Abstract
Due to the success of artificial intelligence (AI) applications in the medical field over the past decade, concerns about the explainability of these systems have increased. The reliability requirements of black-box algorithms for making decisions affecting patients pose a challenge even beyond their accuracy. Recent advances in AI increasingly emphasize the necessity of integrating explainability into these systems. While most traditional AI methods and expert systems are inherently interpretable, the recent literature has focused primarily on explainability techniques for more complex models such as deep learning. This scoping review critically analyzes the existing literature regarding the explainability and interpretability of AI methods within the clinical domain. It offers a comprehensive overview of past and current research trends with the objective of identifying limitations that hinder the advancement of Explainable Artificial Intelligence (XAI) in the field of medicine. Such constraints encompass the diverse requirements of key stakeholders, including clinicians, patients, and developers, as well as cognitive barriers to knowledge acquisition, the absence of standardised evaluation criteria, the potential for mistaking explanations for causal relationships, and the apparent trade-off between model accuracy and interpretability. Furthermore, this review discusses possible research directions aimed at surmounting these challenges. These include alternative approaches to leveraging medical expertise to enhance interpretability within clinical settings, such as data fusion techniques and interdisciplinary assessments throughout the development process, emphasizing the relevance of taking into account the needs of final users to design trustable explainability methods.
1. Introduction
1.1. AI in Medicine: Opportunities and Challenges
Today’s Artificial Intelligence (AI), with its capability to automate and ease almost any kind of task, frequently appearing to surpass human performance, has become a popular and widespread technology for many applications, especially over the last decade, thanks to advances in deep learning (DL), with clinical healthcare being no exception.
Medicine has been one of the most challenging, but also most attention-getting application fields for AI for the past five decades, with diagnostic decision support, the interpretation of medical images and clinical lab tests, drug development, patient management, and others all demonstrating the broad and diverse scope of AI techniques applied to medical issues.
AI methods have promised a range of potential advantages for medical informatics systems. Automating burdensome tasks can be of great help, alleviating clinicians from unnecessary efforts and allowing them to focus on more important issues surrounding patient care. AI systems can perform these tasks with high precision and efficiency, and also, they can assist the extraction of relevant information from the large quantities of data being produced by modern medicine []. AI systems might be particularly beneficial in settings such as developing countries, where advanced medical expertise could not be accessed otherwise.
Therefore, as it has the potential to transform and improve healthcare, there is already plenty of research on AI applied to clinical decision support systems or medical imaging analysis for diagnosis and prognosis [], as well as to other areas such as drug—including vaccines—development or healthcare organisation [].
However, despite the promising opportunities that AI holds, the number of clinical AI applications currently deployed and actually used in real-world healthcare workflow is still relatively limited [], although the US FDA has recently increased the number of approvals for AI-based medical applications []. The reasons behind this “adoption gap” are complex, and depend not only on pure scientific and technological issues, but also on the special components of medical reasoning, ethical issues, organisational aspects, and medical professionals’ training and acceptance of novel procedures and tools, among others.
When a novel AI algorithm is introduced as a tool to support medical decisions that can directly influence people’s lives and wellbeing, a number of questions—including ethical issues—arise concerning the trustworthiness of these systems and the possibilities of making them more predictable [].
These concerns become especially acute when using the classical, so-called “black box” AI algorithms. A recent, singular “mainstream” example is that of deep learning models that cannot provide direct, human understandable insights on the knowledge being used to achieve their final conclusions. Clinicians continue to be reluctant to trust the results of those kinds of systems that cannot easily provide detailed explanations []. Even clinicians with more advanced technological knowledge are also concerned with related issues such as algorithmic biases or more technical aspects—such as overfitting or the quality of the original data—which are much harder to uncover without considerable understanding of the underlying mathematical and computational models, as well as the statistical assumptions behind the actual implementations [].
Finally, it is essential to address the recent public release of ChatGPT (ChatGPT was released 20 November 2022 by OpenAI). It is based on transformer deep neural network (DNN) architecture (GPT-3) that helps to encode and decode the input text, and its main purpose is to extend the input size to allow larger text analysis and extract a latent space that contains the “meaning” of that text. Based on that latent space, the system is able to search for sentence queries or generate related responses. It has a conversational type human–machine interface and language capabilities that allows an easy interaction in natural language, extending the current query capabilities of web search tools. It is still unclear what the main applications are of this technology in medicine (it can be used to access similar previous patients’ reports, take clinical decisions, documentation, summarize, etc.) but from the explainability perspective it can be considered as a technology based on deep neural networks. To show its current potential, in reference [] the authors demonstrate that Large Language Models (LLMs) are able to approach or exceed the required threshold for passing the United States Medical Licensing Examination and its potential to generate novel insights that can assist human learners in a medical education setting. Yet, there are no guarantees or explanations as to why it gives one answer or another [].
1.2. The Emergence of the Field of XAI
Burgeoning expectations about AI, coupled with the need to resolve or skirt around the above problems, have led to the field known as eXplainable Artificial Intelligence (XAI). The latter deals with the different methods and approaches that help to identify the latent knowledge associated with AI models and make them intelligible to users, whether by building and designing more efficient or alternative intrinsically interpretable models, or by providing explanations for the “black box” models relying on statistical and optimisation methods, by using auxiliary techniques []. Concerning medical applications, there is great hope that these XAI-based approaches might be one of the keys to improving understanding, trust, and the final adoption of AI models in clinical practice [,].
Explainability is not a new research topic for AI in medical applications. For consultation and decision support, in particular, the first expert systems, developed in the 1970s, tried to justify the reasoning behind their decisions by showing rules or causal models. Years after these pioneering expert systems, knowledge engineers have not yet been able to facilitate comprehensive and useful explanation facilities for knowledge engineering systems, which are still limited.
Lots of research was conducted on knowledge acquisition and knowledge representation over this first generation of AI systems—also medical systems—and several impediments to creating knowledge bases from human expertise were identified []. These issues are strongly linked to the topic of explainability. In the pursuit of an understandable AI model that provides explanations to its decisions, we have to consider what kind of knowledge, and how it should be represented in the explanation given, in order to be sufficient and useful for the user (in the context of medicine, for a physician).
1.3. Relevance of Explainability in Medical AI
Despite the difficulties, it is clear that there are quite a few potential benefits of explainable and transparent models, motivating the renewed interest in the field (Figure 1). Besides helping increase the acceptance of AI models, enabling clinical professionals to take advantage of their conveniences and addressing ethical concerns, explainability could also the allow better validation of these systems and facilitate troubleshooting during development []. Going even further, it could also be argued that explainable models could assist knowledge discovery in some cases, if we are able to understand the intrinsic relations hidden in the data found by AI models [].
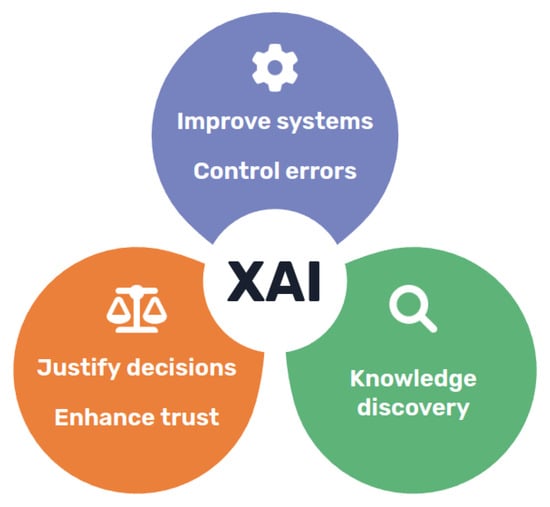
Figure 1.
Main objectives and advantages of XAI approaches, categorised into technical, scientific, and ethical dimensions.
When discussing the potential benefits and relevance of medical XAI, we believe it is also crucial to address the often-overlooked aspect of data quality, closely intertwined with ethical trust, algorithmic bias, and interpretability concerns. The scarcity and often poor quality of medical data pose significant technical and methodological challenges for AI in healthcare, that could potentially be easier to identify and mitigate thanks to XAI. While data-driven models offer the advantage of not requiring explicit expert knowledge, they can become fragile and prone to failure due to low-quality or artifact-ridden data [], leading to issues like overfitting and the hampering of model generalisation. This problem also stems from data biases such as poorly represented populations [,], variations in data collection conditions, and the incorporation of human biases, particularly in the field of medicine, where the individuality and particularities of each patient and clinician will exert great influence []. However, XAI techniques hold promise in helping to detect and address these issues before deploying faulty models [], emphasizing the need for rigorous data curation and comprehensive testing, that should go hand in hand with the development of XAI approaches in healthcare [,,].
In addition, explainability might become a future legal requisite in fields such as medicine, since patients need to be properly informed about how their personal data is being used. Currently, some legal regulations, such as the GDPR (General Data Protection Regulation) in the European Union [], are starting to include the requirement of transparent AI when dealing with sensitive data, pointing out that, in the future, the use of explainability in AI-based systems might be further enforced [].
For those reasons, much of the recent literature is increasingly becoming available, appraising the benefits of XAI and presenting many different new methods which claim to be appropriate solutions. This is especially required in the field of healthcare, where responsibility and understanding become peculiarly important issues. However, there is persistent controversy in the field, since some researchers remain skeptical about current approaches, stating that capabilities of XAI may be overestimated, while several important limitations may be disregarded []. Such a controversy advocates for the rigorous internal and external validation of AI models [].
This scoping review aims to be a starting point for those who want to get a wider and comprehensive view of the field of XAI applied to medicine. There are already a few review articles on this topic, as shown in our preliminary research, either more general [,] or focused in more specific subfields [,,,,,]. However, here we aim to present a broader perspective on the state of the art of XAI methods used in clinical applications, particularly taking into account the lessons learned from pioneering research in medical AI—since the 1970s—spanning until the recent boom of AI around Deep Learning, with the actual participation throughout these years of some of the authors, who have extensive experience in the field of medical AI, with decades of work including pioneering contributions to early medical expert systems such as CASNET []. With such a direct and long perspective in time, we highlight not only the benefits but also the open challenges posed by interpretability based on medical AI models. The review offers a critical perspective, drawing from the lessons learned from this experience, and benefits from an interdisciplinary approach given the diverse backgrounds of the authors in fields such as medicine, life sciences, computer science, and engineering.
2. Objectives
The main goal of this review is to analyse the existing literature dealing with the explainability and interpretability of AI methods applied to the medical field. We aim to provide an overview of current research trends and identify the challenges and limitations that this research area presents, as well as potential research directions and alternative approaches that could help improve trust and transparency for clinical AI applications.
Besides giving an overview of current approaches, this review aims to cover some studies on explainability from classical AI methods as well for applications in medicine and the clinic.
In order to make concrete the objectives of this review, the following research questions are posed:
- RQ1. What are the XAI approaches being used in medical AI applications?
- RQ2. Does any technical limitation to explainability exist that might particularly hinder XAI development in medicine?
- RQ3. What are the current research gaps and directions in the field of XAI that could be of special interest for medical applications?
- RQ4. Is it possible to include some kind of medical validation or knowledge to enhance the explainability of clinical AI systems?
3. Methodology
3.1. Search Strategy
This scoping review has been conducted according to the framework and guidelines proposed by the Joanna Briggs Institute Manual for Evidence Synthesis []. An electronic search over three scientific research databases was conducted, namely Web of Science, PubMed, and Scopus, looking for the relevant literature on the topic of explainability and transparency of AI methods in the field of medicine. This systematic search was conducted in February 2022, thereby encompassing articles published up to February 2022, without establishing a lower temporal limit.
To narrow down the search, several synonyms and related words were included so that each article title contained at least one term associated with medicine, another with artificial intelligence, and another with explainability (Table 1).

Table 1.
Keywords used for the literature search.
In addition to the systematic search, a few articles identified through the references of selected studies, recommended by the authors of this review, or searched by hand for topics not thoroughly covered in the available literature, were also included. Some of them were also selected to include recent advances not covered in the systematic search time range.
3.2. Inclusion and Exclusion Criteria
In order to meet the objectives of this review, the following criteria were defined for source inclusion:
- Directly related to the context of medicine and healthcare.
- Addressing clinical problems with AI algorithms, such as machine learning (ML), DL, computer vision or other kinds of either data- or knowledge-based methods.
- Including a clear focus on any kind of ethical or technical information about explainability, transparency or related issues.
- Published in English and with full text available.
A bottom time limit for the publications included was not established, as one of the objectives of this review was to cover some studies from the first medical AI systems (1970s onwards) to take into account the lessons learned from that period that might be of interest for current and future AI-focused systems. No constraints about the kind of publication were defined either; articles, reviews, and studies of any kind were included as long as they met the inclusion criteria and were of interest for this review.
Articles that did not meet this criteria were excluded, such as those that used other types of computer approaches that do not fall under the scope of AI, those that did not talk about clinical applications or those that only included matters of explainability as a side topic without going into much detail.
3.3. Data Extraction and Analysis
The processes of the sourcing and screening of evidence conducted in this review are shown in Figure 2. After performing the systematic search and excluding duplicates between the results of the different databases, a first stage of selection was performed by analysing the title and abstract of the resulting papers. At this stage, all papers that were clearly not related to the topic of this review, either because they were not related to medicine, because they did not actually focus on AI methods, or because they did not discuss interpretability at all, as well as those that were not published in English, were excluded.
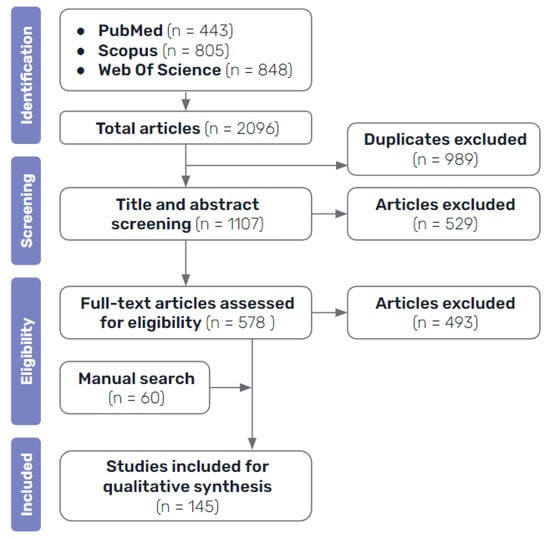
Figure 2.
Diagram illustrating the workflow for the analysis and selection of evidence sources.
Once all the potential candidate articles had been selected, a second stage of selection was performed to reach the final selection of the literature included in this review. A more thorough assessment was made by looking into the full text papers, in order to exclude the ones that were of no interest for this review, those that were too short or general, articles that did not focus sufficiently on the topic of explainability, studies that were too similar to others already included or that employed identical techniques and did not add much value to the synthesis of the review, in addition to ones that had no available full text. This final selection of articles, plus the ones identified by manual search, make up the final bibliography included in this review.
3.4. Literature Search Results
The literature analysis carried out in this review corroborates the rising popularity over the past few years of the topics of explainability and interpretability on medical AI applications. Even though our search shows that some related studies were published in the 1980s, it is not until about 2017 that the number of publications starts to substantially increase, similar to that which other related studies show []. Moreover, as expected, there is a special focus on explainability approaches to DL models, as they are currently the most popular ML methods.
Studies included in this review mainly consist of medical AI applications that employ explainability methods or that are built with interpretable models, but there are also some review papers and ethical commentaries. In what follows, we review the approaches and the methods employed in the reviewed literature to make medical AI models explainable will be presented, as well as other important issues covered in the various articles.
4. XAI Methods in Medicine
4.1. Classification of Explainability Approaches
Several taxonomies have been proposed in the literature to classify XAI methods and approaches, depending on different criteria [,,] (Figure 3).
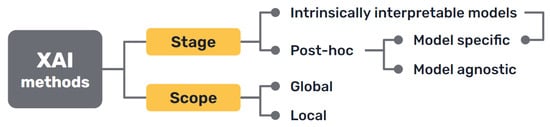
Figure 3.
Most common approaches to the classification of XAI methods.
First, there is a clear distinction between auxiliary techniques that aim to provide explanations for either the model’s prediction or its inner workings, which are commonly called post hoc explainability methods, and AI models that are intrinsically interpretable, either because of their simplicity, the features they use or because they have a straightforward structure that is readily understandable by humans.
Secondly, we can distinguish interpretability techniques by their scope, where explanations provided by an XAI approach can be local, meaning that they refer to particular predictions of the model, or global, if they try to describe the behaviour of the model as a whole.
Other differentiations can be made between interpretability techniques that are model specific, because they have requirements regarding the kind of data or algorithm used, and model agnostic methods, that are general and can be applied in any case. Intrinsically interpretable models are model specific by definition, but post hoc explainability methods can be generally seen as model agnostic, though some of them can have some requisites regarding the data or structure of the model.
More classifications can be made regarding how characteristics of the output explanations are displayed (textual, visual, rules, etc.), the type of input data required, the type of problem they can be applied to [] or how they are produced [].
However, since these classifications overlap [], in the following sections we have chosen to go over the different techniques included in the reviewed literature structured under the most popular taxonomy: interpretable models and post hoc explainability methods (RQ1).
4.2. Intrinsically Interpretable Models
Intrinsically interpretable models are those built using logical relations, statistical or probabilistic frameworks, and similar strategies that represent human-interpretable systems, since they use rules, relationships or probabilities assigned to known variables.
This approach to explainable AI, despite receiving less attention in recent years while the focus has been on DL, is historically the original one, and the perspective taken by knowledge-based systems.
4.2.1. Classical Medical Knowledge-Based Systems
Some knowledge-based systems, commonly known as “expert systems”, are some of the classical AI models that were first developed at the end of the 1960s. Explanations were sometimes introduced as a feature of these first rule-based expert systems by design, as they were needed not only by users, but also by developers, to troubleshoot their code during the design of these models. Thus, the importance of AI explainability has been discussed since the 1970s [,].
In medicine, many of these systems were developed aiming to be an aid for clinicians during the diagnosis of patients and treatment assignment []. We must reiterate that explanations for patient cases are not easily made, in many cases, by human medical professionals. The most widely known of these classical models was MYCIN [], but many more based on causal, taxonomic, and other networks of semantic relations, such as CASNET, INTERNIST, the Present Illiness Program, and others, were designed to support rules by models of underlying knowledge that explained the rules and drove the inferences in clinical decision-making [,,]. Subsequently, the modelling of explanations was pursued explicitly for the MYCIN type of rule-based models []. The role of the explanation was frequently recognised as a major aspect of expert systems [,,].
As shown in Figure 4, expert systems consist of a knowledge base containing the expertise captured from human experts in the field, usually in the form of rules, including both declarative or terminological knowledge and procedural knowledge of the domain []. This knowledge base is consulted by an inference algorithm when the user interacts with the system, and an explanation facility interacts with both the inference system and the knowledge base to construct the corresponding explaining statements [].
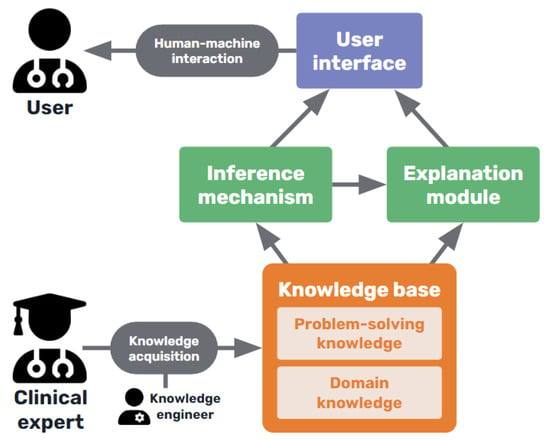
Figure 4.
Basic diagram of a medical knowledge-based expert system.
The explanations provided were pre-modeled [] and usually consisted of tracing the rules used by the inference mechanism to arrive at the final decision and present them in an intelligible way to the user. Such an approach aimed to explain how and why the system produced a diagnosis [] and, in some more sophisticated cases, even the causal evolution of the patient’s clinical status [].
However, these explanations were limited by the knowledge base constructed by the system’s designers; all the justifications for knowledge had to be explicitly captured to produce specific explanations []. Knowledge acquisition (KA) and updating is a challenging task (see Section 6.2) and this was not efficiently resolved, leading KA to become the bottleneck that resulted in the decline of interest in knowledge-based AI systems (as a main example of symbolic AI) in favour of data-based AI and ML systems (which we can call subsymbolic AI).
4.2.2. Interpretable Machine Learning Models
As an alternative to knowledge-based systems, from the early days of medical decision making, statistical Bayesian, Hypothesis-Testing, and linear discriminant models were ML models that can be considered interpretable. They are based on the statistical relationships extracted from clinical databases which allow formal probabilistic inferential methods to be applied. Ledley and Lusted proposed the Bayesian approach in their pioneering article in the journal in 1959 [], with many of the alternatives first discussed in The Diagnostic Process conference []. Logistic regression is an effective statistical approach that can be used for classification and prediction. Generalised Linear Models (GLMs) [] are also used in various problems among the literature [,], while Generalised Additive Models (GAMs), an extension of these, allow the modelling of non-linear relationships and are used for prediction in medical problems as well [,].
Decision trees are considered transparent models because of their hierarchical structure which allows the easy visualisation of the logical processing of data in decision making processes. Moreover, a set of rules can be extracted to formalise that interpretation. They can be used for classification in the medical context [,], however, they sometimes show poor generalisation capabilities, so it is most common to use tree ensembles (like the random forest algorithm []) that show better performance, in combination with post hoc explainability methods, as they lose some interpretability [,,,].
The generalisation of formal models through Bayesian networks [], have become popular for modelling medical prediction problems [,,,,], representing conditional dependencies between variables in the form of a graph, so that evidence can be propagated through the network to update the diagnostic or prognostic states of a patient []. This reasoning process can be easily visualised in a straightforward manner.
Interpretable models can be used by themselves, but another interesting strategy is using them in ensemble models. Ensemble models consist of combining several different ML methods to achieve a better performance and better interpretability than with a black-box model alone []. These approaches can also include these interpretable models in conjunction with DL models such as neural networks [,,,], as well as other post hoc explainability techniques [,], such as the ones that will be presented later in this paper (Section 4.3). However, they pose increasingly, and as yet unresolved, complex interpretation issues, as recently emphasised by Pearl [].
4.2.3. Interpretation of Neural Network Architectures
Despite the fact that neural networks cannot be fully included in the category of intrinsically interpretable models, we can characterize them (DL are also included), such as architectures designed so that they resemble some of the simple neural modelling of brain function and that are used heuristically to recognise images or perform different tasks, and some neural network architectures have been specifically designed to provide interpretability. The first type tries to mimic human decision-making where the decision is based on previously seen examples. In reference [], a prototype learning design is presented to provide the interpretable samples associated with the different types of respiratory sounds (normal, crackle, and wheeze). This technique learns a set of prototypes in a latent space that are used to make a prediction. Moreover, it also allows for a new sample to be compared with the set of prototypes, identifying the most similar and decoding it to its original input representation. The architecture is based on the work in reference [] and it intrinsically provides an automatic process to extract the input characteristics that are related to the associated prototype given that input.
Other methods’ main motivation is to behave in a way more similar to how clinicians diagnose, and provide explanations in the form of relevant features. Among this type, attention maps are widely used. In short, they extract the influence of a feature on the output for a given sample. They are based on the gradients of the learned model and, in [], have been used to provide visual MRI explanations of liver lesions. For small datasets, it is even possible to include some kind of medical knowledge as structural constraint rules over the attention maps during the process design []. Moreover, the attention maps can also be applied at different scales, concatenating feature maps, as proposed in reference [], and being able to identify small structures on retina images.
These approaches are specific to DL but, still, surrogate models or post hoc methods are applicable to add explainability.
4.3. Post Hoc Explainability Methods
Extending the above approach to transparency came with the development of more complex data-based ML methods, such as support vector machines (SVMs), tree ensembles, and, of course, DL techniques. The latter have become popular due to their impressive performance on a huge variety of tasks, sometimes even surpassing human accuracy for concrete applications, but also unfortunately entailing deeper opacity, for instance, than the detailed explanations that classic statistics can provide.
For this reason, different explainability methods have been proposed in order to shed light on the inner workings or algorithmic implementations used in these black-box-like AI models. Because they are implemented as added facilities to these models, executed either over the results or the finished models, they are known as post hoc methods, which produce post hoc explanations, as opposed to the approach of intrinsically interpretable models.
Many of the approaches included in this category, which are also currently the most widely used, as reported in the literature, are model agnostic. Post hoc model agnostic methods are so popular due to their convenience: they are quick and easy to set up, flexible, and well-established. Within this category, there are also some model specific post hoc techniques designed to work only for a particular type of model.
These are less flexible, but tend to be faster and sometimes more accurate due to their specificity, as they can access the model internals and can produce different types of explanations that might be more suitable for some cases [].
Regardless of their range of application, post hoc methods can also be grouped on the basis of their functionalities. Combining the taxonomies proposed in references [] and [], we can broadly differentiate between explanations through simplification (surrogate models), feature relevance methods, visualisation techniques, and example-based explanations. In the following sections, these ideas will be presented as well as some of the most popular and representative methods belonging to each group.
4.3.1. Explanation by Simplification
One way to explain a black-box model is to use a simpler, intrinsically interpretable model for the task of explaining its behaviour.
One method that uses this idea, which is undoubtedly one of the most employed ones throughout all the literature, is LIME (Local Interpretable Model-agnostic Explanations) []. This method builds a simple linear surrogate model to explain each of the predictions of the learned black-box model. The prediction’s input to be explained is locally perturbed creating a new dataset that is used to build the explainable surrogate model. An explanation of instances can help to enforce trust in assisted AI clinical diagnosis within a patient diagnosis workflow [].
Knowledge distillation is another technique included in this category. It was developed to compress neural networks for efficiency purposes, but it can also be used to construct a global surrogate interpretable model []. It consists of using the more complex black-box model as a “teacher” for a simpler model that learns to mimic its output scores. If the “student” model demonstrates sufficient empirical performance, a domain expert may even prefer to use it in place of the teacher model and LIME. The main rationale behind this type of modelling is the assumption that some potential noise and error in the training data may affect the training efficacy of simple models. The authors of reference [] used knowledge distillation to create an interpretable model, achieving a strong prediction performance for ICU outcome prediction.
Under this category, we could also include techniques that attempt to simplify the models by extracting knowledge in a more comprehensive way. For example, rule extraction methods try to approximate the decision-making process of the black-box model, such as a neural network, with a set of rules or decision trees. Some of the methods try decomposing the units of the model to extract these rules [], while others keep treating the original model as a black box and use the outcomes to perform a rule search []. There are also combinations of both approaches [].
4.3.2. Explanation Using Feature Relevance Methods
In the category of feature relevance methods, we can find many popular examples of explainability techniques. These approaches try to find the most relevant variables or features to the model’s predictions, those that most influence the outcome in each case or in general.
The ancestry to these techniques can be found in both statistical and heuristic approaches dating back to the 1930s with Principal Component Analysis (PCA), which explains the weightings of features, or contributions to relevance in terms of their contribution to inter- and intra-population patterns of multinomial variance and covariance []. These techniques were also shown to be central to both dimensionality reduction and its explanation in terms of information content for pattern recognition [] and clinical diagnostic classification and prediction using subspace methods from atomic logic []. Later, related techniques for feature extraction by projection pursuit were developed and applied to clinical decision-making.
More recently, with LIME (that could also be included in this group), SHAP (SHapley Additive exPlanations) is one of the most widely used XAI model agnostic techniques, and it is the main example of the category of feature relevance methods. It is based on concepts from game theory that allow the computing, which are the features that contribute the most to the outcomes of the black-box model, by trying different feature set permutations []. SHAP explanations increase trust by helping to test prior knowledge and can also help to get insights into new ones [].
Other well-known similar examples that measure the importance of different parts of the input by trying different changes is SA (Sensitivity Analysis) [], and LRP (Layer-Wise Relevance Propagation) []. Deep Taylor Decomposition (an evolution of LRP) [] and DeepLIFT [] are other model-specific alternatives for neural networks, that propagate the activation of neurons with respect to the inputs to compute feature importance.
4.3.3. Explanation by Visualisation Techniques
Some of the aforementioned methods can produce visual explanations in some cases. Still, in this section we can mention some other methods that directly visualize the inner workings of the models, like Grad-CAM [], that helps in showing the activation of the layers of a convolutional neural network. In addition, there are other techniques that visualize the inputs and outputs of a model and the relationship between them, such as PDP (Partial Dependence Plots) [] and ICE (Individual Conditional Expectation) plots []. It is worth mentioning that visualisation can help to build explicable interfaces to interact with users, but it is complex to use them as an automatic step of the general explainability process.
4.3.4. Explanations by Examples
Finally, another approach to produce explanations is to provide examples of other similar cases that help in understanding why one instance has been classified as one object or structure or another by the model, or instead, dissimilar instances (counterfactuals) that might provide insights as to why not.
For instance, MMD-critic [] is an unsupervised algorithm that finds prototypes (the most representative instances of a class) as well as criticisms (instances that belong to a class but are not well represented by the prototypes). Another example are counterfactual explanations [] that describe the minimum conditions that would lead to a different prediction by the model.
5. Evaluation of Explainability
Despite the growing body of literature on different XAI methods, and the rising interest in the topics of interpretability, explainability, and transparency, there is still limited research on the field of formal evaluations and measurements for these issues []. Most studies just employ XAI techniques without providing any kind of quantitative evaluation or appraisal of whether the produced explanations are appropriate.
Developing formal metrics and a more systematic evaluation of different methods can be difficult because of the variety of the available techniques and the lack of consensus on the definition of interpretability []. Moreover, contrary to usual performance metrics, there is no ground-truth when evaluating explanations of a black-box model [,]. However, this is a foundational work of great importance, as such evaluation metrics would help towards not only assessing the quality of explanations and somehow measuring if the goal of interpretability is met, but also to compare between techniques and help standardise the different approaches, making it easier to select the most appropriate method for each case [].
In short, there is a need for more robust metrics, standards, and methodologies that help data scientists and engineers to integrate interpretability in medical AI applications in a more detailed, verified, consistent, and comparable way, along the whole methodology, design, and algorithmic development process []. Nevertheless, in the few studies available on this topic, there are some common aspects that establish a starting point for further development, and there are some metrics, such as the robustness, consistency, comprehensibility, and importance of explanations.
A good and useful explanation for an AI model is one that is in accordance with human intuition and easy to understand []. To evaluate this, some qualitative and quantitative intuitions have already been proposed.
- On the one hand, qualitative intuitions include notions about the cognitive form, complexity, and structure of the explanation. For example, what are the basic units that compose the explanation and how many are there (more units mean more complexity), how are they related (rules or hierarchies might be more interpretable for humans), if any uncertainty measure is provided or not, and so on [].
- On the other hand, quantitative intuitions are easier to formally measure and include, for example, notions like identity (for identical instances, explanations should be the same), stability (instances from the same class should have comparable explanations) or separability (distinct instances should have distinct explanations) [,]. The metrics based on these intuitions mathematically measure the similarity between explanations and instances as well as the agreement between the explainer and the black-box model.
Other options to evaluate XAI techniques include factors such as the time needed to output an explanation or the ability to detect bias in the data [].
Another interesting strategy is to quantify the overlap between human intuitions (such as expert annotations) and the explanations obtained [,], or using human ratings by experts on the topic [,,,].
There are also different options regarding the context in which these various metrics can be used. The evaluation of an XAI system can be made either in the context of the final target task with the help of domain experts, in simpler tasks, or using formal definitions []. Depending on the specific characteristics of the problem and available resources, different approaches and metrics can be chosen.
6. Challenges of XAI Approaches
As discussed before, there is an increasing recognition of XAI in recent years, particularly in fields like medicine, rediscovering previous concerns already raised for 50–60 years about statistics, pattern recognition, and earlier AI models, where the ethical requirements for transparency based on the Hippocratic Oath and derived professional ethics of practitioners are even stronger []. But, despite the benefits of using explainable systems being so clear, and with some of the advances already made in the field and its range of available methods, the use of XAI techniques is still not widespread [].
Why, then, do explainability methods still seem insufficient? The issue of AI interpretability is highly intricate, as there are many unresolved technical limitations, related ethical questions, and ongoing controversies surrounding the current approaches.
6.1. Controversy around Current XAI Methods
Currently, one of the frequently discussed issues among researchers in the field of XAI is the choice between different approaches, mainly between post hoc explainability and other model-dependent criteria. There is no consensus about whether it is better to use intrinsically interpretable models or to develop techniques that try to explain the outcomes of black-box models, given the contrasts and complementarities between them that make comparisons incommensurate.
Despite being more understandable (and therefore meeting some of the explainability needs that make other types of systems more problematic), interpretable models are commonly rated as less accurate or efficient than more complex data-based approaches, such as DL (see Figure 5), implying that there is a trade-off between explainability and classification or prediction accuracy [,,] (RQ2).
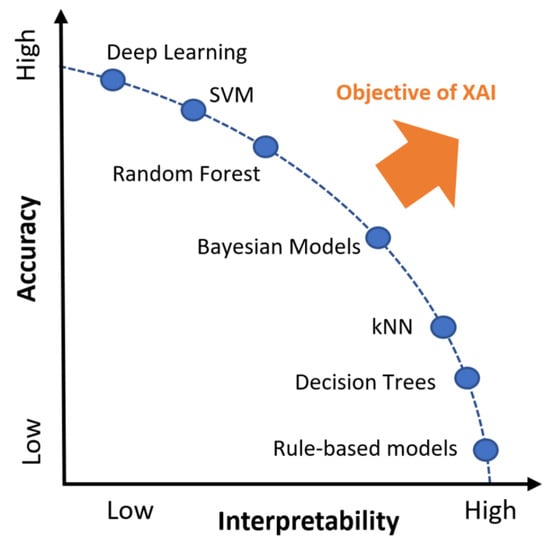
Figure 5.
Trade-off between explainability and performance of different AI methods. Adapted from reference [].
If such a trade-off exists, it could be alleged that it would not be ethical to use a model that does not perform at the best possible level, therefore being more adequate to use top-notch accurate systems, regardless of their black-box nature []. Some researchers maintain that, in fact, explainability might not be so necessary if good accuracy is proven by conducting empirical validations, for example [,], while others disagree [,]. The practical ethical problem is the heterogeneity and lack of “gold standards” of comparable training and testing datasets for any particular problem with statistical and heuristic approaches, which do not take into account the wide range of qualitative differences between different types of data and knowledge, and the implementation of responsible evaluatory judgments by experts which cannot be reduced to simple measures. Complementary approaches for this might be official certifications or criteria for controlling data biases, beside explainability, in order to build trust on these systems [,].
Moreover, some scholars argue that, while it is important that research continues trying to unravel black-box models, we should also focus on pursuing more efficient and stable models, and putting so much emphasis on the interpretability of these models before clinical application might restrain innovation in that direction and might not be that crucial [].
However, from an opposite point of view, other researchers advocate for intrinsically interpretable models, arguing that there is no such trade-off between explainability and accuracy in reality and that these models are more appropriate and safe for high-stakes fields like healthcare, where explanations of black-box models can be misleading and inaccurate, inducing the opposite effects to those intended []. At the same time, some interpretable models have been recognised to require further explanations for non-technical end users such as clinicians or patients [].
While there is no clear agreement on what is the best approach to the problem of transparency and interpretability or if it is even always necessary, some studies show that physicians, unsurprisingly, prefer AI systems that include some form of explainability, while seeming not to have a clear preference between methods [].
In the following section, we will discuss further limitations of the explanations related to this controversy, and that should be taken into account when designing these kinds of models and choosing among explainability approaches.
6.2. Technical Limitations of Explainability
While there is a common trend in favour of employing XAI among researchers, the limitations of these approaches are often underestimated, which can be especially high risk in the field of medicine []. These obstacles might not fully invalidate the use of XAI methods, but researchers and developers should be aware of them, as they can negate the application of these techniques in certain cases, and urgently require study [] (RQ2).
Importantly, it should be noted that explanations and transparency might not translate into understanding underlying biomedical causal relationships [], so we must be very cautious when using explainability as a tool to attempt knowledge discovery. For instance, using a surrogate model to explain a black-box system is only an approximation of the original model, and the produced explanations are unlikely to be faithful to the true inner workings of it []. Beyond this, it cannot be assumed that behind the explanations or decisions of an AI system there is always an equivalent or at least a truly comparable reason that human experts could infer []. For that reason, we should be careful not to build systems that are persuasive rather than transparent [,], by properly analysing, validating, and interpreting the results obtained.
Another point to be taken into account is that, usually, models and explanations that are more simple or compact are more interpretable, but it might not always be that way. Explanations depend on the specific context of the task and expectations of the user [], and, if they are not meaningful, detailed enough or in the needed form, they might not be useful and may only complicate explanations of decision pathways [].
It should also be considered that there can also be serious cognitive limits to explainability, as was acknowledged about knowledge acquisition in the development of expert systems in the first decades of AI development. When human experts, like clinicians, become proficient in an area, they perform their tasks in a kind of automated, or at least compiled, way, effortlessly and efficiently. At this point, the knowledge needed for performing these tasks has become tacit, meaning that it is compiled in their mind and not available for conscious access and sharing []. For that reason, sometimes they cannot completely explain what the exact reasons are behind their decisions.
This issue of after-the-fact justificatory explanations by experts makes it really hard to accurately or veridically model expert knowledge when the problem to solve is complex or vaguely defined, which is typical for the medical domain []. Unlike scientific disciplines such as biology, in clinical practice where reproducible experimentation is approximately feasible at least, there is a considerable uniquely patient-and-expert-encounter-specific practical expertise, and a lack of generalizable knowledge for many practical problem-solving situations []. In consequence, it is really difficult to elicit reproducible and useful models of physician reasoning in the form of concrete rules [].
This is one of the reasons that have contributed to making it impossible to fully capture the knowledge needed for building expert systems [], and, ironically, it presents parallels to the problems with black-box AI heuristic or statistical models [], so maybe it is unrealistic to expect comprehensive explanations from these systems as we might desire, in the same way that we do not entirely understand much of human neural processes within their complex biochemical, genetic, and developmental living contexts. Moreover, one must take into account that post hoc explanations frequently suffer from the same problems of interpretability as human-expert explanations [].
7. Research Opportunities for Medical XAI
The research limitations and shortcomings surrounding XAI discussed in this review, include the need for more robust evaluations (Section 5) and for more studies about cognitive limitations of explainability (Section 6), as well as efforts for improving data quality (Section 1.3). Here, we discuss explainability, as it is especially related to the field of medicine (RQ3).
Before specifying, it ought to be emphasised that much more interdisciplinary work would benefit the building of XAI systems. Not only do users’ needs have to be taken into account more explicitly, but detecting errors in models that are related to a particular field of application has to be learned and imbued by technical designers, in order to improve current explainability techniques and develop and implement novel and more effective ones.
Lately, most of the studies in the field of interpretability tend to focus and develop some particular techniques that are more popular at the moment (for example, feature extractions or deep learning model visualisations), giving less attention to other categories (such as interpretable models, for instance). Popular purely technological techniques are hardly the most appropriate solutions for every case and, moreover, combining ideas from different fields could lead to really interesting advances to achieve more effective explanations [].
7.1. Alternative Ways to Incorporate Medical Knowledge
We have already commented on the difficulties of capturing and modelling knowledge from experts, especially in healthcare, due to the uncertainty and incompleteness of knowledge in clinical practice []. However, it is undeniable that knowledge would help in building more robust AI systems, with enhanced clinical interpretability. For that reason, research on alternative ways of embedding clinicians’ expertise in AI models will be most useful (RQ3 and RQ4).
One way of incorporating medical knowledge can involve human-in-the-loop evaluations of these systems []. Interaction with medical experts during development, or discussing the results of XAI models, can help detect errors, validate these systems, and identify possible underlying causes of the model’s behaviour that would go unnoticed by technical developers. The use of the novel technique ChatGPT could also be helpful to automatize this process or part of it.
Another interesting research path is information fusion. Combining different types of multimodal data, such as medical images, family histories, genomic data or electronic health records can help specify, define, and incorporate clinical context into a model, improving not only its diagnostic accuracy, but also its interpretability [,,]. Moreover, using data collected from different centres can also help with domain variability and shift and enhance AI models [].
There are already some studies on how to achieve this fusion of different kinds of data [] as well as studies taking this kind of approach [], however, more research and comprehensive data collection and annotation should be made to facilitate the building of these enhanced AI systems [].
7.2. Tailoring of Explanations to Diverse User Needs
Choosing the right XAI approach for each case is not a straightforward task, as the decision depends on many factors, such as characteristics of the concrete problem, of the application environment or of the available data, as different applications have different interpretability needs []. However, the most important aspect involves the requirement of the actual end-users of a system: clinicians and, in some cases, patients.
The questions from the users will vary depending on their expertise and knowledge of the domain, and their views on an application problem. A clinician using an AI system to confirm a diagnosis who wants to know if the model is working properly but has no technical knowledge will need different explanations than the system builder who wants to check the models’ performance, or a patient who is using an AI system by themselves. For these reasons, the kind and extent of explanations need to be adjusted to the specific type of user needs in order to build trust [], without falling into over-trust [].
Depending on the prospective end users of the explanations, whether patients, clinicians or technical designers, it might be preferable to design different types of explainability, about the exhaustive workings of the model or about the relevant features being used, for example []. It should be identified in each case why users want explanations from the model, what information they need that said explanations contain, and how they prefer them to be presented [,] (RQ3).
If the factors above are not considered, accurate explanations will not match the needs of the users and be informative and understandable to them []. Achieving this user understanding might also need interaction between the system and its human user, in order to obtain further answers to different questions [].
To summarise, in order to enable collaboration between humans and AI, XAI outcomes have to be appropriately tailored to different end users, so more attention has to focus on these aspects of research: human–machine interaction and users’ mental models. It is most likely that general solutions will not be feasible, so the context of the problem has to be taken into account, preferably with interdisciplinary collaboration, and combining different types of explainability to fulfill users’ needs [].
8. Conclusions
AI has the potential to transform and improve healthcare; nevertheless, without explainable and trustworthy systems, its application will continue to be limited.
In this paper, we have reviewed the precedents and background with the state of the art of XAI as applied in medicine. Figure 6 shows the interdisciplinary of the work and the main concepts explored in this review. However, there are some limitations of this review that are worth noting. The field of XAI, much like AI in general, is subject to constant change and evolution. Our study is limited in scope, extending only up to early 2023. Given the rapid pace of developments in these fields, some of the latest advances, especially in ML techniques, might not be comprehensively reflected in this review. Furthermore, it is important to understand the nature of this review. As a scoping review, rather than a meta-analysis, its purpose is to provide an overview of the current state of the field, but it is not exhaustive in coverage. Instead, we aimed to present a critical and conceptual perspective, focusing on key aspects and highlighting the important directions and challenges for future research.
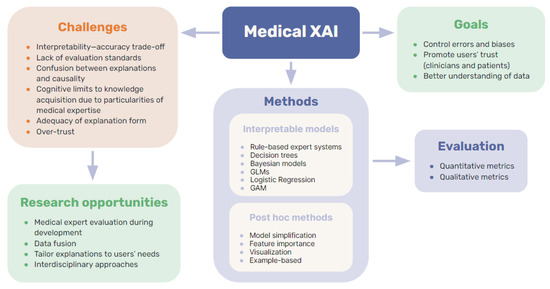
Figure 6.
Summary of key concepts explored in this review.
Still, several popular approaches and techniques have been discussed providing an overview of the available options in including explainability as an aspect of clinical AI systems (RQ1).
The benefits of explainable systems could be considerable: promoting trust, enabling better interpretation of the data and predictions or enhancing the detection of errors, for example. However, the considerable challenges identified in this review need to be overcome in the field of medical XAI. Besides the high dimensionality and black-box nature of many AI models, in medicine, the problem of data quality is especially serious, if we are to be able to develop accurate XAI techniques without the risk of being influenced by unrepresentative or poorly selected or curated and filtered data. Moreover, cognitive limitations to knowledge acquisition regarding clinicians’ reasoning are also related to the extent of how far XAI methods might be able to prove useful (RQ2).
In order to develop and consolidate further robust explainability methods and interpretable models, making them a useful tool for clinicians to trust medical AI applications and, therefore, supporting their implementation in the real world, future research on this topic should be focused on overcoming these challenges, as well as better analysing user needs, enhancing human–system interaction and studying alternatives such as data fusion or clinicians’ feedback to include medical validation and knowledge in different ways without the need to explicitly model it. Moreover, involving medical experts in the process of designing and developing these systems would also help in building more robust models and improving user understanding (RQ3 and RQ4).
All in all, the prospects for XAI methods in clinical applications is that they are essential in many ways, but that further research is needed to overcome the current limitations enhancing these techniques in order to build secure, trustworthy, and efficient systems that benefit both patients and clinicians.
Author Contributions
Conceptualisation, E.G.-C., V.M. and C.A.K.; methodology, R.G.-A., E.G.-C., R.G.-A. and V.M.; formal analysis, R.G.-A. and E.G.-C.; investigation, V.M., C.A.K., E.G.-C. and R.G.-A.; writing—original draft preparation, R.G.-A.; writing—review and editing, C.A.K., V.M. and E.G.-C.; supervision, V.M. and E.G.-C.; funding acquisition, V.M. and E.G.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work has been supported by the IAX ‘Inteligencia Artificial eXplicable’ grant from the Comunidad de Madrid through the call Universidad Politénica de Madrid-Jóvenes Investigadores 2022/2024, and the Proyecto colaborativo de integración de datos genómicos (CICLOGEN) (No. PI17/01561), funded by the Carlos III Health Institute from the Spanish National Plan for Scientific and Technical Research and Innovation 2017–2020 and the European Regional Development Fund (FEDER).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Asan, O.; Bayrak, A.E.; Choudhury, A. Artificial intelligence and human trust in healthcare: Focus on clinicians. J. Med. Internet Res. 2020, 22, e15154. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A. Explainable AI and Multi-Modal Causability in Medicine. i-com 2020, 19, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Adadi, A.; Berrada, M. Explainable AI for healthcare: From black box to interpretable models. In Embedded Systems and Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2020; pp. 327–337. [Google Scholar]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef]
- Joshi, G.; Jain, A.; Adhikari, S.; Garg, H.; Bhandari, M. FDA approved Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices: An updated 2022 landscape. medRxiv 2023. [Google Scholar] [CrossRef]
- Han, C.; Rundo, L.; Murao, K.; Nemoto, T.; Nakayama, H. Bridging the gap between AI and healthcare sides: Towards developing clinically relevant AI-powered diagnosis systems. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, León, Spain, 14–17 June 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 320–333. [Google Scholar]
- Diprose, W.; Buist, N.; Hua, N.; Thurier, Q.; Shand, G.; Robinson, R. Physician understanding, explainability, and trust in a hypothetical machine learning risk calculator. J. Am. Med. Inform. Assoc. 2020, 27, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Kerasidou, A. Ethics of artificial intelligence in global health: Explainability, algorithmic bias and trust. J. Oral Biol. Craniofac. Res. 2021, 11, 612–614. [Google Scholar] [CrossRef]
- Kung, T.H.; Cheatham, M.; Medenilla, A.; Sillos, C.; De Leon, L.; Elepaño, C. Performance of ChatGPT on USMLE: Potential for AI-assisted medical education using large language models. PLoS Digit. Health 2023, 2, e0000198. [Google Scholar] [CrossRef]
- Lee, P.; Goldberg, C.; Kohane, I. The AI Revolution in Medicine: GPT-4 and Beyond; Pearson: London, UK, 2023. [Google Scholar]
- Adadi, A.; Berrada, M. Peeking inside the black-box: A survey on explainable artificial intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Sethi, T.; Kalia, A.; Sharma, A.; Nagori, A. Interpretable artificial intelligence: Closing the adoption gap in healthcare. In Artificial Intelligence in Precision Health: From Concept to Applications; Elsevier: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Yoon, C.H.; Torrance, R.; Scheinerman, N. Machine learning in medicine: Should the pursuit of enhanced interpretability be abandoned? J. Med. Ethics 2021, 48, 581–585. [Google Scholar] [CrossRef]
- Musen, M.A. An overview of knowledge acquisition. Second. Gener. Expert Syst. 1993, 405–427. [Google Scholar] [CrossRef]
- Cruz, H.F.d.; Pfahringer, B.; Schneider, F.; Meyer, A.; Schapranow, M.P. External validation of a “black-box” clinical predictive model in nephrology: Can interpretability methods help illuminate performance differences? In Proceedings of the Conference on Artificial Intelligence in Medicine in Europe, Poznan, Poland, 26–29 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 191–201. [Google Scholar]
- Vilone, G.; Longo, L. Explainable artificial intelligence: A systematic review. arXiv 2020, arXiv:2006.00093. [Google Scholar]
- Quinn, T.P.; Senadeera, M.; Jacobs, S.; Coghlan, S.; Le, V. Trust and medical AI: The challenges we face and the expertise needed to overcome them. J. Am. Med. Inform. Assoc. 2021, 28, 890–894. [Google Scholar] [CrossRef] [PubMed]
- Subbaswamy, A.; Saria, S. From development to deployment: Dataset shift, causality, and shift-stable models in health AI. Biostatistics 2020, 21, 345–352. [Google Scholar] [CrossRef]
- Brady, A.; Neri, E. Artificial Intelligence in Radiology-Ethical Considerations. Diagnostics 2020, 10, 231. [Google Scholar] [CrossRef] [PubMed]
- Maojo, V. Domain-specific particularities of data mining: Lessons learned. In Proceedings of the International Symposium on Biological and Medical Data Analysis, Barcelona, Spain, 18–19 November 2004; Springer: Berlin/Heidelberg, Germany, 2004; pp. 235–242. [Google Scholar]
- Caruana, R.; Lou, Y.; Gehrke, J.; Koch, P.; Sturm, M.; Elhadad, N. Intelligible models for healthcare: Predicting pneumonia risk and hospital 30-day readmission. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Sydney, NSW, Australia, 10–13 August 2015; pp. 1721–1730. [Google Scholar]
- Gudivada, V.; Apon, A.; Ding, J. Data quality considerations for big data and machine learning: Going beyond data cleaning and transformations. Int. J. Adv. Softw. 2017, 10, 1–20. [Google Scholar]
- Chandler, C.; Foltz, P.W.; Elvevåg, B. Using machine learning in psychiatry: The need to establish a framework that nurtures trustworthiness. Schizophr. Bull. 2020, 46, 11–14. [Google Scholar] [CrossRef]
- Papadimitroulas, P.; Brocki, L.; Chung, N.C.; Marchadour, W.; Vermet, F.; Gaubert, L.; Eleftheriadis, V.; Plachouris, D.; Visvikis, D.; Kagadis, G.C.; et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys. Medica 2021, 83, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Goodman, B.; Flaxman, S. European Union regulations on algorithmic decision-making and a “right to explanation”. AI Mag. 2017, 38, 50–57. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.; Precise4Q Consortium. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef]
- Lipton, Z.C. The Mythos of Model Interpretability: In machine learning, the concept of interpretability is both important and slippery. Queue 2018, 16, 31–57. [Google Scholar] [CrossRef]
- Ghassemi, M.; Oakden-Rayner, L.; Beam, A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 2021, 3, e745–e750. [Google Scholar] [CrossRef] [PubMed]
- Antoniadi, A.; Du, Y.; Guendouz, Y.; Wei, L.; Mazo, C.; Becker, B.; Mooney, C. Current Challenges and Future Opportunities for XAI in Machine Learning-Based Clinical Decision Support Systems: A Systematic Review. Appl. Sci. 2021, 11, 5088. [Google Scholar] [CrossRef]
- Abdullah, T.; Zahid, M.; Ali, W. A Review of Interpretable ML in Healthcare: Taxonomy, Applications, Challenges, and Future Directions. Symmetry 2021, 13, 2439. [Google Scholar] [CrossRef]
- Payrovnaziri, S.; Chen, Z.; Rengifo-Moreno, P.; Miller, T.; Bian, J.; Chen, J.; Liu, X.; He, Z. Explainable artificial intelligence models using real-world electronic health record data: A systematic scoping review. J. Am. Med. Inform. Assoc. 2020, 27, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Gulum, M.; Trombley, C.; Kantardzic, M. A Review of Explainable Deep Learning Cancer Detection Models in Medical Imaging. Appl. Sci. 2021, 11, 4573. [Google Scholar] [CrossRef]
- Nazar, M.; Alam, M.; Yafi, E.; Su’ud, M. A Systematic Review of Human-Computer Interaction and Explainable Artificial Intelligence in Healthcare with Artificial Intelligence Techniques. IEEE Access 2021, 9, 153316–153348. [Google Scholar] [CrossRef]
- Salahuddin, Z.; Woodruff, H.; Chatterjee, A.; Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2022, 77, 29–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Weng, Y.; Lund, J. Applications of Explainable Artificial Intelligence in Diagnosis and Surgery. Diagnostics 2022, 12, 237. [Google Scholar] [CrossRef]
- Weiss, S.M.; Kulikowski, C.A.; Amarel, S.; Safir, A. A model-based method for computer-aided medical decision-making. Artif. Intell. 1978, 11, 145–172. [Google Scholar] [CrossRef]
- Peters, M.D.; Godfrey, C.; McInerney, P.; Munn, Z.; Tricco, A.C.; Khalil, H. Updated methodological guidance for the conduct of scoping reviews. JBI Evid. Synth. 2020, 18, 2119–2126. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Gite, S. A Brief Bibliometric Survey of Explainable AI in Medical Field. Libr. Philos. Pract. 2021, 2021, 1–27. [Google Scholar]
- Stiglic, G.; Kocbek, P.; Fijacko, N.; Zitnik, M.; Verbert, K.; Cilar, L. Interpretability of machine learning-based prediction models in healthcare. Wiley Interdiscip. Rev.-Data Min. Knowl. Discov. 2020, 10, e1379. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Vilone, G.; Longo, L. Classification of explainable artificial intelligence methods through their output formats. Mach. Learn. Knowl. Extr. 2021, 3, 615–661. [Google Scholar] [CrossRef]
- Arya, V.; Bellamy, R.K.; Chen, P.Y.; Dhurandhar, A.; Hind, M.; Hoffman, S.C.; Houde, S.; Liao, Q.V.; Luss, R.; Mojsilović, A.; et al. One explanation does not fit all: A toolkit and taxonomy of ai explainability techniques. arXiv 2019, arXiv:1909.03012. [Google Scholar]
- Biran, O.; Cotton, C. Explanation and justification in machine learning: A survey. In Proceedings of the IJCAI-17 Workshop on Explainable AI (XAI), Melbourne, Australia, 19–25 August 2017; Volume 8, pp. 8–13. [Google Scholar]
- Preece, A. Asking ‘Why’ in AI: Explainability of intelligent systems–perspectives and challenges. Intell. Syst. Account. Financ. Manag. 2018, 25, 63–72. [Google Scholar] [CrossRef]
- Vourgidis, I.; Mafuma, S.J.; Wilson, P.; Carter, J.; Cosma, G. Medical expert systems—A study of trust and acceptance by healthcare stakeholders. In Proceedings of the UK Workshop on Computational Intelligence, Nottingham, UK, 5–7 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 108–119. [Google Scholar]
- Shortliffe, E.H.; Davis, R.; Axline, S.G.; Buchanan, B.G.; Green, C.C.; Cohen, S.N. Computer-based consultations in clinical therapeutics: Explanation and rule acquisition capabilities of the MYCIN system. Comput. Biomed. Res. 1975, 8, 303–320. [Google Scholar] [CrossRef]
- Miller, R.A.; Pople, H.E., Jr.; Myers, J.D. Internist-I, an experimental computer-based diagnostic consultant for general internal medicine. In Computer-Assisted Medical Decision Making; Springer: Berlin/Heidelberg, Germany, 1985; pp. 139–158. [Google Scholar]
- Long, W.; Naimi, S.; Criscitiello, M.; Pauker, S.; Szolovits, P. An aid to physiological reasoning in the management of cardiovascular disease. In Proceedings of the 1984 Computers in Cardiology Conference, IEEE Computer Society, Long Beach, CA, USA, 14–18 February 1984; pp. 3–6. [Google Scholar]
- Clancey, W.J.; Shortliffe, E.H. Readings in Medical Artificial Intelligence: The First Decade; Addison-Wesley Longman Publishing Co., Inc.: Boston, MA, USA, 1984. [Google Scholar]
- Ford, K.M.; Coffey, J.W.; Cañas, A.; Andrews, E.J. Diagnosis and explanation by a nuclear cardiology expert system. Int. J. Expert Syst. 1996, 9, 4. [Google Scholar]
- Hogan, W.R.; Wagner, M.M. The use of an explanation algorithm in a clinical event monitor. In Proceedings of the AMIA Symposium. American Medical Informatics Association, Washington, DC, USA, 6–10 November 1999; p. 281. [Google Scholar]
- Darlington, K. Using explanation facilities in healthcare expert systems. In Proceedings of the HEALTHINF 2008: Proceedings of the First International Conference on Health Informatics, Funchal, Madeira, Portugal, 28–31 January 2008; Volume 1. [Google Scholar]
- Darlington, K.W. Designing for explanation in health care applications of expert systems. Sage Open 2011, 1, 2158244011408618. [Google Scholar] [CrossRef]
- Rennels, G.D.; Shortliffe, E.H.; Miller, P.L. Choice and explanation in medical management: A multiattribute model of artificial intelligence approaches. Med. Decis. Mak. 1987, 7, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Molino, G.; Console, L.; Torasso, P. Causal expert systems supporting medical decision making and medical education: Explanations based on simulated situations. In Proceedings of the Images of the Twenty-First Century, Annual International Engineering in Medicine and Biology Society; IEEE: Seattle, WA, USA, 1989; pp. 1827–1828. [Google Scholar]
- Ledley, R.S.; Lusted, L.B. Reasoning foundations of medical diagnosis: Symbolic logic, probability, and value theory aid our understanding of how physicians reason. Science 1959, 130, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Jacquez, J. The Diagnostic Process: Proceedings of a Conference Held at the University of Michigan; Malloy Lithographing, Inc.: Ann Arbor, MI, USA, 1963. [Google Scholar]
- Nelder, J.A.; Wedderburn, R.W. Generalized linear models. J. R. Stat. Soc. Ser. A 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Meacham, S.; Isaac, G.; Nauck, D.; Virginas, B. Towards explainable AI: Design and development for explanation of machine learning predictions for a patient readmittance medical application. In Proceedings of the Intelligent Computing-Proceedings of the Computing Conference, London, UK, 16–17 July 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 939–955. [Google Scholar]
- Banegas-Luna, A.J.; Peña-García, J.; Iftene, A.; Guadagni, F.; Ferroni, P.; Scarpato, N.; Zanzotto, F.M.; Bueno-Crespo, A.; Pérez-Sánchez, H. Towards the interpretability of machine learning predictions for medical applications targeting personalised therapies: A cancer case survey. Int. J. Mol. Sci. 2021, 22, 4394. [Google Scholar] [CrossRef] [PubMed]
- Karatekin, T.; Sancak, S.; Celik, G.; Topcuoglu, S.; Karatekin, G.; Kirci, P.; Okatan, A. Interpretable machine learning in healthcare through generalized additive model with pairwise interactions (GA2M): Predicting severe retinopathy of prematurity. In Proceedings of the 2019 International Conference on Deep Learning and Machine Learning in Emerging Applications (Deep-ML), Boca Raton, FL, USA, 26–28 August 2019; IEEE: New York City, NY, USA, 2019; pp. 61–66. [Google Scholar]
- Wang, H.; Huang, Z.; Zhang, D.; Arief, J.; Lyu, T.; Tian, J. Integrating co-clustering and interpretable machine learning for the prediction of intravenous immunoglobulin resistance in kawasaki disease. IEEE Access 2020, 8, 97064–97071. [Google Scholar] [CrossRef]
- Itani, S.; Rossignol, M.; Lecron, F.; Fortemps, P. Towards interpretable machine learning models for diagnosis aid: A case study on attention deficit/hyperactivity disorder. PLoS ONE 2019, 14, e0215720. [Google Scholar] [CrossRef]
- Brito-Sarracino, T.; dos Santos, M.R.; Antunes, E.F.; de Andrade Santos, I.B.; Kasmanas, J.C.; de Leon Ferreira, A.C.P. Explainable machine learning for breast cancer diagnosis. In Proceedings of the 2019 8th Brazilian Conference on Intelligent Systems (BRACIS), Salvador, Brazil, 15–18 October 2019; IEEE: New York City, NY, USA, 2019; pp. 681–686. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Mattogno, P.P.; Caccavella, V.M.; Giordano, M.; D’Alessandris, Q.G.; Chiloiro, S.; Tariciotti, L.; Olivi, A.; Lauretti, L. Interpretable Machine Learning–Based Prediction of Intraoperative Cerebrospinal Fluid Leakage in Endoscopic Transsphenoidal Pituitary Surgery: A Pilot Study. J. Neurol. Surg. Part Skull Base 2022, 83, 485–495. [Google Scholar] [CrossRef]
- Alsinglawi, B.; Alshari, O.; Alorjani, M.; Mubin, O.; Alnajjar, F.; Novoa, M.; Darwish, O. An explainable machine learning framework for lung cancer hospital length of stay prediction. Sci. Rep. 2022, 12, 607. [Google Scholar]
- El-Sappagh, S.; Alonso, J.M.; Islam, S.; Sultan, A.M.; Kwak, K.S. A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021, 11, 2660. [Google Scholar] [CrossRef] [PubMed]
- Pearl, J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference; Morgan Kaufmann: Cambridge, MA, USA, 1988. [Google Scholar]
- Chang, C.C.; Cheng, C.S. A Bayesian decision analysis with fuzzy interpretability for aging chronic disease. Int. J. Technol. Manag. 2007, 40, 176–191. [Google Scholar] [CrossRef]
- Casini, L.; McKay Illari, P.; Russo, F.; Williamson, J. Recursive Bayesian nets for prediction, explanation and control in cancer science. Theoria 2011, 26, 495–4548. [Google Scholar]
- Kyrimi, E.; Marsh, W. A progressive explanation of inference in ‘hybrid’ Bayesian networks for supporting clinical decision making. In Proceedings of the Conference on Probabilistic Graphical Models. PMLR, Lugano, Switzerland, 6 September 2016; pp. 275–286. [Google Scholar]
- Xie, W.; Ji, M.; Zhao, M.; Zhou, T.; Yang, F.; Qian, X.; Chow, C.Y.; Lam, K.Y.; Hao, T. Detecting symptom errors in neural machine translation of patient health information on depressive disorders: Developing interpretable bayesian machine learning classifiers. Front. Psychiatry 2021, 12, 771562. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Basak, M.; Han, M.M. Bayesian rule modeling for interpretable mortality classification of COVID-19 patients. Cmc-Comput. Mater. Continua 2021, 2827–2843. [Google Scholar] [CrossRef]
- Kyrimi, E.; Mossadegh, S.; Tai, N.; Marsh, W. An incremental explanation of inference in Bayesian networks for increasing model trustworthiness and supporting clinical decision making. Artif. Intell. Med. 2020, 103, 101812. [Google Scholar] [CrossRef] [PubMed]
- Kanda, E.; Epureanu, B.I.; Adachi, T.; Tsuruta, Y.; Kikuchi, K.; Kashihara, N.; Abe, M.; Masakane, I.; Nitta, K. Application of explainable ensemble artificial intelligence model to categorization of hemodialysis-patient and treatment using nationwide-real-world data in Japan. PLoS ONE 2020, 15, e0233491. [Google Scholar] [CrossRef]
- Chen, J.; Dai, X.; Yuan, Q.; Lu, C.; Huang, H. Towards interpretable clinical diagnosis with Bayesian network ensembles stacked on entity-aware CNNs. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020; pp. 3143–3153. [Google Scholar]
- Ahmed, Z.U.; Sun, K.; Shelly, M.; Mu, L. Explainable artificial intelligence (XAI) for exploring spatial variability of lung and bronchus cancer (LBC) mortality rates in the contiguous USA. Sci. Rep. 2021, 11, 24090. [Google Scholar] [CrossRef]
- Singh, R.K.; Pandey, R.; Babu, R.N. COVIDScreen: Explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Appl. 2021, 33, 8871–8892. [Google Scholar] [CrossRef]
- Yu, T.H.; Su, B.H.; Battalora, L.C.; Liu, S.; Tseng, Y.J. Ensemble modeling with machine learning and deep learning to provide interpretable generalized rules for classifying CNS drugs with high prediction power. Briefings Bioinform. 2022, 23, bbab377. [Google Scholar] [CrossRef]
- Peng, J.; Zou, K.; Zhou, M.; Teng, Y.; Zhu, X.; Zhang, F.; Xu, J. An explainable artificial intelligence framework for the deterioration risk prediction of hepatitis patients. J. Med. Syst. 2021, 45, 61. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jeon, E.T.; Yu, S.; Oh, K.; Kim, C.K.; Song, T.J.; Kim, Y.J.; Heo, S.H.; Park, K.Y.; Kim, J.M.; et al. Interpretable machine learning for early neurological deterioration prediction in atrial fibrillation-related stroke. Sci. Rep. 2021, 11, 20610. [Google Scholar]
- Pearl, J.; Mackenzie, D. The Book of Why: The New Science of Cause and Effect; Hachette Basic Books: New York City, NY, USA, 2018. [Google Scholar]
- Ren, Z.; Nguyen, T.T.; Nejdl, W. Prototype learning for interpretable respiratory sound analysis. In Proceedings of the ICASSP 2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 22–27 May 2022; pp. 9087–9091. [Google Scholar]
- Li, O.; Liu, H.; Chen, C.; Rudin, C. Deep learning for case-based reasoning through prototypes: A neural network that explains its predictions. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Wan, Y.; Zheng, Z.; Liu, R.; Zhu, Z.; Zhou, H.; Zhang, X.; Boumaraf, S. A Multi-Scale and Multi-Level Fusion Approach for Deep Learning-Based Liver Lesion Diagnosis in Magnetic Resonance Images with Visual Explanation. Life 2021, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, M.; Liu, H.; Yang, H.; Wang, H.; Lu, S.; Liang, T.; Li, X.; Xu, M.; Li, L.; et al. A hierarchical deep learning approach with transparency and interpretability based on small samples for glaucoma diagnosis. NPJ Digit. Med. 2021, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.; Zou, B.; Zhao, R.; Chen, Y.; He, Z.; Zhou, M. Clinical interpretable deep learning model for glaucoma diagnosis. IEEE J. Biomed. Health Inform. 2019, 24, 1405–1412. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Cconference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Magesh, P.R.; Myloth, R.D.; Tom, R.J. An explainable machine learning model for early detection of Parkinson’s disease using LIME on DaTSCAN imagery. Comput. Biol. Med. 2020, 126, 104041. [Google Scholar] [CrossRef]
- Tan, S.; Caruana, R.; Hooker, G.; Lou, Y. Distill-and-compare: Auditing black-box models using transparent model distillation. In Proceedings of the 2018 AAAI/ACM Conference on AI, Ethics, and Society, New Orleans, LA, USA, 2–3 February 2018; pp. 303–310. [Google Scholar]
- Che, Z.; Purushotham, S.; Khemani, R.; Liu, Y. Interpretable deep models for ICU outcome prediction. In Proceedings of the AMIA annual symposium proceedings. American Medical Informatics Association, Chicago, IL, USA, 16 November 2016; Volume 2016, p. 371. [Google Scholar]
- Krishnan, R.; Sivakumar, G.; Bhattacharya, P. A search technique for rule extraction from trained neural networks. Pattern Recognit. Lett. 1999, 20, 273–280. [Google Scholar] [CrossRef]
- Etchells, T.A.; Lisboa, P.J. Orthogonal search-based rule extraction (OSRE) for trained neural networks: A practical and efficient approach. IEEE Trans. Neural Netw. 2006, 17, 374–384. [Google Scholar] [CrossRef]
- Barakat, N.; Diederich, J. Eclectic rule-extraction from support vector machines. Int. J. Comput. Intell. 2005, 2, 59–62. [Google Scholar]
- Fisher, R.A. The logic of inductive inference. J. R. Stat. Soc. 1935, 98, 39–82. [Google Scholar] [CrossRef]
- Kaminuma, T.; Takekawa, T.; Watanabe, S. Reduction of clustering problem to pattern recognition. Pattern Recognit. 1969, 1, 195–205. [Google Scholar] [CrossRef]
- Kulikowski, C.A. Pattern recognition approach to medical diagnosis. IEEE Trans. Syst. Sci. Cybern. 1970, 6, 173–178. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4768–4777. [Google Scholar]
- Weis, C.; Cuénod, A.; Rieck, B.; Dubuis, O.; Graf, S.; Lang, C.; Oberle, M.; Brackmann, M.; Søgaard, K.K.; Osthoff, M.; et al. Direct antimicrobial resistance prediction from clinical MALDI-TOF mass spectra using machine learning. Nat. Med. 2022, 28, 164–174. [Google Scholar] [CrossRef]
- Saltelli, A. Sensitivity analysis for importance assessment. Risk Anal. 2002, 22, 579–590. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef]
- Montavon, G.; Lapuschkin, S.; Binder, A.; Samek, W.; Müller, K.R. Explaining nonlinear classification decisions with deep taylor decomposition. Pattern Recognit. 2017, 65, 211–222. [Google Scholar] [CrossRef]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 3145–3153. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Goldstein, A.; Kapelner, A.; Bleich, J.; Pitkin, E. Peeking inside the black box: Visualizing statistical learning with plots of individual conditional expectation. J. Comput. Graph. Stat. 2015, 24, 44–65. [Google Scholar] [CrossRef]
- Kim, B.; Khanna, R.; Koyejo, O.O. Examples are not enough, learn to criticize! criticism for interpretability. Adv. Neural Inf. Process. Syst. 2016, 29. [Google Scholar]
- Wachter, S.; Mittelstadt, B.; Russell, C. Counterfactual explanations without opening the black box: Automated decisions and the GDPR. Harv. JL Tech. 2017, 31, 841. [Google Scholar] [CrossRef]
- Doshi-Velez, F.; Kim, B. Considerations for Evaluation and Generalization in Interpretable Machine Learning. In Explainable and Interpretable Models in Computer Vision and Machine Learning. The Springer Series on Challenges in Machine Learning; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Markus, A.; Kors, J.; Rijnbeek, P. The role of explainability in creating trustworthy artificial intelligence for health care: A comprehensive survey of the terminology, design choices, and evaluation strategies. J. Biomed. Inform. 2021, 113, 103655. [Google Scholar] [CrossRef] [PubMed]
- Kaur, D.; Uslu, S.; Durresi, A.; Badve, S.; Dundar, M. Trustworthy Explainability Acceptance: A New Metric to Measure the Trustworthiness of Interpretable AI Medical Diagnostic Systems. Complex Intell. Softw. Intensive Syst. 2021, 278. [Google Scholar] [CrossRef]
- Kolyshkina, I.; Simoff, S. Interpretability of Machine Learning Solutions in Public Healthcare: The CRISP-ML Approach. Front. Big Data 2021, 4, 660206. [Google Scholar] [CrossRef] [PubMed]
- ElShawi, R.; Sherif, Y.; Al-Mallah, M.; Sakr, S. Interpretability in healthcare: A comparative study of local machine learning interpretability techniques. Comput. Intell. 2021, 37, 1633–1650. [Google Scholar] [CrossRef]
- Honegger, M.R. Shedding light on black box machine learning algorithms: Development of an axiomatic framework to assess the quality of methods that explain individual predictions. arXiv 2018, arXiv:1808.05054. [Google Scholar]
- Muddamsetty, S.M.; Jahromi, M.N.; Moeslund, T.B. Expert level evaluations for explainable AI (XAI) methods in the medical domain. In Proceedings of the International Conference on Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 35–46. [Google Scholar]
- de Souza, L.; Mendel, R.; Strasser, S.; Ebigbo, A.; Probst, A.; Messmann, H.; Papa, J.; Palm, C. Convolutional Neural Networks for the evaluation of cancer in Barrett’s esophagus: Explainable AI to lighten up the black-box. Comput. Biol. Med. 2021, 135, 104578. [Google Scholar] [CrossRef] [PubMed]
- Kumarakulasinghe, N.B.; Blomberg, T.; Liu, J.; Leao, A.S.; Papapetrou, P. Evaluating local interpretable model-agnostic explanations on clinical machine learning classification models. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020; pp. 7–12. [Google Scholar]
- Singh, A.; Balaji, J.; Rasheed, M.; Jayakumar, V.; Raman, R.; Lakshminarayanan, V. Evaluation of Explainable Deep Learning Methods for Ophthalmic Diagnosis. Clin. Ophthalmol. 2021, 15, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Deperlioglu, O.; Kose, U.; Gupta, D.; Khanna, A.; Giampaolo, F.; Fortino, G. Explainable framework for Glaucoma diagnosis by image processing and convolutional neural network synergy: Analysis with doctor evaluation. Future Gener. Comput. Syst. 2022, 129, 152–169. [Google Scholar] [CrossRef]
- Kulikowski, C.A. Pandemics: Historically slow “learning curve” leading to biomedical informatics and vaccine breakthroughs. Yearb. Med. Inform. 2021, 30, 290–301. [Google Scholar] [CrossRef]
- Došilović, F.K.; Brčić, M.; Hlupić, N. Explainable artificial intelligence: A survey. In Proceedings of the 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 21–25 May 2018; pp. 210–215. [Google Scholar]
- Durán, J.M.; Jongsma, K.R. Who is afraid of black box algorithms? On the epistemological and ethical basis of trust in medical AI. J. Med. Ethics 2021, 47, 329–335. [Google Scholar] [CrossRef]
- McCoy, L.G.; Brenna, C.T.; Chen, S.S.; Vold, K.; Das, S. Believing in black boxes: Machine learning for healthcare does not need explainability to be evidence-based. J. Clin. Epidemiol. 2022, 142, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Véliz, C.; Prunkl, C.; Phillips-Brown, M.; Lechterman, T.M. We might be afraid of black-box algorithms. J. Med. Ethics 2021, 47, 339–340. [Google Scholar] [CrossRef]
- Reimer, U.; Maier, E.; Tödtli, B. Going Beyond Explainability in Medical AI Systems. In Proceedings of the Modellierung (Companion), Vienna, Austria, 19–21 February 2020; pp. 185–191. [Google Scholar]
- Reimer, U.; Tödtli, B.; Maier, E. How to Induce Trust in Medical AI Systems. In Proceedings of the International Conference on Conceptual Modeling, Hyderabad, India, 7–20 October 2020; pp. 5–14. [Google Scholar]
- Wang, F.; Kaushal, R.; Khullar, D. Should health care demand interpretable artificial intelligence or accept “black box” medicine? Ann. Intern. Med. 2020, 172, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Babic, B.; Gerke, S.; Evgeniou, T.; Cohen, I.G. Beware explanations from AI in health care. Science 2021, 373, 284–286. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Muller, H. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev.-Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef]
- Petch, J.; Di, S.; Nelson, W. Opening the black box: The promise and limitations of explainable machine learning in cardiology. Can. J. Cardiol. 2021, 38, 204–213. [Google Scholar] [CrossRef]
- Tjoa, E.; Guan, C. A Survey on Explainable Artificial Intelligence (XAI): Toward Medical XAI. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 4793–4813. [Google Scholar] [CrossRef]
- Herman, B. The promise and peril of human evaluation for model interpretability. arXiv 2017, arXiv:1711.07414. [Google Scholar]
- Gilpin, L.H.; Bau, D.; Yuan, B.Z.; Bajwa, A.; Specter, M.; Kagal, L. Explaining explanations: An overview of interpretability of machine learning. In Proceedings of the 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy, 1–3 October 2018; pp. 80–89. [Google Scholar]
- London, A. Artificial Intelligence and Black-Box Medical Decisions: Accuracy versus Explainability. Hastings Cent. Rep. 2019, 49, 15–21. [Google Scholar] [CrossRef]
- Huang, S.C.; Pareek, A.; Seyyedi, S.; Banerjee, I.; Lungren, M.P. Fusion of medical imaging and electronic health records using deep learning: A systematic review and implementation guidelines. NPJ Digit. Med. 2020, 3, 136. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, A.; Dehmer, M.; Emmert-Streib, F.; Cucchiara, R.; Augenstein, I.; Del Ser, J.; Samek, W.; Jurisica, I.; Diaz-Rodriguez, N. Information fusion as an integrative cross-cutting enabler to achieve robust, explainable, and trustworthy medical artificial intelligence. Inf. Fusion 2022, 79, 263–278. [Google Scholar] [CrossRef]
- Kamal, M.S.; Northcote, A.; Chowdhury, L.; Dey, N.; Crespo, R.G.; Herrera-Viedma, E. Alzheimer’s patient analysis using image and gene expression data and explainable-ai to present associated genes. IEEE Trans. Instrum. Meas. 2021, 70, 1–7. [Google Scholar] [CrossRef]
- Larasati, R.; De Liddo, A.; Motta, E. AI Healthcare System Interface: Explanation Design for Non-Expert User Trust. In Proceedings of the ACMIUI-WS 2021: Joint Proceedings of the ACM IUI 2021 Workshops, CEUR Workshop Proceedings, Online, 13–17 April 2021; Volume 2903. [Google Scholar]
- Barda, A.J.; Horvat, C.M.; Hochheiser, H. A qualitative research framework for the design of user-centered displays of explanations for machine learning model predictions in healthcare. BMC Med. Inform. Decis. Mak. 2020, 20, 257. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, T.; Lee, H.; Byun, S. A Clinical Decision Support System for Sleep Staging Tasks with Explanations From Artificial Intelligence: User-Centered Design and Evaluation Study. J. Med. Internet Res. 2022, 24, e28659. [Google Scholar] [CrossRef]
- Cutillo, C.; Sharma, K.; Foschini, L.; Kundu, S.; Mackintosh, M.; Mandl, K.; Beck, T.; Collier, E.; Colvis, C.; Gersing, K.; et al. Machine intelligence in healthcare-perspectives on trustworthiness, explainability, usability, and transparency. NPJ Digit. Med. 2020, 3, 47. [Google Scholar] [CrossRef]
- Sariyar, M.; Holm, J. Medical Informatics in a Tension Between Black-Box AI and Trust. Stud. Health Technol. Inform. 2022, 289, 41–44. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).