotta, G.; Pérez-Pulido, A.J.; Garonna, A.; Facella, P.; Daddiego, L. The transcriptional response to the olive fruit fly (Bactrocera oleae) reveals extended differences between tolerant and susceptible olive (Olea europaea L.) varieties. PLoS ONE 2017, 12, e0183050. [Google Scholar] [CrossRef] [PubMed]
- Iannotta, N.; Scalercio, S. Susceptibility of Cultivars to Biotic Stresses. In Olive Germplasm; Innocenzo, M., Ed.; IntechOpen: Rijeka, Croatia, 2012; Chapter 5. [Google Scholar]
- Medjkouh, L.; Costa, A.; Tamendjari, A.; Bekdouche, F.; Bouarroudj, K.; Oliveira, M.B.P. Susceptibility of eight Algerian olive cultivars to Bactrocera oleae infestation—A pomological and nutritional quality perspective. Phytoparasitica 2018, 46, 595–605. [Google Scholar] [CrossRef]
- Corrado, G.; Alagna, F.; Rocco, M.; Renzone, G.; Varricchio, P.; Coppola, V.; Coppola, M.; Garonna, A.; Baldoni, L.; Scaloni, A. Molecular interactions between the olive and the fruit fly Bactrocera oleae. BMC Plant Biol. 2012, 12, 86. [Google Scholar] [CrossRef]
- Alagna, F.; Kallenbach, M.; Pompa, A.; De Marchis, F.; Rao, R.; Baldwin, I.T.; Bonaventure, G.; Baldoni, L. Olive fruits infested with olive fly larvae respond with an ethylene burst and the emission of specific volatiles. J. Integr. Plant Biol. 2016, 58, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Garantonakis, N.; Varikou, K.; Markakis, E.; Birouraki, A.; Sergentani, C.; Psarras, G.; Koubouris, G.C. Interaction between Bactrocera oleae (Diptera: Tephritidae) infestation and fruit mineral element content in Olea europaea (Lamiales: Oleaceae) cultivars of global interest. Appl. Entomol. Zool. 2016, 51, 257–265. [Google Scholar] [CrossRef]
- Rebora, M.; Salerno, G.; Piersanti, S.; Gorb, E.; Gorb, S. Role of fruit epicuticular waxes in preventing Bactrocera oleae (Diptera: Tephritidae) attachment in different cultivars of Olea europaea. Insects 2020, 11, 189. [Google Scholar] [CrossRef]
- Nardi, F.; Carapelli, A.; Boore, J.; Roderick, G.; Dallai, R.; Frati, F. Domestication of olive fly through a multi-regional host shift to cultivated olives: Comparative dating using complete mitochondrial genomes. Mol. Phylogenetics Evol. 2010, 57, 678–686. [Google Scholar] [CrossRef]
- Ehrlich, P.R.; Raven, P.H. Butterflies and plants: A study in coevolution. Evolution 1964, 18, 586–608. [Google Scholar] [CrossRef]
- Futuyma, D.J.; Agrawal, A.A. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. USA 2009, 106, 18054–18061. [Google Scholar] [CrossRef]
- Doledec, S.; Chessel, D. Co-inertia analysis: An alternative method for studying specie-environment relationships. Freshw. Biol. 1994, 31, 277–294. [Google Scholar] [CrossRef]
- Corrado, G.; Imperato, A.; La Mura, M.; Perri, E.; Rao, R. Genetic diversity among olive varieties of Southern Italy and the traceability of olive oil using SSR markers. J. Hortic. Sci. Biotechnol. 2011, 86, 461–466. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 1989. [Google Scholar]
- Sefc, K.; Lopes, M.; Mendonça, D.; Santos, M.R.D.; Machado, M.L.D.C.; Machado, A.D.C. Identification of microsatellite loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 2000, 9, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, G.; Marrazzo, M.; Marconi, R.; Cimato, A.; Testolin, R. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor. Appl. Genet. 2002, 104, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Carriero, F.; Fontanazza, G.; Cellini, F.; Giorio, G. Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 2002, 104, 301–307. [Google Scholar] [CrossRef]
- Manco, R.; Basile, B.; Capuozzo, C.; Scognamiglio, P.; Forlani, M.; Rao, R.; Corrado, G. Molecular and phenotypic diversity of traditional European plum (Prunus domestica L.) germplasm of Southern Italy. Sustainability 2019, 11, 4112. [Google Scholar] [CrossRef]
- Corrado, G.; Forlani, M.; Rao, R.; Basile, B. Diversity and relationships among neglected apricot (Prunus armeniaca L.) landraces using morphological traits and SSR markers: Implications for agro-biodiversity conservation. Plants 2021, 10, 1341. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef]
- Robert, P.; Escoufier, Y. A unifying tool for linear multivariate statistical methods: The RV-coefficient. J. R. Stat. Soc. Ser. C: Appl. Stat. 1976, 25, 257–265. [Google Scholar] [CrossRef]
- Corrado, G.; Garonna, A.; Cabanás, C.G.-L.; Gregoriou, M.; Martelli, G.P.; Mathiopoulos, K.D.; Mercado-Blanco, J.; Saponari, M.; Tsoumani, K.T.; Rao, R. Host response to biotic stresses. In The Olive Tree Genome; Springer: Cham, Switzerland, 2016; pp. 75–98. [Google Scholar]
- Malheiro, R.; Casal, S.; Pinheiro, L.; Baptista, P.; Pereira, J. Olive cultivar and maturation process on the oviposition preference of Bactrocera oleae (Rossi) (Diptera: Tephritidae). Bull. Entomol. Res. 2019, 109, 43–53. [Google Scholar] [CrossRef]
- Fletcher, B.; Pappas, S.; Kapatos, E. Changes in the ovaries of olive flies (Dacus oleae (Gmelin)) during the summer, and their relationship to temperature, humidity and fruit availability. Ecol. Entomol. 1978, 3, 99–107. [Google Scholar] [CrossRef]
- Vibhute, A.; Bodhe, S.K. Applications of image processing in agriculture: A survey. Int. J. Comput. Appl. 2012, 52, 34–40. [Google Scholar] [CrossRef]
- Pontikakos, C.M.; Tsiligiridis, T.A.; Drougka, M.E. Location-aware system for olive fruit fly spray control. Comput. Electron. Agric. 2010, 70, 355–368. [Google Scholar] [CrossRef]
- Miranda, M.Á.; Barceló, C.; Valdés, F.; Feliu, J.F.; Nestel, D.; Papadopoulos, N.; Sciarretta, A.; Ruiz, M.; Alorda, B. Developing and implementation of Decision Support System (DSS) for the control of olive fruit fly, Bactrocera oleae, in mediterranean olive orchards. Agronomy 2019, 9, 620. [Google Scholar] [CrossRef]
- Belaj, A.; Satovic, Z.; Cipriani, G.; Baldoni, L.; Testolin, R.; Rallo, L.; Trujillo, I. Comparative study of the discriminating capacity of RAPD, AFLP and SSR markers and of their effectiveness in establishing genetic relationships in olive. Theor. Appl. Genet. 2003, 107, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, I.; Ojeda, M.A.; Urdiroz, N.M.; Potter, D.; Barranco, D.; Rallo, L.; Diez, C.M. Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet. Genomes 2014, 10, 141–155. [Google Scholar] [CrossRef]
- Menkir, A.; Adetimirin, V.O.; Yallou, C.G.; Gedil, M. Relationship of genetic diversity of inbred lines with different reactions to Striga hermonthica (Del.) Benth and the performance of their crosses. Crop Sci. 2010, 50, 602–611. [Google Scholar] [CrossRef]
- Dray, S. On the number of principal components: A test of dimensionality based on measurements of similarity between matrices. Comput. Stat. Data Anal. 2008, 52, 2228–2237. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Canonical analysis. In Developments in Environmental Modelling; Elsevier: Amsterdam, The Netherlands, 2012; Volume 24, pp. 625–710. [Google Scholar]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Scarpati, M.L.; Scalzo, R.L.; Vita, G.; Gambacorta, A. Chemiotropic behavior of female olive fly (Bactrocera oleae Gmel.) on Olea europaea L. J. Chem. Ecol. 1996, 22, 1027–1036. [Google Scholar] [CrossRef]
- Malheiro, R.; Casal, S.; Cunha, S.C.; Baptista, P.; Pereira, J.A. Olive volatiles from Portuguese cultivars Cobrançosa, Madural and Verdeal Transmontana: Role in oviposition preference of Bactrocera oleae (Rossi) (Diptera: Tephritidae). PLoS ONE 2015, 10, e0125070. [Google Scholar] [CrossRef]
- Li, W.; Zheng, T.; Yang, Z.; Li, M.; Sun, C.; Yang, X. Classification and detection of insects from field images using deep learning for smart pest management: A systematic review. Ecol. Inform. 2021, 66, 101460. [Google Scholar] [CrossRef]

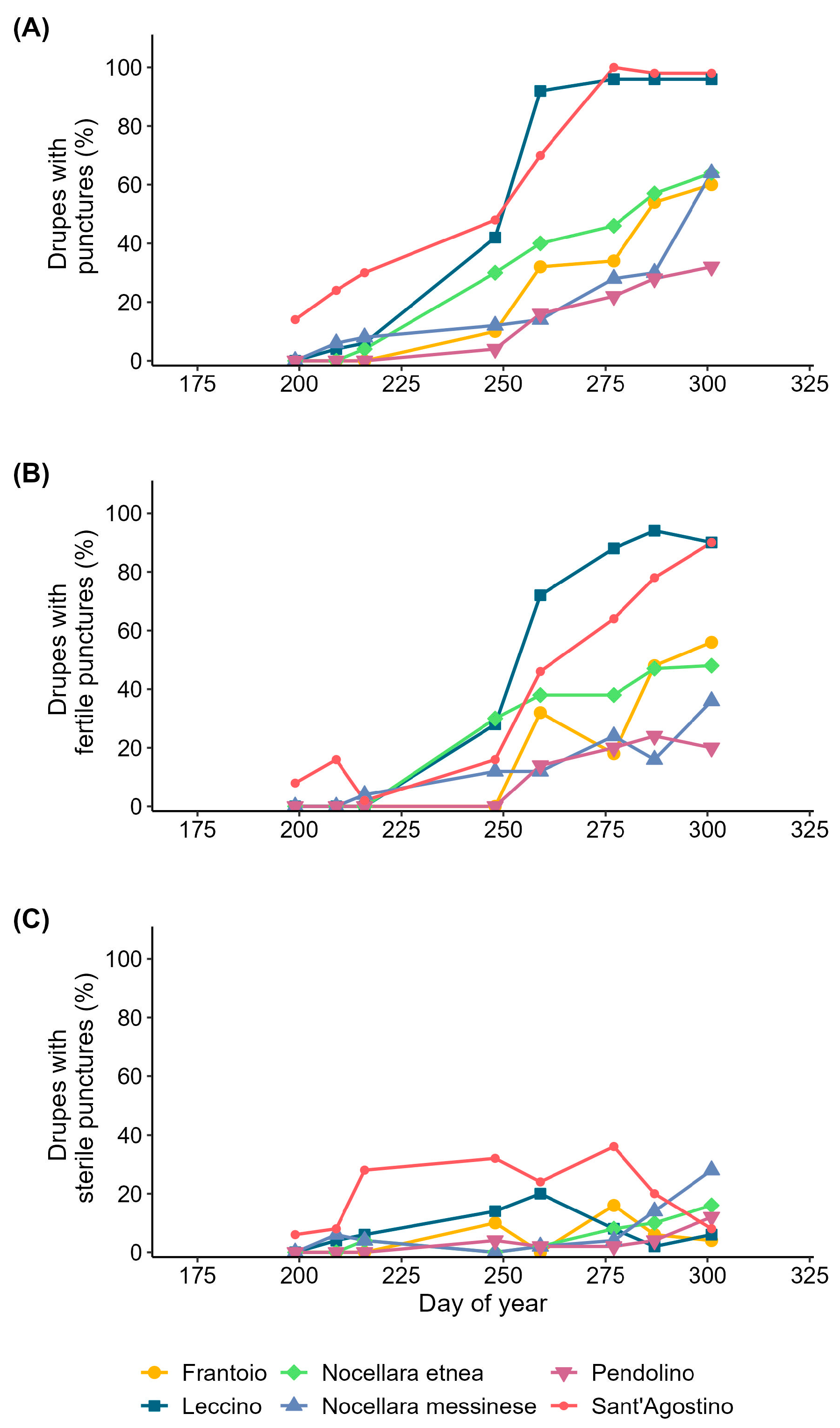
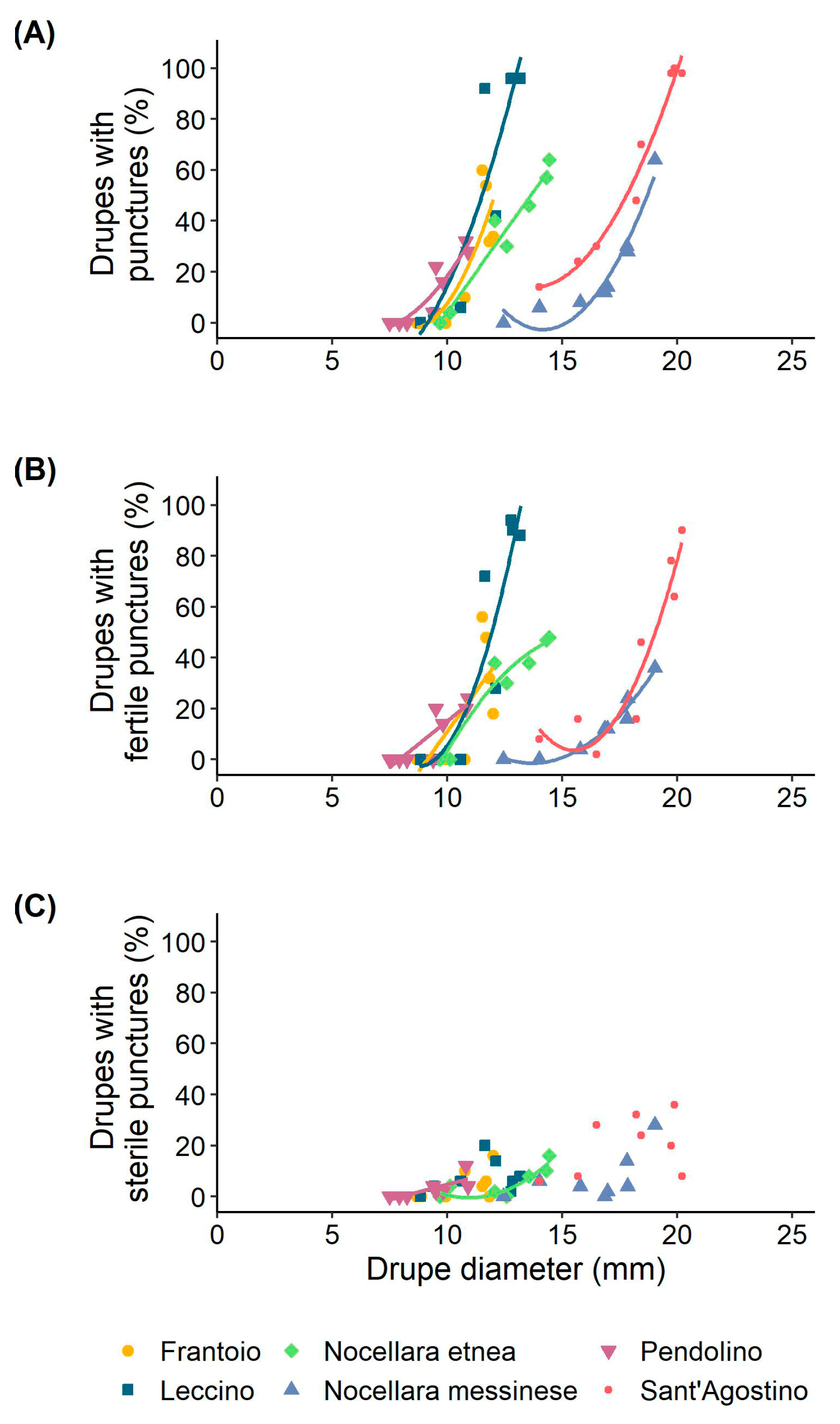
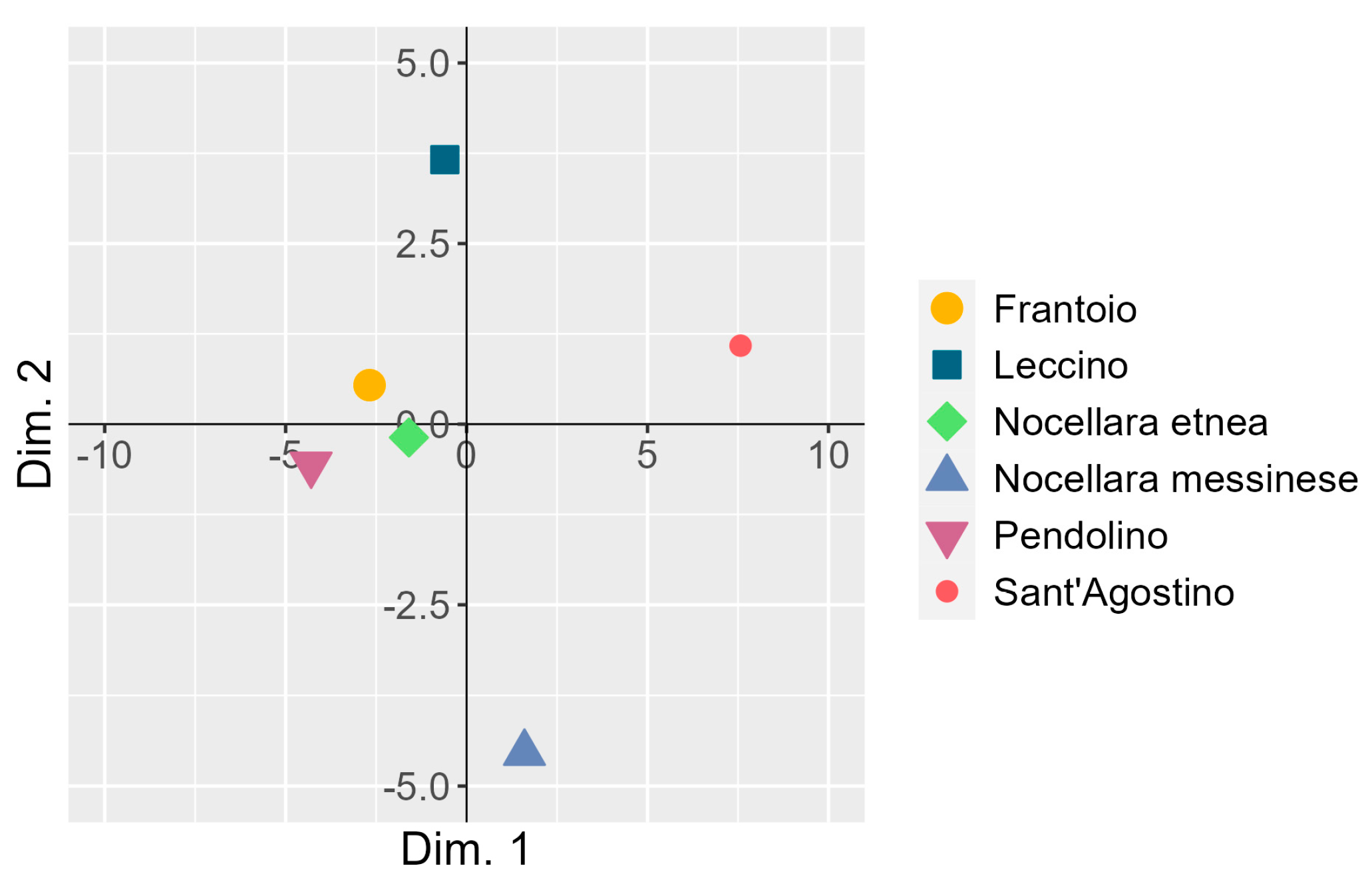
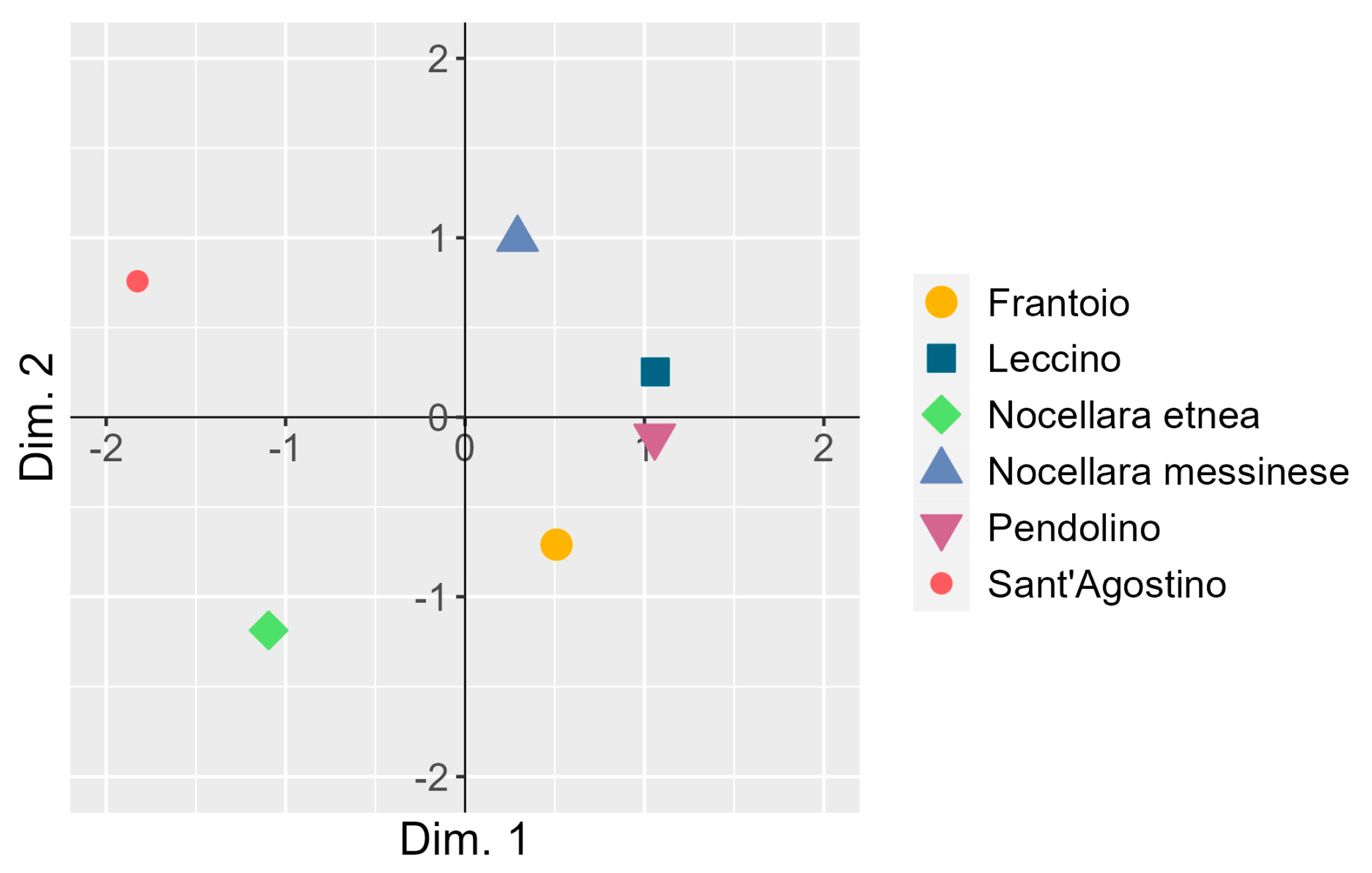
| Source of Variation | Marginal Mean of Drupe Diameter (mm) | df | SS | MS | F Value | p-Value |
|---|---|---|---|---|---|---|
| Cultivar (CV) | 5 | 16,047 | 3209 | 3038.6 | <0.001 | |
| ‘Frantoio’ | 10.4 ± 0.1 e | |||||
| ‘Leccino’ | 11.0 ± 0.1 d | |||||
| ‘Nocellara etnea’ | 11.6 ± 0.1 c | |||||
| ‘Nocellara messinese’ | 15.9 ± 0.2 b | |||||
| ‘Pendolino’ | 9.0 ± 0.1 f | |||||
| ‘Sant’Agostino’ | 17.2 ± 0.2 a | |||||
| Time (T) | 9 | 6315 | 702 | 664.4 | <0.001 | |
| DOY 188 | 9.0 ± 0.1 h | |||||
| DOY 199 | 10.2 ± 0.2 g | |||||
| DOY 209 | 10.9 ± 0.2 f | |||||
| DOY 216 | 11.9 ± 0.2 e | |||||
| DOY 235 | 12.7 ± 0.2 d | |||||
| DOY 248 | 13.3 ± 0.2 c | |||||
| DOY 259 | 13.5 ± 0.2 c | |||||
| DOY 277 | 14.3 ± 0.3 b | |||||
| DOY 287 | 14.5 ± 0.3 ab | |||||
| DOY 301 | 14.8 ± 0.3 a | |||||
| CV × T | 45 | 627 | 14 | 13.2 | <0.001 | |
| Residuals | 1740 | 1838 | 1 |
| Principal Component | Eigenvalue | Variance Explained (%) | Cumulated Variance Explained (%) |
|---|---|---|---|
| Dim. 1 | 14.74 | 61.4 | 61.4 |
| Dim. 2 | 5.98 | 24.9 | 86.3 |
| Dim. 3 | 2.27 | 9.5 | 95.8 |
| Dim. 4 | 0.63 | 2.6 | 98.4 |
| Dim. 5 | 0.38 | 1.6 | 100.0 |
| Locus | Na | ASR (bp) | I | Ho | PIC | E5 |
|---|---|---|---|---|---|---|
| DCA3 | 5 | 228–250 | 1.31 | 0.83 | 0.67 | 0.73 |
| DCA4 | 5 | 128–160 | 1.42 | 0.83 | 0.72 | 0.82 |
| DCA5 | 3 | 198–206 | 0.72 | 0.50 | 0.40 | 0.64 |
| DCA9 | 5 | 162–206 | 1.55 | 1.00 | 0.78 | 0.95 |
| DCA16 | 6 | 124–176 | 1.47 | 0.83 | 0.69 | 0.68 |
| DCA17 | 5 | 102–140 | 1.59 | 1.00 | 0.79 | 0.97 |
| DCA18 | 6 | 171–185 | 1.63 | 0.83 | 0.78 | 0.85 |
| GAPU47 | 6 | 187–203 | 1.70 | 1.00 | 0.81 | 0.92 |
| GAPU71b | 4 | 124–144 | 1.31 | 1.00 | 0.71 | 0.90 |
| UDO31 | 7 | 106–150 | 1.79 | 0.67 | 0.81 | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corrado, G.; Mataffo, A.; Garonna, A.P.; Rao, R.; Basile, B. Investigation of Fruit Growth Patterns, Olive Fly Bactrocera oleae (Rossi) Infestation, and Genetic Diversity in Italian Olive Cultivars. Appl. Sci. 2023, 13, 9929. https://doi.org/10.3390/app13179929
Corrado G, Mataffo A, Garonna AP, Rao R, Basile B. Investigation of Fruit Growth Patterns, Olive Fly Bactrocera oleae (Rossi) Infestation, and Genetic Diversity in Italian Olive Cultivars. Applied Sciences. 2023; 13(17):9929. https://doi.org/10.3390/app13179929
Chicago/Turabian StyleCorrado, Giandomenico, Alessandro Mataffo, Antonio P. Garonna, Rosa Rao, and Boris Basile. 2023. "Investigation of Fruit Growth Patterns, Olive Fly Bactrocera oleae (Rossi) Infestation, and Genetic Diversity in Italian Olive Cultivars" Applied Sciences 13, no. 17: 9929. https://doi.org/10.3390/app13179929
APA StyleCorrado, G., Mataffo, A., Garonna, A. P., Rao, R., & Basile, B. (2023). Investigation of Fruit Growth Patterns, Olive Fly Bactrocera oleae (Rossi) Infestation, and Genetic Diversity in Italian Olive Cultivars. Applied Sciences, 13(17), 9929. https://doi.org/10.3390/app13179929







