1. Introduction
By 2019, 523 million people have had a cardiovascular disease, and 18.6 million related deaths have been reported [
1]. High blood pressure is a direct cause of death from cardiovascular disease (CVD) [
2,
3]. Therefore, accurate blood pressure (BP) measurements are necessary for diagnosing hypertension. BP monitoring has become vital for people with CVD, particularly elderly people who are living alone. Rapid changes in BP in these patients can indicate an underlying severe illness. Furthermore, BP varies owing to intrinsic physiological changes for various reasons, such as food intake, environmental temperature, exercise, disease, and stress. Thus, the precision and uncertainty of BP measurements induced by physiological parameters [
4] have been a constant concern for clinicians and practitioners [
5,
6,
7]. However, using BP monitoring devices for estimating BP uncertainty is currently impossible. This necessitates a standard protocol for confidence intervals (CIs), representing the uncertainty of BP monitors [
5,
8,
9]. Most BP monitors offer only single-point estimates without CIs [
5,
8,
9]. Therefore, patients, nurses, and physicians may be unable to differentiate intrinsic variations owing to the estimate’s statistical variation and physiological processes [
7,
8,
9].
Machine learning (ML) algorithms are commonly used for BP estimation [
5,
9,
10,
11,
12]. ML algorithms, including multiple linear regression (MLR) [
13,
14], artificial neural networks (ANNs) [
14,
15,
16], and support vector machine (SVM) [
17,
18,
19] have been utilized for estimating BP [
14,
20,
21,
22]. Wang et al. [
15] introduced a BP estimation method using photoplethysmography (PPG) signals using novel ANNs. Nandi et al. [
23] proposed a new long short-term memory (LSTM) and convolutional neural network using cuffless BP estimation based on PPG and electrocardiogram (ECG) signals. Multichannel PPG was introduced using an SVM-ensemble-based continuous BP estimation in [
19]. Qiu et al. [
14] proposed a new method for estimating BP using a window-function-based piecewise neural network. This study evaluated a random-forest-based regression network, three-layer ANN-based regression network, and SVM model using PPG signals. Many studies on PPG-signal-based cuffless BP estimation have been conducted [
13,
15,
21,
22,
23]. By contrast, studies on CIs estimation are limited. However, there have been studies on the uncertainty of estimating a few BP data based on conventional oscilloscope BP measurements [
5,
6,
7,
8,
9]. The two most used methods for cuffless BP estimation were obtained using the extracted features and pulse transit time (PTT) from PPG signal pulses [
14,
24,
25,
26]. The feature extraction method using PTT effectively estimates BP because PTT is closely correlated with BP [
27]. According to this principle, arterial pressure can be determined by measuring pulse wave velocity (PWV) at pulse wave speed. This is because changes in PTT correspond to changes in PWV at a fixed distance and indicates a change in BP [
26,
28,
29,
30].
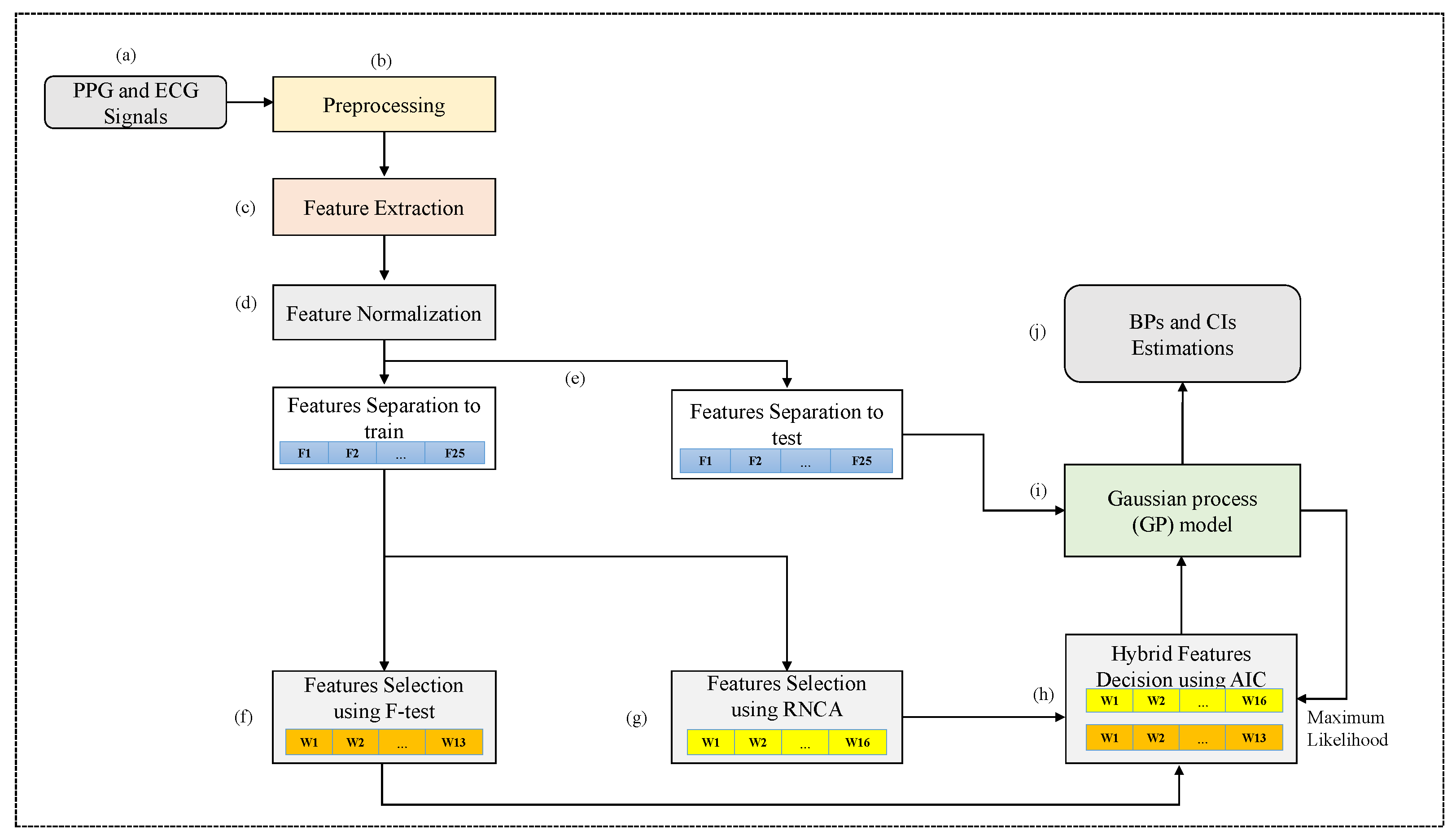
Accordingly, this study proposed a new methodology for simultaneously estimating BP and CIs using cuffless BP measurement through a hybrid feature selection and decision based on the Gaussian process (GP). This study aimed to estimate BP and CIs, representing the uncertainty of cuffless BP estimation. Moreover, the GP can directly generate uncertainty estimates [
31,
32,
33], such as providing a distribution of estimates rather than a single value. Another advantage of GP algorithms is that, similar to other kernel methods, they can be optimized precisely for given hyperparameter values [
34]. Therefore, they perform well owing to the well-optimized parameter values, particularly with limited datasets [
34]. However, the CIs automatically calculated by the GP algorithm are too wide to effectively represent the uncertainty. Therefore, CIs are estimated by applying the bootstrap method [
8] using the results of the GP algorithm [
31] to represent the uncertainty in the cuffless BP estimation.
The proposed hybrid feature selection and decision based on the GP algorithm can provide a means for distinguishing between estimation errors (statistical variance of estimates) and changes in estimates owing to physiological variability [
5,
7]. This study obtained CIs using a bootstrap algorithm [
10,
35] to determine the uncertainty (physiological variability) in cuffless BP estimation. Specifying CIs for cuffless measurements is beneficial because CIs measurement are necessary for estimating BP. If the BP measurement’s CIs are too broad, healthcare workers may misjudge a patient’s health status. Therefore, establishing CIs based on accurate BP estimates allows for more accurate and faster meaningful determinations of BP measurement CIs [
9]. However, studies on determining the uncertainty of physiological measurements [
4] using oscillometric BP signals [
5,
6,
7,
8] are limited. Therefore, a new method for evaluating and representing the uncertainty in BP measurement should be developed by providing an estimated range for cuffless BP measurements. Consequently, repeatable, irregular, and broad CIs based on aggregated statistical data can provide patients, clinicians, and families a warning system for BPs outside the normal range [
5,
9,
10]. Another problem in improving the performance of ML algorithms is the choice of features to replace the original ones and using them as input data. Feature selection is an essential part of the learning algorithm’s performance; it selects a subset of features with higher weights for the response variable and eliminates duplicate features [
36,
37]. Thus, the proposed methodology uses a hybrid F-test [
38] and robust neighbor component analysis (RNCA) [
39] to select weighted features from the original features. The best feature set is selected using Akaike’s information criterion (AIC) [
40] based on the maximum likelihood from the GP algorithm [
31]. First, the F-test is used to acquire weighted features to compute a feature’s within- and between-group variance ratios [
38]. Second, the weighted features are obtained using RNCA [
39], which selects the high-weighted features from the original features. The importance of the BP estimation performance can be corrected by repeating the estimation experiment several times. Therefore, this study proposes an adaptive AIC of automatically calibrated likelihoods based on the GP algorithm to determine the best feature subset as a model selection problem, yielding an excellent performance with negligible computational overhead after calibration.
This study provides uncertainty for cuffless BP measurements and introduces a method for reducing the error of BP estimates based on PPG and ECG signals. As previously mentioned, the proposed study estimates the exact BPs and CIs concurrently, representing the uncertainty of cuffless BP estimation. To the best of the authors’ knowledge, this study is the first to propose a GP-based feature selection and decision process (GFSDP) algorithm to simultaneously estimate cuffless BPs and CIs. Although some CI studies on conventional oscillometric BP estimation methods have been performed [
5,
6,
7,
8], studies on CI estimation in cuffless BP measurements are limited, as summarized in
Table 1. The contributions of the study to BP and CI estimations are as follows:
CIs are estimated using a bootstrap based on the GP algorithm to express uncertainty in cuffless BP estimation.
The proposed methodology uses a hybrid F-test and RNCA to select the weighted features among the original features.
An adaptive AIC of automatically calibrated likelihoods is proposed based on the GP algorithm to determine the best feature subset as a model selection problem.
5. Discussion
This study is the first to propose a GFSDP algorithm to concurrently estimate cuffless BPs and CIs.
Table 4 lists the high-scoring features selected using the F-test and RNCA algorithms [
38,
39]. The ranked features relied on the selection method. They also changed according to the SBP or DBP target variables. Therefore, the final decision process using the AIC algorithm [
40] for the proposed hybrid-feature-selected sets was more significant for improving the performance of ML than using a fixed subset.
Table 6 confirms that the proposed GFSDP algorithm was more complex than the GP algorithm in computational complexity. This indicates that computing resources were consumed during the HFD process to weigh the features and finalize the assigned weights. Nevertheless, in terms of estimation accuracy, the proposed GFSDP algorithm exhibited the lowest MAE for the SBP (7.66 mmHg) and DBP (5.47 mmHg) compared with those of the ANN, MLR, SVM, LSTM, and GP algorithms. In particular, compared with the conventional GP algorithm, the accuracy of the SBP and DBP estimation was 30.94% and 22.49%, respectively, confirming that the hybrid feature selection and decision effectively improved the accuracy of estimating SBP and DBP. In addition, the proposed GFSDP algorithm exhibited an improved performance of 48.43% and 32.18% for the SBP and DBP estimations, respectively, compared with the SVM algorithm. The SDEs of the MAEs in all algorithms showed stable values, as shown in
Table 7.
The proposed GFSDP algorithm exhibited a slight performance loss compared with the BHS protocol [
46], as shown in
Table 8. The MAEs were 49.49% (≤5 mmHg), 73.01% (≤10 mmHg), and 84.97% (≤15 mmHg) for the SBP and 60.26% (≤5 mmHg), 85.39% (≤10 mmHg), and 93.74% (≤15 mmHg) for the DBP, as shown in
Table 8. Therefore, the proposed GFSDP obtained classes C and B for evaluating SBP and DBP. Furthermore, the proposed GFSDP algorithm was more accurate than conventional algorithms for cuffless BP estimation.
The SDE of the ME was also evaluated according to the AAMI protocol [
47]. The proposed GFSDP algorithm for the SBP (10.83 mmHg) and DBP (8.31 mmHg) had a lower SDE of the ME than conventional algorithms; however, all algorithms failed to meet the AAMI criteria, as shown in
Table 9. Thus, the AAMI standards are stringent.
Figure 5 shows the SDE results of the ME of the proposed GFSDP and SVM algorithms. These results indicate that the results of the SVM were slightly more spread out than those of the proposed algorithm. This further proves the superiority of the proposed algorithm, in addition to its low MAE. Moreover, the proposed GFSDP algorithm is more favorable because it correctly estimated errors based on the weighted feature subset. Therefore, the GFSDP process accurately estimated cuffless BP values.
Cuffless BP-measuring devices typically provide single-point predictions without CIs. Therefore, predicting CIs for cuffless BP measurements improves reliability. Thus, this study predicted CIs using the GP and GFSDP algorithms to express uncertainties in cuffless BP estimation. This study extracted four to eight PPG segment waveforms from each patient’s record. A small sample size of each patient affected the accuracy of the bootstrap CI estimation. The results confirmed that the CIs obtained from the proposed
for SBP and DBP were also wide. The CIs derived from the conventional algorithm using the Student’s t-distribution (ST) were broader than those obtained from the proposed method for both SBP and DBP, as shown in
Table 10. The CI estimates based on the ST distribution are adequate when the sample size from each patient is large (at least 30) [
8]. The results confirmed that the proposed GFSDP algorithm was more accurate than the conventional algorithms for cuffless BP and CI estimations. As previously mentioned, GP algorithms predict distributions rather than single-response values. Therefore, CIs were provided along with the predicted responses to represent the uncertainty. However, even with the GP algorithm, few samples for each patient resulted in wide CIs [
8].