Abstract
The precise control of potato diseases is an urgent demand in smart agriculture, with one of the key aspects being the accurate identification and segmentation of potato leaf diseases. Some disease spots on potato leaves are relatively small, and to address issues such as information loss and low segmentation accuracy in the process of potato leaf disease image segmentation, a novel approach based on an improved UNet network model is proposed. Firstly, the incorporation of ResNet50 as the backbone network is introduced to deepen the network structure, effectively addressing problems like gradient vanishing and degradation. Secondly, the unique characteristics of the UNet network are fully utilized, using UNet as the decoder to ingeniously integrate the characteristics of potatoes with the network. Finally, to better enable the network to learn disease spot features, the SE (squeeze and excitation) attention mechanism is introduced on top of ResNet50, further optimizing the network structure. This design allows the network to selectively emphasize useful information features and suppress irrelevant ones during the learning process, significantly enhancing the accuracy of potato disease segmentation and identification. The experimental results demonstrate that compared with the traditional UNet algorithm, the improved RS-UNet network model achieves values of 79.8% and 88.86% for the MIoU and Dice metrics, respectively, which represent improvements of 8.96% and 6.33% over UNet. These results provide strong evidence for the outstanding performance and generalization ability of the RS-UNet model in potato leaf disease spot segmentation, as well as its practical application value in the task of potato leaf disease segmentation.
1. Introduction
Potatoes are a crucial crop in many countries, profoundly impacting both local and global agricultural economies. They are a significant source of income for farmers and provide ample opportunities for the food processing and trading industries [1]. Consequently, the stable development of the potato industry is paramount for enhancing farmer incomes and promoting socio-economic growth. Presently, the detection of potato leaf diseases primarily relies on visual inspection by farmers or experts, identifying disease symptoms by observing the potato plant’s leaves [2]. While this method offers convenience in field operations, it requires professional knowledge and experience for accurate disease identification [3]. If leaf diseases are not promptly detected and managed, adverse effects on potato yield and quality can occur, harming farmers’ economic interests [4,5]. Deep learning models, trained on extensive labeled samples, can achieve automated, efficient, and precise detection of leaf diseases [6]. The application of such technology aids in the early detection of diseases, allowing farmers to take timely and appropriate preventive measures, safeguarding crop health, and improving yields and agricultural productivity.
Numerous researchers have extensively studied plant leaf segmentation and identification in recent years. J. Sund et al. [7] proposed a leaf segmentation network, ULeaf-Net, based on a U-shaped symmetric encoder–decoder architecture. This network employs cross-layer feature fusion to enhance segmentation results. Zhida Jia et al. [8] introduced a UNet persimmon leaf disease image segmentation based on a self-attention mechanism and discernible convolution. They used UNet as the primary network, applied variable convolution networks for downsampling to enhance feature extraction, and utilized self-attention mechanisms to acquire more spatial and contextual information, achieving an MIOU value of 83.58%. Agarwal M [9] and his colleagues suggested differential evolution for segmenting diseased regions on leaves, aiming to reduce training time and space. Chen Congping [10] and team put forward the application of Deeplab v3+ for segmenting potato leaves and then combined texture features with VGG16 for feature extraction to build a convolutional neural network for disease identification. Chen Peng et al. [11] presented a wheat Fusarium head blight semantic segmentation network model, UNetA, that integrated convolutional neural networks and attention mechanisms. They used a weighted cross-entropy loss function to measure the disparity between predicted and actual values while mitigating sample imbalance issues, achieving an MIoU value of 83.90%. Mao Wanjing et al. [12] introduced an improved U-Net strawberry semantic segmentation model based on the attention mechanism. They incorporated a CNN–Transformer hybrid structure in the encoder and replaced the traditional upsampling with a dual upsampling module in the decoder, achieving a segmentation accuracy of 92.56%. However, there were challenges with high computational demands and extended training times. U-Net, which combines low-detail and high-semantic information, has impressive performance in biomedical image segmentation [13,14]. Deep residual units make deep networks easy to train, and in-network skip connections assist in transmitting information without degradation [15,16]. Inspired by the advantages of U-Net and MultiResUNet [17], an improved UNet (RS-Net) was developed for the image segmentation of diseased potato leaves.
Despite the impressive performance of various advanced models in plant disease recognition research, significant challenges persist. Existing models exhibit diversity in segmentation performance when dealing with different datasets, tiny disease spot features, fuzzy boundaries, and morphological variations. To address these challenges, this paper proposes a potato leaf disease segmentation method based on an improved UNet network. It employs ResNet50 as the feature extraction front-end and incorporates the SE attention mechanism, resulting in SE-ResNet, which serves the purpose of feature extraction, to highlight potato leaf disease spot information while suppressing redundant information. Simultaneously, UNet is employed as the back-end network. The experimental results demonstrate that the proposed model meets the requirements in terms of metrics such as Dice and MIoU, achieving favorable segmentation outcomes for potato leaf disease spots.
2. Materials and Methods
2.1. Data Collection and Expansion
In this research experiment, the resources from the “PlantVillage dataset” were utilized, which contains samples of images of disease on potato leaves, including select images of potato leaf diseases, as shown in Figure 1. Owing to the uneven distribution of data labels, three types of potato leaf disease samples were chosen: late blight, early blight, and anthracnose. A total of 486 relevant dataset images were selected.

Figure 1.
Partial Potato Leaf Disease Dataset.
Considering the relatively limited dataset available, a series of image preprocessing techniques were employed to enhance the useful information within the images. This encompassed operations such as Gaussian blur, brightness adjustments, contrast enhancement, image rotation, and flipping, with the effects of some of these operations shown in Figure 2. The dataset for potato diseases is comparatively smaller than larger datasets, making it susceptible to overfitting during training. To address this issue, data augmentation was applied to increase the dataset size and mitigate overfitting. Data augmentation not only enhances the characteristics of disease spots in the sample images, but also improves the model’s ability to learn these spots, thereby enhancing model accuracy. Consequently, the original set of 486 images of late blight, early blight, and anthracnose on potato leaves underwent augmentation, resulting in a total of 2000 augmented images, with 1600 designated for the training set and 400 for the validation set.

Figure 2.
Data Augmentation Examples.
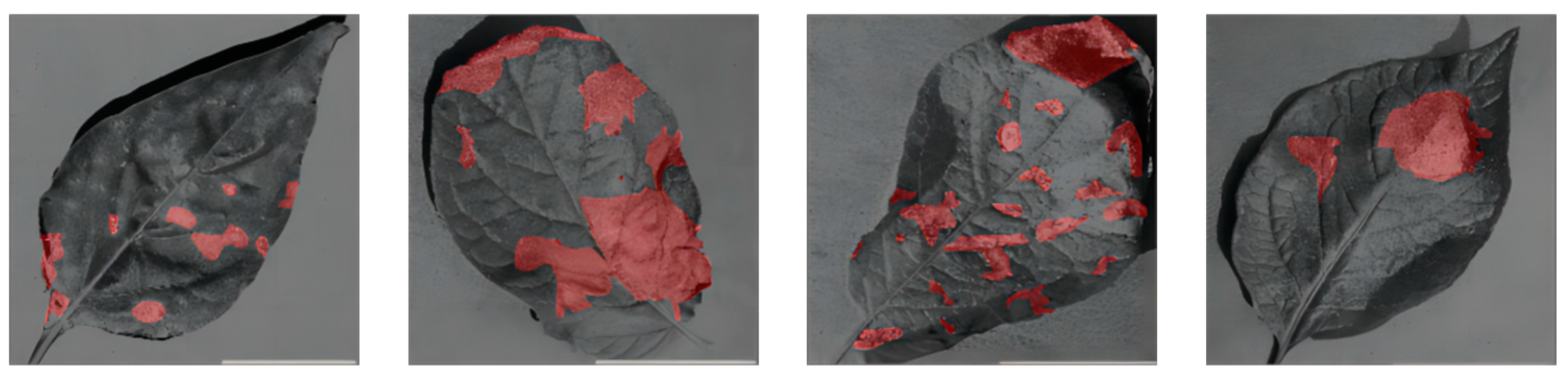
Finally, the images were normalized to a resolution of 256 × 256 pixels, and the disease spots on potato leaves in these images were annotated and analyzed. The “Labelme” tool was utilized for marking the disease spot regions on the potato leaves in the images. These annotation details were stored in a format known as JSON. Subsequently, the “Labelme_to_dataset” command was employed to convert these label data into binary PNG images for further processing. Figure 3 presents examples of annotated images, with the disease spot regions marked in red, while black represents the background.

Figure 3.
Annotated Image Examples.
2.2. Methods
2.2.1. UNet Network Model
The UNet architecture has wide applications in fields such as medical image segmentation and natural image segmentation. Its primary objective is a pixel-wise classification of input images to identify distinct objects or regions. It comprises two crucial components, the encoder and decoder, connected through skip connections, as depicted in Figure 4. This connection mechanism enables the decoder to effectively utilize feature information from different levels of the encoder, thereby enhancing segmentation accuracy [18]. This mechanism’s role is to merge shallow and deep-level features, enabling the network to simultaneously learn local and global image information. UNet’s design enables outstanding performance, especially in domains with limited data, such as medical imaging [19]. In each downsampling step, the feature dimensions double. Conversely, in the expansion or decoder path, upsampling is employed to reduce the feature dimensions and restore the image size to that of the original input image. Each upsampling step halves the feature dimensions. To ensure the retention of critical details, connections to features from the downsampling path are considered during the upsampling process. Given the limitations of the original UNet architecture concerning the potato leaf disease spot image dataset and the diverse and small morphology of potato leaf disease spots, there is a risk of spatial information loss and incomplete recovery during upsampling. This can result in feature extraction that is blurry and insensitive to edge details in the leaf disease spot regions. Hence, improvements were made to UNet to fully leverage its capabilities and effectively combine shallow and deep-level features [20].
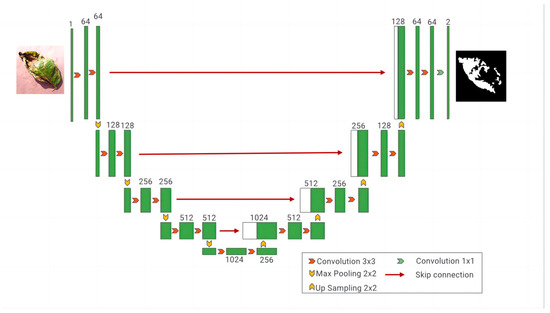
Figure 4.
The UNet network model.
2.2.2. Residual Network
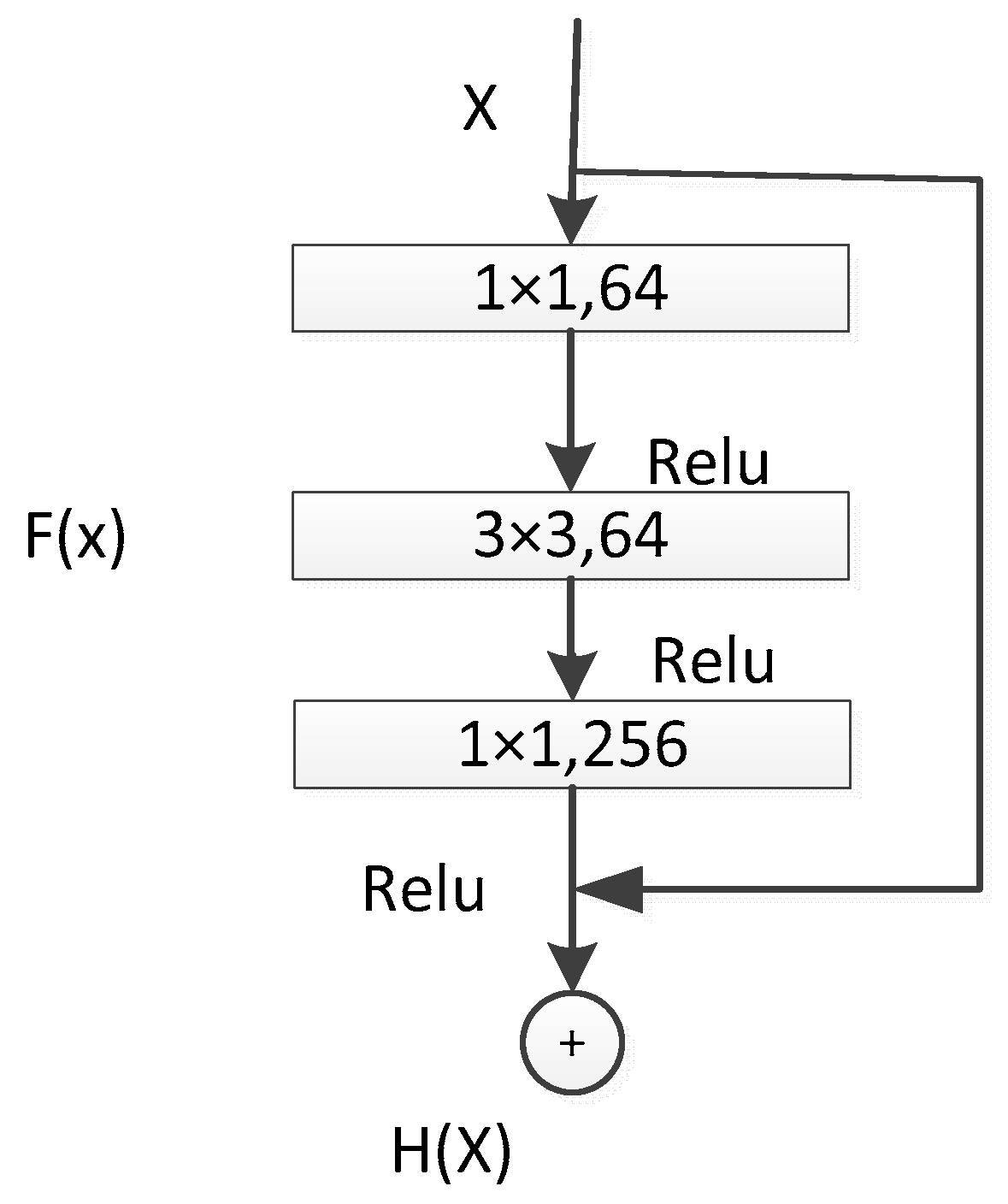
The performance of the shallow layers of the UNet network may degrade. To address this issue, the concept of residual networks was introduced, which consists of identity mapping and residual blocks. In this context, the residual block used comprised three consecutive convolutional layers (1 × 1, 3 × 3, 1 × 1 convolutions), as illustrated in Figure 5. Unlike traditional sequential network structures, residual units introduce skip connections that add the input and output, compensating for feature information loss during convolution [21,22]. The introduction of residual units makes the feature mapping more sensitive to output changes, facilitating easier gradient training. Even when the residual is zero, indicating that the stacked layers only achieve identity mapping, the network performance does not degrade.
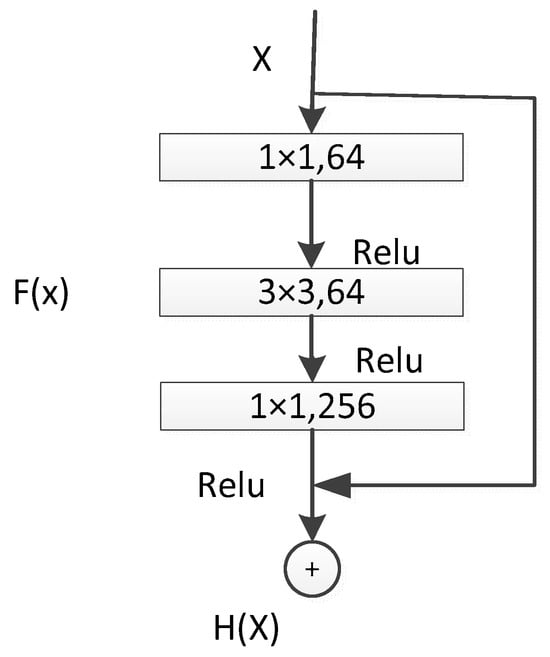
Figure 5.
ResNet50 Residual Unit.
In the deep ResNet50 + UNet network, a deeper ResNet50 network was chosen as the front-end feature-extraction component [23]. A 1 × 1 convolutional layer was added for dimensionality reduction before computation, followed by 3 × 3 convolution, and then 1 × 1 convolution for restoration, reducing computational complexity. This network structure effectively utilizes feature information obtained through downsampling and introduces residual networks to enhance feature extraction from potato leaf disease spot images. This combination has two main advantages: first, the introduction of residual units simplifies the entire network training process; second, the skip connections within residual units and between high and low layers promote information propagation within the network, further improving the performance of neural networks with fewer parameters in semantic segmentation tasks [24].
2.2.3. SE Attention Mechanism
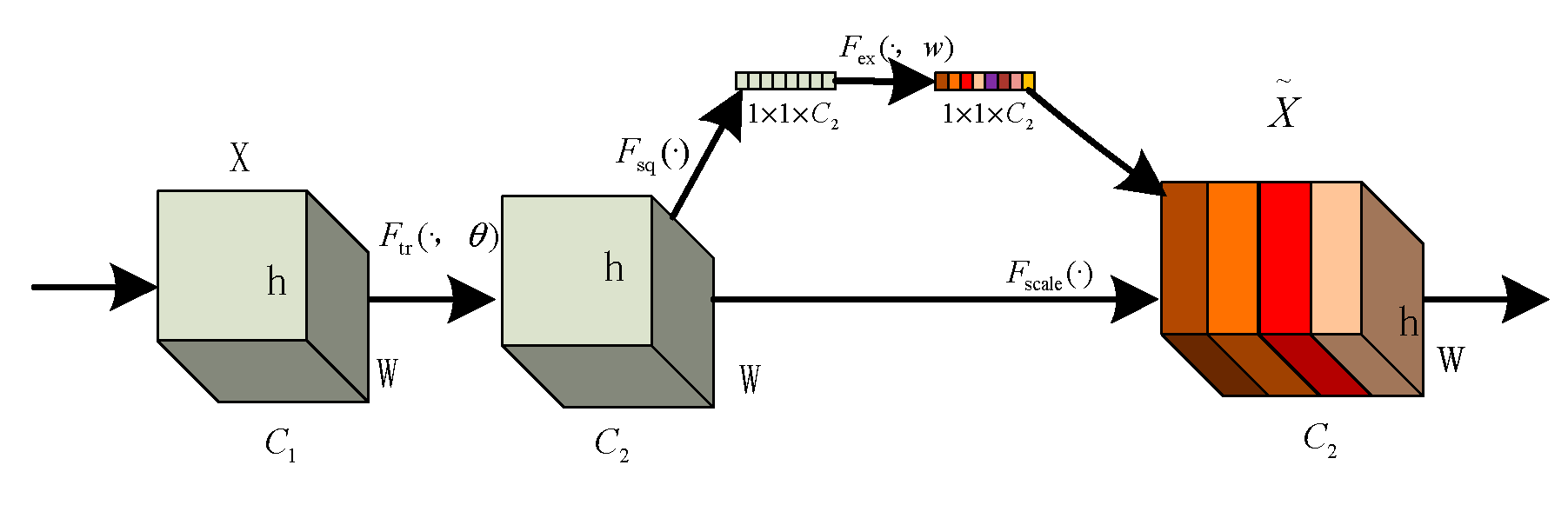
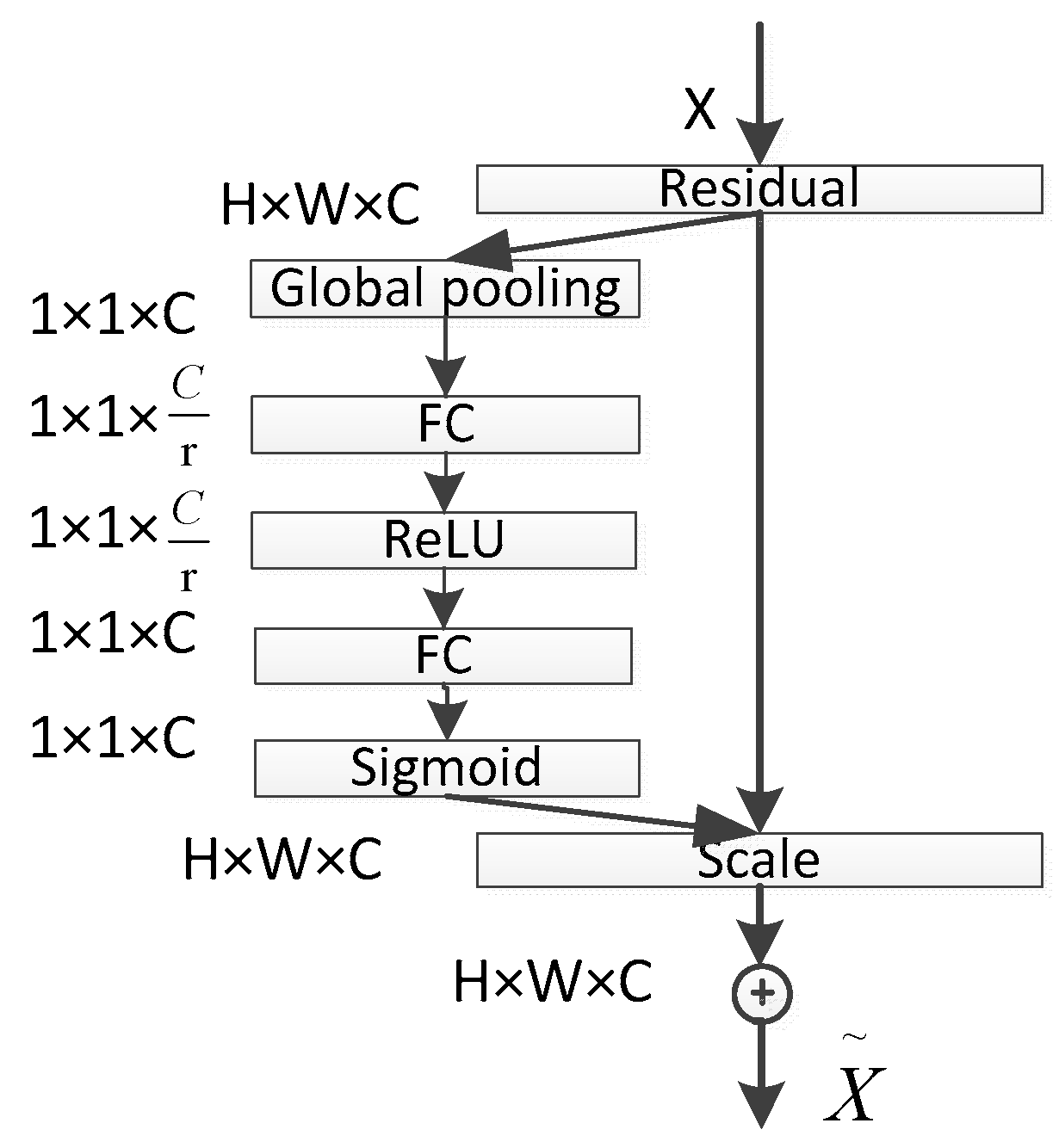
The core idea of the SE module lies in first performing a squeeze operation on the feature maps obtained through convolution to capture global features across the entire channel hierarchy. Subsequently, through the excitation operation, the model learns relationships between different channels and derives weights for each channel [25]. Finally, these weights are applied to the original feature maps of potato leaf disease spots to obtain the ultimate feature maps, with the output size of potato leaf images remaining unchanged. In practice, the SE module operates on the channel dimension by performing attention or gating operations. It expands the feature maps, originally in a low-dimensional space, to a higher-dimensional space using fully connected layers and recalculates the weights for each feature map. These weights are then aggregated to construct a distribution reflecting the importance of feature maps, as illustrated in Figure 6 of the SE attention mechanism. Different colors represent various values used to assess the importance of channels. The SE attention mechanism enables the network to adaptively select and strengthen important feature channels of potato leaf disease spots while suppressing unimportant ones, thereby improving the network’s ability to represent input data features more distinctly. Moreover, given the small and diverse morphology of potato leaf disease spots, traditional feature extraction methods can negatively impact segmentation results.
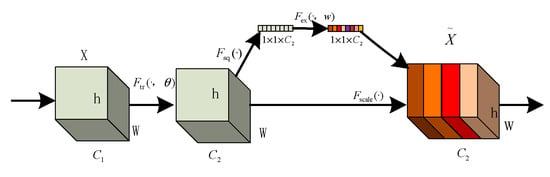
Figure 6.
SE structure.
The squeeze operation effectively entails a form of global average pooling on the feature maps. Applying the squeeze operation to a feature map with c channels and an H × W size compresses the map into a c-channel of 1 × 1 size. During this process, the information in the original feature map is compressed into a representation that encapsulates global information. The squeeze operation achieves this by employing global average pooling, aggregating channel information from the original feature map into a single channel. Each channel retains only one average value, which encapsulates the global information of the entire feature map.
In this context, δ represents the ReLU (rectified linear unit) function.
The excitation operation essentially involves two fully connected layers. Firstly, it performs a fully connected operation on the global pooling result obtained through the previous squeeze operation (which can already be considered as a C-dimensional vector). This operation yields a C/r-dimensional vector, followed by the application of a ReLU activation function. Subsequently, this C/r-dimensional vector undergoes another fully connected operation, restoring the vector dimension to C, and is then passed through a sigmoid activation function (ensuring that the values range between 0 and 1) to obtain the final weight matrix. Throughout this process, critical channel features receive higher weighting, while less important channel features receive lower weighting. Ultimately, the channel features are multiplied by their respective weights and combined with the output of the SE module.
In the SE block, the transformed feature maps are considered as the non-identity branch of the residual module. Both the squeeze and excitation operations are performed before the summation with the identity branch of the residual module. As networks tend to overfit during training, resulting in suboptimal segmentation performance on the test set, the SE attention mechanism is introduced into ResNet50 to alleviate the impact of overfitting. The network structure is depicted in Figure 7. The core idea of the ResNet module is to learn the residual between the input and output, denoted as H(X-X). This residual module enables the network to better fit an identity mapping, facilitating the network’s ability to capture small target disease spots or edge details in images. This enhances the detection of small target disease spots, significantly improves model performance and generalization, and reduces model complexity and training costs, making it more suitable for potato leaf disease spot segmentation tasks. Additionally, this module directs the network’s focus toward important features and facilitates better generalization to new data, thereby enhancing the model’s performance in potato leaf disease spot segmentation tasks.
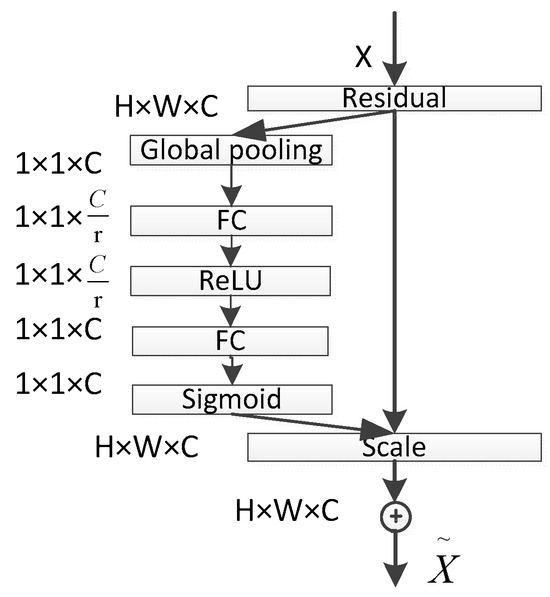
Figure 7.
SE-ResNet Network Architecture.
2.3. Improved U-Net Network Model
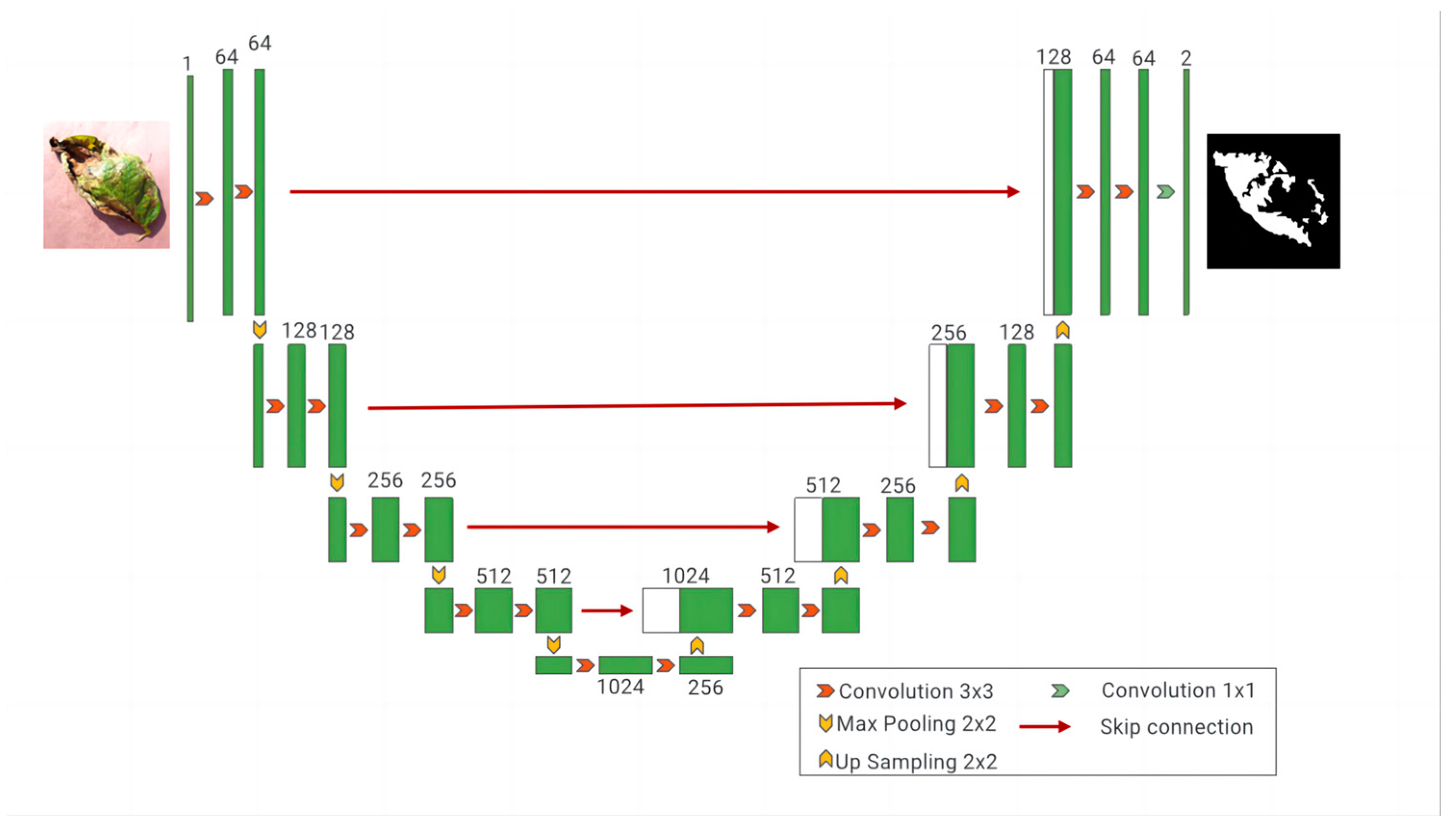
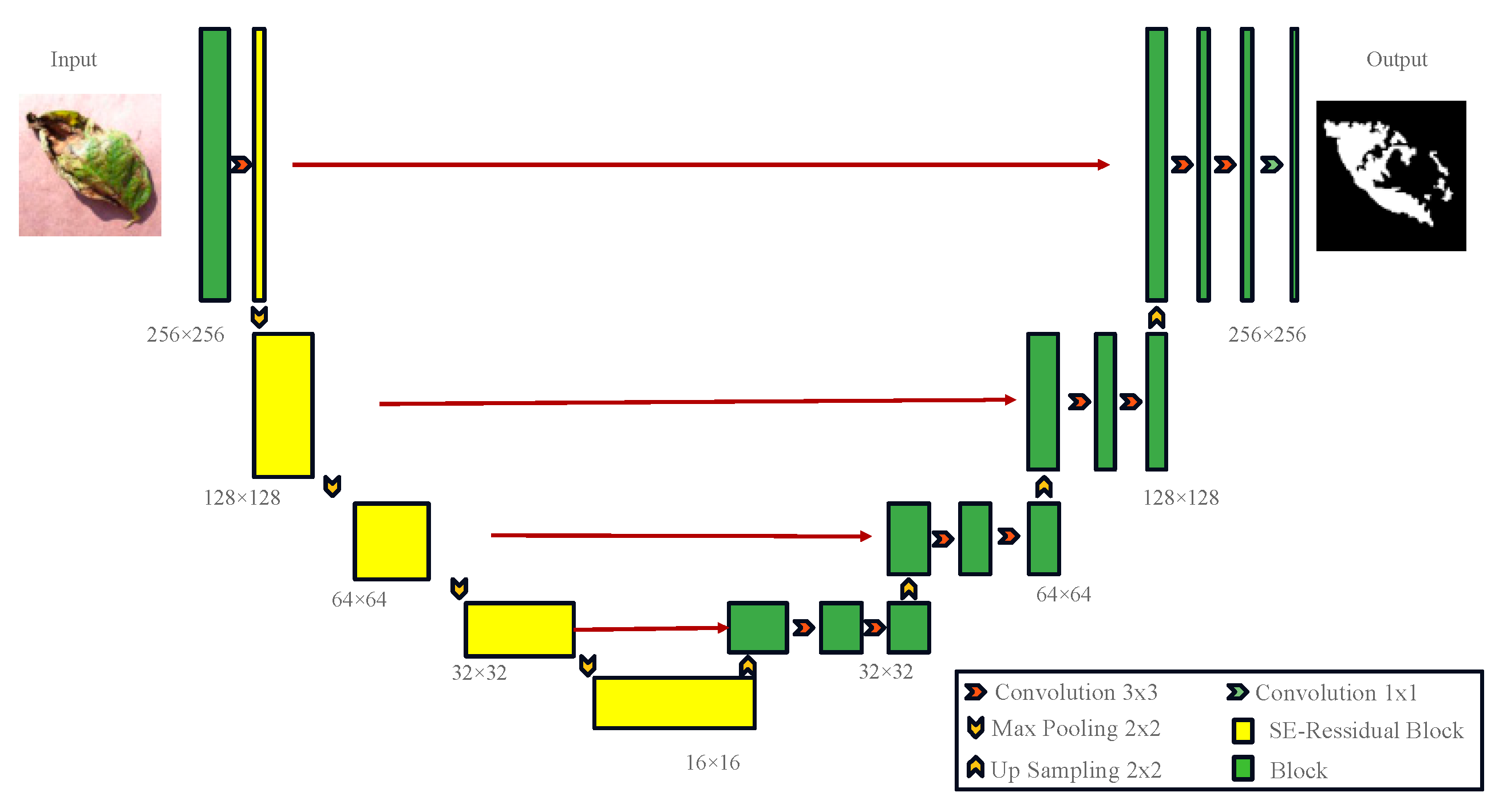
UNet was originally widely applied to small-sample segmentation tasks in medical imaging, showing remarkable performance. However, when applied to the segmentation of complex potato leaf images, it faces challenges due to the diverse morphologies, colors, and sizes of potato leaf disease spots, necessitating a more robust feature-extraction mechanism to enhance segmentation accuracy [26,27]. Therefore, improvements were made to UNet by incorporating an encoder, ResNet50, and introducing the SE (squeeze and excitation) attention mechanism, while retaining the UNet structure as the decoder, resulting in the RS-UNet architecture as illustrated in Figure 8. Using ResNet50 as the encoder: Traditional UNet models employ convolution and pooling layers in the encoder section to progressively extract and reduce the features of input images. However, when dealing with complex and diverse image data, a traditional encoder may not fully capture complex features in the image [28,29]. Thus, ResNet50 was chosen as the encoder, known for its superior feature extraction capabilities for potato leaf disease spots. ResNet50 introduces residual blocks during training, which can effectively train deep networks, avoid gradient vanishing issues, and enhance feature representation. Introducing the SE attention mechanism: The SE attention mechanism is a mechanism used to enhance feature representation capabilities. Applying the SE attention mechanism to the feature maps of ResNet50 allows the network to focus more on important feature channels while ignoring less important ones. This mechanism adaptively adjusts the importance of different channels in the feature maps by learning channel weights. In disease spot segmentation tasks, different disease spot morphologies and colors in potato leaves may be reflected in different channels, and the SE attention mechanism helps the network to concentrate on these key features, thereby improving segmentation accuracy. Retaining UNet as the decoder: UNet is a common architecture used for image segmentation [30], and its decoder section can effectively restore the spatial information of feature maps, generating prediction results. In the improvement, the decoder structure of UNet was retained, ensuring that the network can preserve detailed information when generating prediction results. The decoder retains skip connections, connecting the feature maps of the encoder and decoder, thus facilitating the fusion of multi-level features and improving segmentation accuracy.
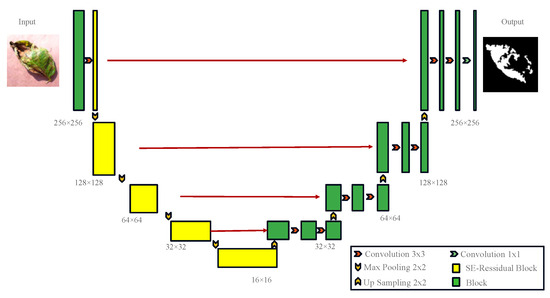
Figure 8.
RS-UNet Network Model.
The network structure is shown in Figure 8. In the encoding phase, input images of potato leaf disease spots with a size of 256 × 256 undergo convolutional layers and pooling layers in ResNet50, resulting in a series of feature maps containing potato leaf disease spots. The SE attention mechanism is applied to each feature map to enhance the representation of important features [31]. The output features are enhanced by the SE attention mechanism. The encoding stage includes a 3 × 3 convolution operation, followed by a Resblock and Max-pooling operation, using maximum pooling with a stride of 2 to reduce the Image size to 128 × 128, and eventually, after four convolutions, the size is reduced to 16 × 16, reaching a minimum size. After each downsampling, the traditional convolution module is replaced by the SE-Resnet structure to address the overfitting issues caused by deep networks. The decoder adopts the UNet structure, taking the improved feature maps as input into the UNet encoder section. The encoder consists of a series of convolutional layers and pooling layers, progressively extracting features and reducing the size of feature maps. The output feature maps of the encoder are connected to the corresponding input feature maps of the decoder through skip connections to preserve more detailed information. In the decoding phase, four sets of 2 × 2 upsampling and 3 × 3 convolution operations are used to restore the size of feature maps. The components are convolved layer by layer from the fourth level to the first level, and the output is resized to the original 256 × 256 size through a 1 × 1 convolutional layer. The decoder section consists of a series of convolutional layers and upsampling layers, gradually restoring the size of feature maps and generating the final prediction results. The final prediction results are subjected to the BCELoss and Dice loss linear combination through an activation function to obtain binary segmentation results.
2.4. Optimize the Loss Function
UNet is employed for pixel-level segmentation tasks, with the most common loss function being the cross-entropy loss. To enhance performance, a linear combination of cross-entropy loss (BCELoss) and Dice loss is utilized [32]. In segmentation tasks, the number of background pixels often far outweighs that of defect samples. Relying solely on cross-entropy loss could lead the network to converge to local minima. The Dice loss function measures the similarity between samples, focusing on the accuracy of foreground image classification while emphasizing background pixels [33]. This approach effectively mitigates the issue of foreground–background sample imbalance. A non-linear loss function approach is adopted to address extreme data imbalance by combining the Dice loss with the cross-entropy loss. This method not only considers the overall image loss, but also pays greater attention to variations in target object loss, thus counteracting the influence of feature region size on segmentation accuracy. The objective is to emphasize accurate loss computation for target regions while considering the entire image context, aiming to improve segmentation precision.
BCELoss function’s loss rate (LBce) is calculated as follows:
The Dice loss function’s loss rate (LDice) can be calculated as follows:
where |X∩Y| represents the intersection between sets X and Y, |X| and |Y| respectively represent the number of pixels in sets X and Y, N denotes the total number of pixels in the segmented image, n stands for the number of training iterations, xn corresponds to the independent variable used during loss function training, and yn signifies the dependent variable of gradient changes.
3. Results
3.1. Experimental Environment Configuration
In the experiments, the following hardware environment was used for training: an Intel(R) Core(TM) i7-10750H CPU, an NVIDIA Tesla T4 GPU (with 16 GB of VRAM), 32 GB of RAM, and a Windows operating system. To implement deep learning, PyTorch 1.5.0 was chosen as the framework, coupled with Python 3.9.7, CUDA 11.7 for the computing framework, and CUDNN 10.2 for the deep neural network acceleration library.
3.2. Training Setup
During the training process, the image sizes were uniformly adjusted to 215 × 215 pixels. By applying the finalized data augmentation strategy, the original dataset was expanded to a total of N images. To effectively assess model performance and prevent overfitting, all images were randomly allocated into the training and validation sets. To enhance convergence and training stability, a learning rate (lr) of 1 × 10−5 was set, with a batch size of 1 for a total of 300 iterations. To accommodate the model’s requirements at different training stages, the stochastic gradient descent (SGD) [34] algorithm was used for optimization, allowing the adjustment of network parameters to minimize the loss function and accelerate model convergence. Such experimental settings facilitated the training and optimization of deep learning models within the selected hardware and software environment.
3.3. Model Evaluation Index
Evaluation metrics are a direct reflection of assessing the quality of an algorithm. In this experiment, the primary evaluation metrics employed on potato leaf images include the Dice coefficient, intersection over union (IoU), and, considering the presence of three types of potato disease spots, accuracy (Acc) is also used as an evaluation metric. The specific calculation formulas for these evaluation metrics are as follows:
The Dice coefficient metric is utilized to measure the similarity between predicted results and expert-annotated ground truth labels:
where N represents the number of potato disease categories, TP represents the true positive instances of potato disease correctly predicted, and FP represents the false positive instances of potato disease incorrectly predicted. FN corresponds to false negative instances of potato disease not detected in predictions.
Pixel accuracy (Acc) is the ratio of correctly classified pixels to the total number of pixels.
where N represents the number of potato disease classes, TP stands for truly predicted potato disease samples, FP represents falsely predicted potato disease samples, and FN represents the predicted samples of potato disease that were not detected.
Intersection over Union (IoU) is the most crucial metric for assessing the effectiveness of semantic segmentation. It calculates the intersection over the union of two sets: the ground truth and the predicted segmentation, respectively. It is a ratio of the intersection to the union of these two sets.
where N represents the number of potato disease categories, TP stands for true positive, which represents the correctly predicted instances of potato disease, FP represents false positive, indicating the instances that were incorrectly predicted as potato disease, and FN represents false negative, indicating the instances of potato disease that were not detected by the prediction.
3.4. Experimental Results and Analysis
This study employed an improved UNet network model to train on the potato dataset, yielding a well-trained model. To validate the effectiveness of this model, a batch size of 1 and 300 iterations were selected for the training process. This accelerated the model’s training speed and significantly improved its data-fitting capabilities, contributing to enhanced accuracy in potato lesion segmentation on a limited dataset. Ultimately, the model achieved an impressive Dice coefficient of 88.86% in the segmentation task, which is a notable result. During training, the model was saved every five iterations, and the stochastic gradient descent (SGD) algorithm was utilized for optimization, aiding in the gradual fine-tuning of network parameters to optimize model performance. Through these steps, a trained and optimized model was established, capable of accurate potato leaf lesion segmentation in practical applications.
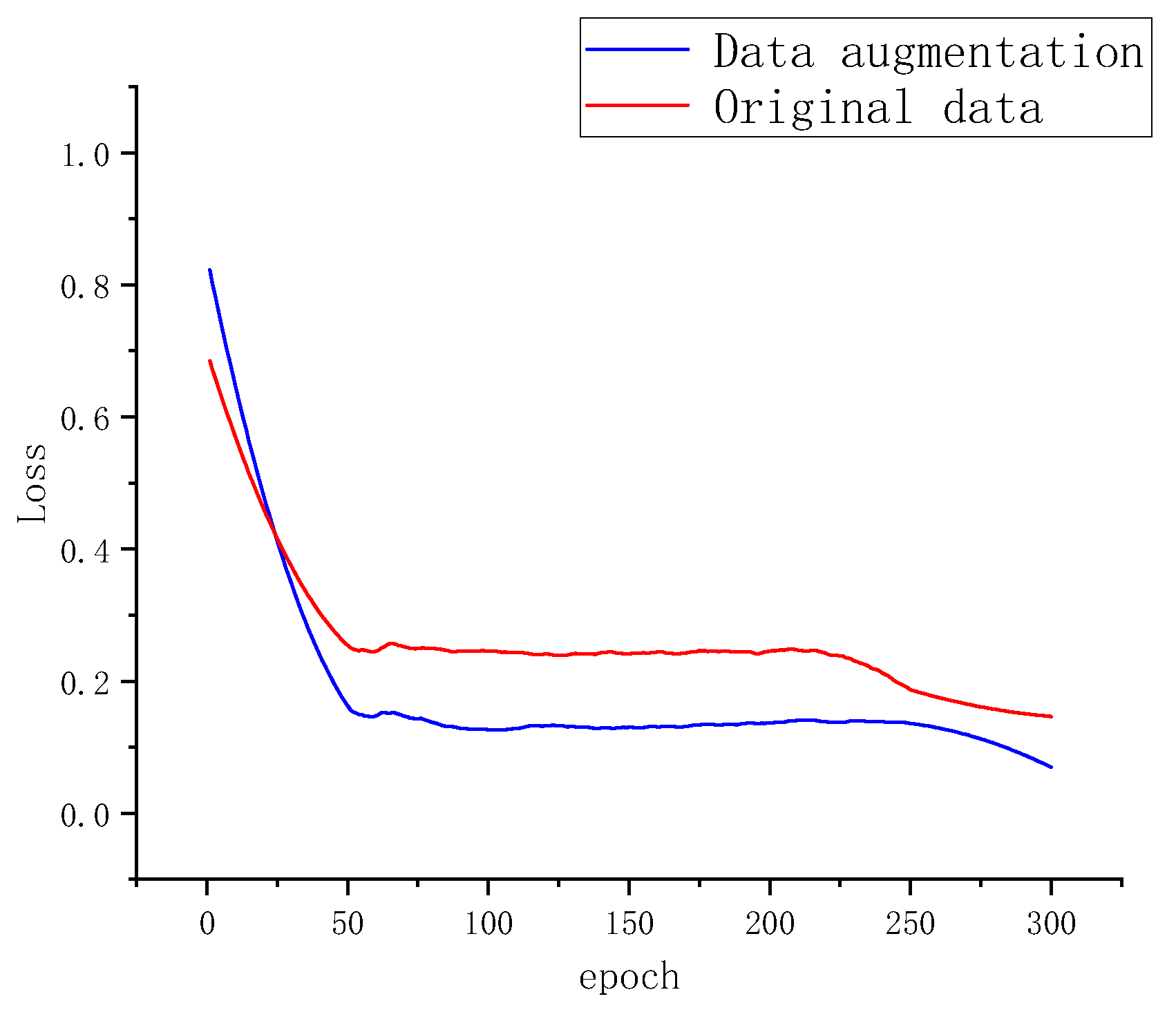
3.4.1. Comparison between Raw Data and Data Augmentation
This study systematically investigated the impact of data augmentation on reducing the misclassification rate in potato leaf disease segmentation. As depicted in Figure 9, to demonstrate the effectiveness of the augmented dataset, comparative experiments were conducted between the raw data and the data-augmented dataset, resulting in misclassification rates during training.
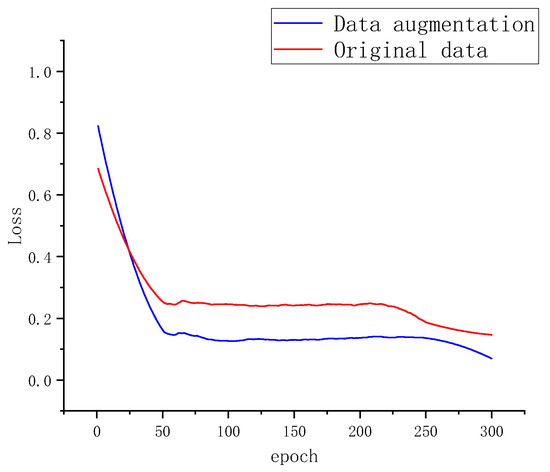
Figure 9.
Segmentation Loss of Two Datasets.
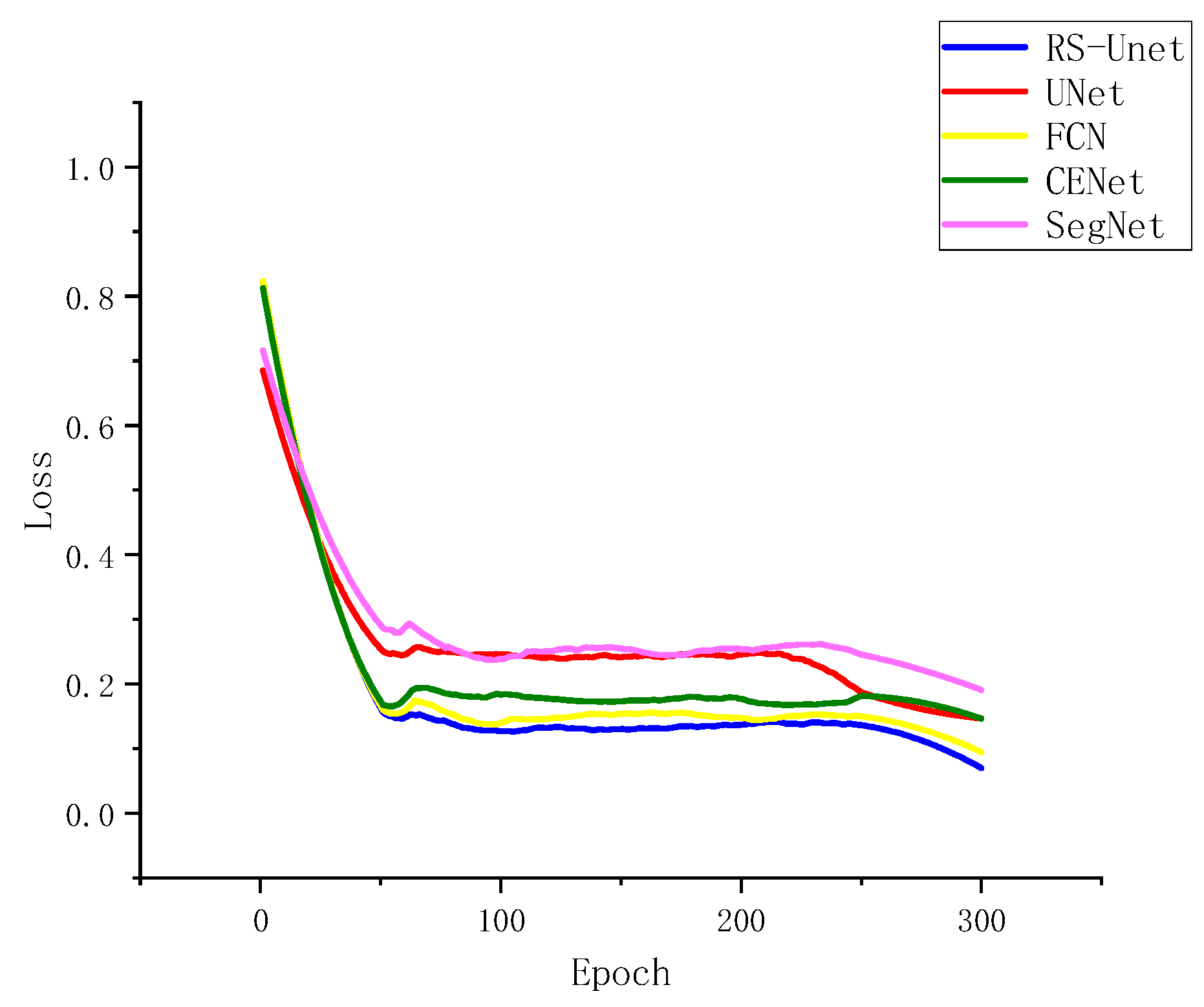
3.4.2. Comparison of Experiments with Different Models
To test the improved RS-UNet network alongside UNet, SegNet, FCN, and CENet on augmented datasets, the relationship between loss and iteration count during convergence was examined, as depicted in Figure 10. The five network models exhibited a rapid decrease in loss within the initial 50 iterations and stabilized around 80 iterations. Figure 10 also illustrates that the improved UNet network outperformed the other four models, yielding the best results after 300 iterations. This improvement could be attributed to the adoption of ResNet50 as the encoder in the enhanced UNet network, which led to enhanced training speed and segmentation performance.
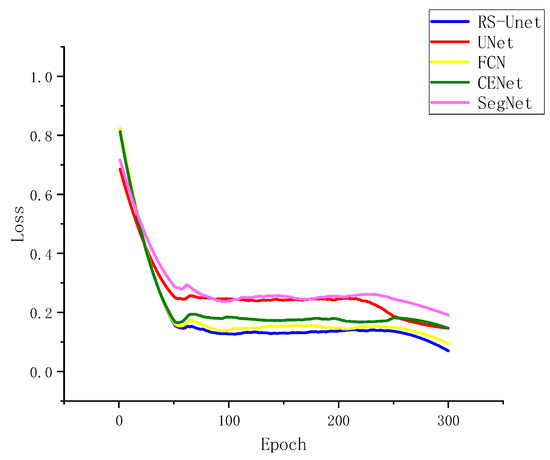
Figure 10.
Segmentation Loss of Different Network Architectures.
3.4.3. Comparison of Experimental Results between Different Network Models
To visually validate the model’s accuracy in potato disease spot recognition, Table 1 compares the recognition accuracy results of four different network models (UNet, CENe, SegNet, and FCN) with the proposed network. As shown in Table 1, the improved model achieved good accuracy in identifying potato early blight, anthracnose, and late blight diseases. The accuracy of the improved model on the potato dataset was higher than that of the other three networks, indicating that the improved network model had good generalization capabilities for potato disease datasets.

Table 1.
Precision of 5 Types of Diseases Under Different Models.
In the same cycle, a comparison was conducted among the improved RS-UNet, UNet, SegNet, CENet, and FCN network models regarding potato disease spot segmentation effectiveness, as shown in Table 2. The improved RS-UNet outperformed other network models regarding the Dice coefficient, IoU, and training time. Notably, the SegNet segmentation time was twice that of the improved UNet, and the increased segmentation time did not lead to higher IoU or Dice values. The Dice value of RS-UNet (88.86%) was 11.22% higher than that of SegNet (77.64%). The time required for UNet segmentation was 0.84 h faster than the improved UNet segmentation. However, the segmentation results exhibited a lower Dice value of 6.57% and a lower IoU value of 8.96% for UNet. Comparatively, the time required for FCN segmentation was 2.19 h slower than that of the improved RS-UNet segmentation. The segmentation results of FCN showed a lower Dice value of 2.82% and a lower IoU value of 3.84%. The improved U-Net network model demonstrated significant advantages regarding the Dice coefficient, IoU, and accuracy.

Table 2.
Segmentation Performance of Different Network Models.
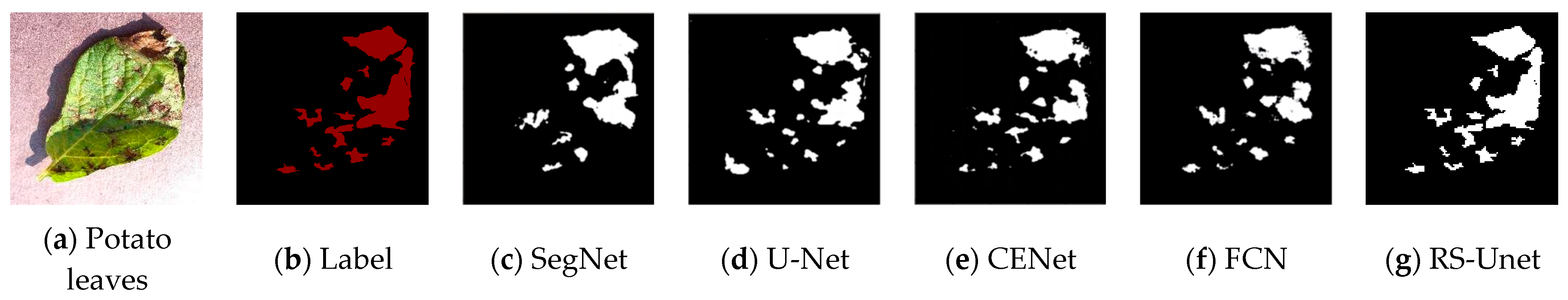
To visually compare the differences in segmentation results of different networks, four randomly selected images of potato leaf diseases were visualized, as shown in Figure 11, with results from various network models. In the images, black represents the background, while white represents the segmented lesions. The first column displays images of potato leaf diseases, the second column showcases manually annotated disease spot images in red, and each subsequent column illustrates the white regions, which represent the segmentation results produced by different network models. From Figure 11, it is evident that different diseases exhibited distinct colors, shapes, and noticeable differences. In the case of potato leaf disease spots, both SegNet and UNet network models could segment the approximate areas of the lesions. However, due to the small size of some potato leaf lesions, there were inaccuracies and missed segments in some areas. FCN and CENet showed significant improvements in the segmentation results, with their potato disease spot segmentation results being closer to the labels. However, CENet exhibited errors in the segmentation of edge areas and other lesion regions, while the FCN network model had less clear edges. For the improved RS-UNet network, some lesions closely matched the labeled areas, and even tiny lesions within the diseased leaves could be accurately extracted. This demonstrates that the improved RS-UNet could accurately segment disease leaf images, including lesion areas and their edges. This advantage could be attributed to the use of a deep residual network for feature extraction during re-segmentation, along with the incorporation of the SE attention mechanism on top of the ResNet, which highlights the disease spot’s feature information while suppressing irrelevant features. Therefore, the improved network model achieved higher segmentation efficiency. In summary, compared with other networks, RS-UNet was more suitable as a model for potato leaf disease segmentation.
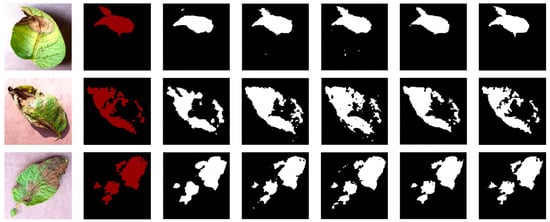
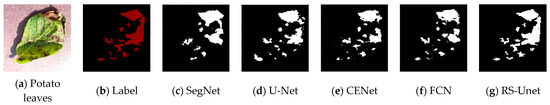
Figure 11.
Comparison of Segmentation Results from Different Network Models.
4. Discussion
Considering the present repercussions of potato leaf diseases on potato crop yields, precise identification through deep learning assumes considerable significance within the potato industry. It can facilitate farmers in the timely detection and proactive mitigation of potato leaf diseases, providing swifter and more convenient strategies for the preservation of potato crop well-being [35,36].
In this study, our focus was on potato leaf disease images, and we successfully trained and validated the improved UNet network model, RS-UNet. This network is designed to handle the task of potato leaf disease image segmentation under a single background and has been compared with various other segmentation network models. Based on these findings, each network had its pros and cons in terms of performance on the potato leaf disease segmentation task. To effectively reduce the loss of small-scale object information and enhance the integration capability of lesion area edge details, innovations were introduced into the UNet architecture. By employing ResNet50 as the encoder, the network model was deepened, and the SE attention mechanism was introduced to improve the extraction of relevant disease spot information while suppressing irrelevant information. Furthermore, the combination of cross-entropy loss and Dice loss improved the accuracy of potato leaf disease spot identification and localization.
Nonetheless, it is prudent to acknowledge that, as a network model becomes progressively deeper and parameters increase in magnitude, the network’s complexity amplifies, thereby introducing greater intricacies in the training process. Interestingly, while our RS-UNet exhibited a shorter training duration in comparison with the basic UNet, it surpassed it in segmentation precision. Moreover, our RS-UNet demonstrated superior performance compared with SegNet, UNet, CENet, and FCN across various metrics, including MIoU, Dice indices, and the quality of the segmented result images.
Although the current network model exhibited remarkable performance in our present task, we recognize that there exists substantial room for future improvements. Our research was limited to a single background scenario. To this end, we can further optimize the network architecture or explore more refined semantic segmentation models [37,38,39], as exemplified by Wang Hao et al. [40], who proposed a segmentation network tailored to the detection of pear leaf diseases in complex natural environments. Simultaneously, the creation of additional potato datasets could be contemplated to augment the model’s robustness and precision.
5. Conclusions
This study proposes an improved convolutional neural network, RS-UNet, based on UNet. By incorporating deep residual network modules to increase the depth of the UNet network while keeping fewer parameters and integrating the SE attention mechanism to enhance the disease area, UNet was used as the backend network for feature fusion. While deepening the network model to prevent degradation, it could fully utilize UNet’s effective combination of low-resolution and high-resolution features, achieving good results in the segmentation and recognition of potato leaf disease spots. In addition, a combination optimization algorithm was used, and under the premise of ensuring segmentation accuracy, it effectively shortened the network training time, making the segmentation results closer to the actual disease spots. Compared with the original UNet, the improved network model demonstrated superior segmentation performance. Specifically, this method achieved 79.8% MIoU and 88.86% Dice scores. Finally, by using a combination optimization algorithm, the network’s Dice coefficient was improved from 82.29% to 88.86%, resulting in a nearly 6.57% increase in recognition accuracy.
While this method was effective in segmenting potato leaf diseases, the proposed model was not able to achieve segmentation in complex backgrounds. Future research will focus on segmenting potato leaf diseases in complex backgrounds, simplifying the model’s new strategies, and calculating the severity of the disease, integrating the solution into the latest disease detection platforms. This approach aims to achieve higher segmentation accuracy and efficiency, making it more suitable for practical production applications.
Author Contributions
Writing—original draft preparation, J.F.; resources, Y.Z.; supervision, G.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The experimental data used to support the findings of this study are available from the authors upon request.
Acknowledgments
The authors would like to show sincere thanks to those technicians who have contributed to this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Branch, M. A multi layer perceptron neural network trained by invasive weed optimization for potato color image segmentation. Trends Appl. Sci. Res. 2012, 7, 445–455. [Google Scholar]
- Hou, C.; Zhuang, J.; Tang, Y.; He, Y.; Miao, A.; Huang, H.; Luo, S. Recognition of early blight and late blight diseases on potato leaves based on graph cut segmentation. J. Agric. Food Res. 2021, 5, 100154. [Google Scholar] [CrossRef]
- Li, X.; Zhou, Y.; Liu, J.; Wang, L.; Zhang, J.; Fan, X. The detection method of potato foliage diseases in complex background based on instance segmentation and semantic segmentation. Front. Plant Sci. 2022, 13, 899754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, G.; Lu, C.; Chen, A.; Wang, Y.; Li, L.; Cai, W. MMDGAN: A fusion data augmentation method for tomato-leaf disease identification. Appl. Soft. Comput. 2022, 123, 108969. [Google Scholar] [CrossRef]
- Dayang, P.; Meli, A.S.K. Evaluation of image segmentation algorithms for plant disease detection. Int. J. Image Graph. Signal Process. 2021, 13, 14–26. [Google Scholar] [CrossRef]
- Johnson, J.; Sharma, G.; Srinivasan, S.; Masakapalli, S.K.; Sharma, S.; Sharma, J.; Dua, V.K. Enhanced field-based detection of potato blight in complex backgrounds using deep learning. Plant Phenomics 2021, 2021, 9835724. [Google Scholar] [CrossRef]
- Sun, J.; Zhao, J.; Ding, Z. ULeaf-Net: Leaf segmentation network based on u-shaped symmetric encoder-decoder architecture. In Proceedings of the 2021 International Symposium on Computer Science and Intelligent Controls (ISCSIC), Rome, Italy, 12–14 November 2021; pp. 109–113. [Google Scholar] [CrossRef]
- Jia, Z.; Shi, A.; Xie, G.; Mu, S. Image segmentation of persimmon leaf diseases based on UNet. In Proceedings of the 2022 7th International Conference on Intelligent Computing and Signal Processing (ICSP), Xi’an, China, 15–17 April 2022; pp. 2036–2039. [Google Scholar] [CrossRef]
- Agarwal, M.; Gupta, S.K.; Biswas, K.K. Plant leaf disease segmentation using compressed UNet architecture. In Trends and Applications in Knowledge Discovery and Data Mining: PAKDD 2021 Workshops, WSPA, MLMEIN, SDPRA, DARAI, and AI4EPT, Delhi, India, May 11, 2021 Proceedings 25; Springer International Publishing: Cham, Switzerland, 2021; pp. 9–14. [Google Scholar]
- Chen, C.P.; Niu, J.W.; Ding, K.; Jintao, J. Intelligent Recognition of Potato Diseases Based on Deep Learning. Comput. Simul. 2023, 40, 214–217+222. [Google Scholar]
- Chen, P.; Ma, Z.H.; Zhang, J.; Yi, X.; Bing, W.; Dong, L. Wheat Rust Semantic Segmentation Network Based on Fusion Attention Mechanism. Trans. Chin. Soc. Agric. Mach. 2023, 44, 145–152. [Google Scholar] [CrossRef]
- Mao, W.J.; Ruan, J.Q.; Liu, S. Strawberry Disease Semantic Segmentation Based on Improved UNet with Attention Mechanism. Comput. Syst. Appl. 2023, 32, 251–259. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Quan, X.R. DUNet++: Skin Lesion Segmentation Network Based on Improved UNet++. Comput. Knowl. Technol. 2023, 19, 95–97. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Metlek, S.; Çetıner, H. ResUNet+: A New Convolutional and Attention Block-Based Approach for Brain Tumor Segmentation. IEEE Access 2023, 11, 69884–69902. [Google Scholar] [CrossRef]
- Chong, Y.; Xie, N.; Liu, X.; Pan, S. P-TransUNet: An improved parallel network for medical image segmentation. BMC Bioinform. 2023, 24, 285. [Google Scholar] [CrossRef]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Shan, L.; Tang, M.; Liu, Y.; Bai, X.; Meng, F. Brain Tumor Image Segmentation Algorithm Based on Improved Res-Unet. Autom. Instrum. 2022, 9, 13–18+23. [Google Scholar] [CrossRef]
- Kumar, V.V.N.S.; Reddy, G.H.; GiriPrasad, M.N. A novel glaucoma detection model using Unet++-based segmentation and ResNet with GRU-based optimized deep learning. Biomed. Signal Process. Control. 2023, 86, 105069. [Google Scholar] [CrossRef]
- Deng, Y.; Xi, H.; Zhou, G.; Chen, A.; Wang, Y.; Li, L.; Hu, Y. An effective image-based tomato leaf disease segmentation method using MC-UNet. Plant Phenomics 2023, 5, 0049. [Google Scholar] [CrossRef]
- Kaushik, M.; Prakash, P.; Ajay, R.; Veni, S. Tomato leaf disease detection using convolutional neural network with data augmentation. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 1125–1132. [Google Scholar]
- Zhang, R.; Zhu, Y.; Ge, Z.; Mu, H.; Qi, D.; Ni, H. Transfer learning for leaf small dataset using improved ResNet50 network with mixed activation functions. Forests 2022, 13, 2072. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Vuola, A.O.; Akram, S.U.; Kannala, J. Mask-RCNN and U-Net ensembled for nuclei segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019. [Google Scholar]
- El-Assiouti, H.S.; El-Saadawy, H.; Al-Berry, M.N.; Tolba, M.F. Lite-SRGAN and Lite-UNet: Towards Fast and Accurate Image Super-Resolution, Segmentation, and Localization for Plant Leaf Diseases. IEEE Access 2023, 11, 67498–67517. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, L.; Zhang, H.; Xiao, X. Breast cancer histopathological image classification using convolutional neural networks with small SE-ResNet module. PLoS ONE 2019, 14, e0214587. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Albanie, S.; Sun, G.; Wu, E. Squeeze-and-excitation networks. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Mukti, I.Z.; Biswas, D. Transfer learning based plant diseases detection using ResNet50. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 20–22 December 2019; pp. 1–6. [Google Scholar]
- Botev, Z.I.; Kroese, D.P.; Rubinstein, R.Y.; L’ecuyer, P. The Cross-Entropy Method for Optimization in Handbook of Statistics; Elsevier: Amsterdam, The Netherlands, 2013; Volume 31, pp. 35–59. [Google Scholar]
- Keskar, N.S.; Socher, R. Improving generalization performance by switching from Adam to SGD. arXiv 2017, arXiv:1712.07628. [Google Scholar]
- Tiwari, D.; Ashish, M.; Gangwar, N.; Sharma, A.; Patel, S.; Bhardwaj, S. Potato leaf diseases detection using deep learning. In Proceedings of the 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 13–15 May 2020; pp. 461–466. [Google Scholar]
- Sharma, R.; Singh, A.; Dutta, M.K.; Riha, K.; Kriz, P. Image processing based automated identification of late blight disease from leaf images of potato crops. In Proceedings of the 2017 40th International Conference on Telecommunications and Signal Processing (TSP), Barcelona, Spain, 5–7 July 2017; pp. 758–762. [Google Scholar]
- Rashid, J.; Khan, I.; Ali, G.; Almotiri, S.H.; AlGhamdi, M.A.; Masood, K. Multi-level deep learning model for potato leaf disease recognition. Electronics 2021, 10, 2064. [Google Scholar] [CrossRef]
- Yang, K.; Zhong, W.; Li, F. Leaf segmentation and classification with a complicated background using deep learning. Agronomy 2020, 10, 1721. [Google Scholar] [CrossRef]
- Bhagat, S.; Kokare, M.; Haswani, V.; Hambarde, P.; Kamble, R. Eff-UNet++: A novel architecture for plant leaf segmentation and counting. Ecol. Inform. 2022, 68, 101583. [Google Scholar] [CrossRef]
- Divyanth, L.G.; Ahmad, A.; Saraswat, D. A two-stage deep-learning based segmentation model for crop disease quantification based on corn field imagery. Smart Agric. Technol. 2023, 3, 100108. [Google Scholar] [CrossRef]
- Wang, H.; Ding, J.; He, S.; Feng, C.; Zhang, C.; Fan, G.; Wu, Y.; Zhang, Y. MFBP-UNet: A Network for Pear Leaf Disease Segmentation in Natural Agricultural Environments. Plants 2023, 12, 3209. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).