1. Introduction
Apical periodontitis is the consequence of root canal infection by bacteria that is manifested as a periapical bone resorption that develops as a response of the host’s defense against bacterial infection [
1]. Apical periodontitis affects about 33 to 62% of the adult population and it can have detrimental effects on both oral and systemic health [
2]. Thus, this condition should be diagnosed and treated without delay. Failure to treat might lead to the spread of disease to the surrounding tissues, resulting in serious complication for the patient [
3]. While an initial diagnosis of acute apical periodontitis may be made clinically, the detection of chronic apical periodontitis is made by radiographs used to reveal characteristic periapical radiolucencies that are usually called apical lesions [
4]. These apical lesions appear as a widened periodontal ligament space and are detected by radiographic investigation of endodontically treated teeth [
5,
6].
Detection of apical lesions can be performed with several available radiological options, e.g., cone beam computerized tomography (CBCT), periapical radiographs and panoramic radiographs. CBCT demonstrates significantly higher discriminatory ability than periapical radiographs [
7], however, the associated costs and high radiation are major limitations that restrict its use to very few indications. Periapical radiographs are usually considered as the gold standard imaging techniques for diagnosis of apical lesions [
8]. However, there could be inconsistency across dentists in their interpretation of such radiographs, and due to radiation concerns, they cannot be routinely used for screening the entire dentition. The radiographic appearance of endodontic pathosis in a periapical radiographs could be subjective and they have shown limited discriminatory ability when compared against histopathological analysis [
6,
9,
10]. A previous study showed that interpreters were only able to reach a 50% level of agreement on the assessment periapical lesions on periapical radiographs. In addition, re-evaluation of radiographs by the same clinician showed different interpretation of their own original diagnosis [
10]. Thus, image interpretation by dentists could sometimes be inconsistent [
11].
Other techniques such as CBCT, MRI and echography can also be useful [
8,
12]. However, these methods cannot be used for routine screening because CBCT requires too much radiation, MRI is very expensive and time consuming, and echography is ineffective in lesions not affecting the cortical bone. In this context, OPG are better suited for screening. However, even though dentists are supposed to make accurate screenings of periapical lesions on OPGs, human errors occur and dentists can often miss obvious periapical lesions. A tool to automate detection can help minimize these errors.
Panoramic radiographs are routinely used in dental practice because despite having lower resolution, they are able to capture an extensive area of the oral cavity with significantly lower doses of radiation compared to CBCT imaging [
13,
14]. They allow the easy examination of the complete dentition including the alveolar bone, temporomandibular joints, and adjacent structures providing a valuable screening opportunity [
15,
16]. Regarding the diagnosis of periapical lesions on panoramic radiographs, experienced clinicians can achieve high specificity (95.8%) but a low sensitivity (34.2%) compared to the use of CBCT [
17]. Moreover, there is considerable variability in the dental professionals’ abilities to read panoramic radiographs, which is affected by their individual skills, experience, and biases [
18,
19]. These limitations in the assessment of panoramic radiographs may lead to misdiagnosis or mistreatment [
18,
20].
Recent years have seen an increased use of artificial intelligence in all branches of medicine and dentistry. These computer programs can take over human tasks imitating intelligent human behavior, performing complex activities such as decision-making, solving problems, and even recognizing objects and words [
21,
22]. Neural networks (NNs) are a type of artificial intelligence algorithm which, through a process of deep learning with extensive amounts of data, can enable a computer to present with the capacity to learn to think on its own and make decisions and solve problems in a similar way to humans [
23,
24].
Convolutional neural networks have been used to detect periapical lesions on different radiographic modalities [
11,
25] including panoramic radiographs. This could make the interpretation more objective, and help the dentists save time and focus more on the treatment and identifying the problem at an early stage and avoid further complications. However, even though the performance of these algorithms is promising, there is need for improvement in order to meet the requirements for clinical application [
5,
26,
27]. Thus, the aim of this study was to develop an AI able to detect PL on panoramic radiographs. This was done using CNN in a two-step approach that involved the use of a detector followed by a classifier. We hypothesized that an AI tool trained on healthy and non-healthy periapical areas in panoramic radiographs could detect the periapical regions of the teeth and classify them into healthy apices and apices with periapical lesions.
2. Materials and Methods
2.1. Dataset and Preprocessing
This retrospective diagnostic cohort study was conducted after obtaining approval from the Institutional Research Board (IRB) of the University of Sao Paolo. The approved protocol was given the Ethics Committee number 3.239.265. We collected 713 panoramic radiographs (width: 2879, height: 1563 pixels) from patients treated by Dr Claudio Costa and Dr Arthur Cortes using a panoramic imaging system (Cranex D, Soredex, Tuusula, Finland) set-up with the following parameters: 85 kVp, 10 mA, exposure time 17.6 s, CCD sensor size 48 micrometer, and focal spot size 0.5 mm. We de-identified patients’ information by cropping the panoramic radiographs to size 2250, 1000 pixels) to include only the dental radiograph and exclude patient information registered on the image. We excluded images with unacceptable quality, containing severe artifacts, and radiographs of mixed dentition and edentulous patients. Three examiners (A. D., E.A., and R. BH) independently annotated the Periapical Root Areas (PRAs) as having Periapical Lesion (PL) or not having periapical lesion (Healthy (H)) in duplicate; a fourth examiner (S.O.) settled discrepancies between examiners. At the time of the conduction of the study all above-mentioned examiners had more than 15 years of clinical experience. The examiners were calibrated on 10 OPGs to address discrepancies between them, and then, recalibrated using 20 OPGs and a Kappa index for inter examiner agreement in detecting PLs of 90% was achieved. Upon labeling, only 5% of the cases need the consultation of the fourth examiner to settle discrepancies. We labelled 18,618 PRAs, which contained 1732 unhealthy PRAs (PL), and 16,886 of healthy PRA (H).
The exact location of the PRAs and its labeling as PL or H was done by drawing a bounding box at each PRA using labelImg, an open-source annotation tool that allows annotations in xml Pascal VOC format. We converted the annotation format from xml to coco format as a compatibility requirement to use the Detectron2 framework. In all experiments, we shuffled our dataset once and followed 80–20 sampling split for training and testing datasets, respectively. We dedicated 10% of the training dataset for validation to prevent overfitting when tuning the network hyper-parameters.
Table 1 summarizes the applied data preparation methods.
2.2. Proposed Method
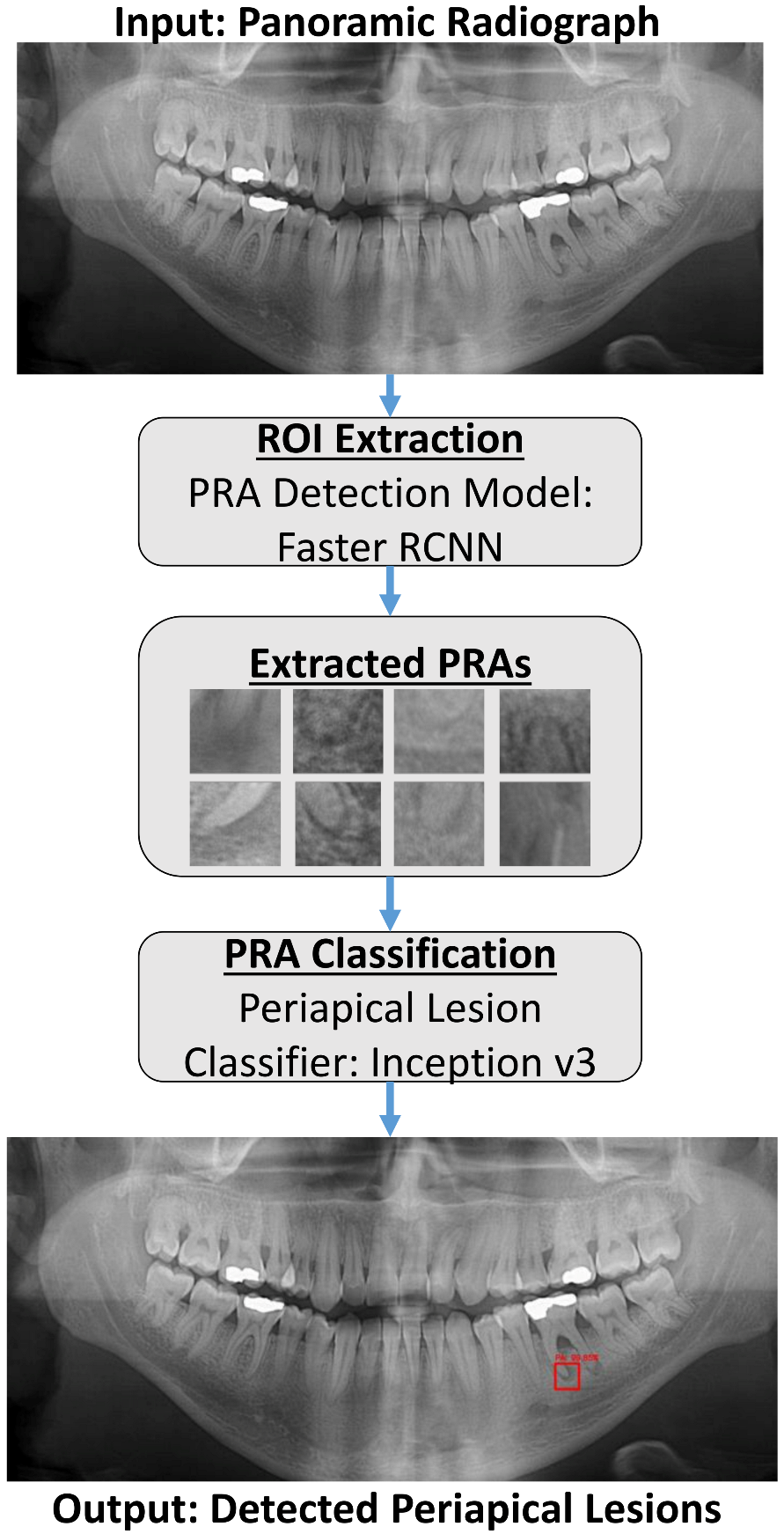
The proposed method consisted of two main CNNs: a detector that we called Periapical Root Area Detection Model, and a classifier that we called Periapical Lesion Classification Model. Our model accepts a panoramic radiograph as input and outputs the location of the detected periapical lesion on the given panoramic image. The detector localized PRAs using a bounding-box-based object detection model, while the classifier classified the extracted PRAs as PL or H. We developed our model on GoogleColab PRO + notebook using Python.
The overall proposed system workflow is shown in
Figure 1. The entire panoramic radiograph is used as an input to the presented system. The PRA Detection Model with Faster R-RCNN processes the panoramic radiograph to detect and define the boundaries of each PRA by a bounding box. The proposed system then crops all predicted bounding boxes, which are the PRAs extracted by the system, and then feeds them to the PL Classifier with Inception v3. The PL Classifier in turn classifies each cropped PRA as Periapical Lesion (PL) or Healthy (H). The system finally outputs in red only the bounding boxes corresponding to Periapical Lesions with a confidence score. A confidence score is a percentage calculated by the AI, indicating how sure the AI is that the output is correct.
2.2.1. Periapical Root Area (PRA) Detection Model
We employed the Faster R-CNN object detection model using the Detectron2 detection platform for localizing the region of interest (ROI), which is the PRA on a panoramic radiograph. Detectron2 provides state-of-the-art detection algorithms, such as object detection, semantic, instance, and panoptic segmentation. We tested Detectron2 implementation of Faster R-CNN using different backbone models and configurations. The results showed that Faster R-CNN ResNet X101 base model achieved the highest detection rates on panoramic radiographs.
We performed training, validation and testing experiments with NVIDIA Tesla P100 and T4 on GoogleColab notebook. We used Detectron2’s default data preprocessing settings for image flipping and resizing (to 1333/800 pixels). Aspect ratios were maintained while resizing. We used 513 panoramic radiographs for training, 57 images for validation, and 143 images for testing. We selected a batch size of 4 due to the high-resolution images. We initialized the Faster R-CNN base model from the ResNet101 pretrained ImageNet classifier. We initially used a learning rate of 0.001, but we decreased the learning by half at epoch 1000 because learning stagnated. We trained until convergence. The model achieved the best performance on iteration 1500.
During the inference time, we fed the entire panoramic radiograph, with 2250 × 1000 resolution, as input to our detection model. The detector then localized all PRAs on a given panoramic radiograph by drawing a bounding box on each detected PRA. We used Detectron2 default Intersection over Union (IoU) and Non-Maximum Suppression (NMS) thresholds, which by default set to 0.5. On average, it took 0.57 s per image using NVIDIA Tesla T4 to detect all PRAs in a given panoramic image. We developed a custom function that returned the detected PRA coordinates, crops, and processes each detected PRA to prepare it for the lesion classification task.
2.2.2. Periapical Lesion Classification Model
Before feeding the cropped PRA images into the classification model for training purpose, we applied the below data preprocessing augmentation techniques to increase our training dataset since we have limited number of unhealthy samples:
Normalized pixel values of each image to range (0, 1)
Resize images to 75 × 75
Random image rotation between 0–20°
Random zoom ranging between 0.8 to 1.2
Shuffle images
We employed a fine-tuned Inception v3 classification model for classifying detected PRAs as healthy (H) or lesion (PL) using Keras framework. We used a balanced subset of the original dataset, which included 3249 PRAs, 1593 of unhealthy PRAs (PLs) and 1656 of healthy PRAs (H). We divided this dataset into training, and testing following the 80:20 ratio. We initialized the model from an ImageNet pretrained inception weights excluding the classification layer. For feature extraction, we froze all layers, stacked and trained only the classification layer. We optimized the network parameters using the Adam optimizer with an initial learning rate of 1 × 10− 5. We decreased the learning rate by half when learning stagnated. We selected a dropout rate of 0.2 to avoid overfitting. We trained for 100 epochs. We then unfroze and re-trained the top layers to fine-tune the network. We optimized the network using Adam optimizer with learning rate of 1 × 10− 7 and trained for another 100 epochs.
During inference time, we fed the detected PRAs as input to the classifier; which in turn labeled each PRA as H or PL. On average, it took 5 milliseconds to classify a PRA from our test dataset. We developed a custom visualization function to visualize PRAs, which our classifier labeled as PL by drawing a bounding box around the detected periapical lesion.
2.3. Evaluation Criteria
We divided the evaluation metrics of the proposed method into two main groups: accuracy of PRA detection and accuracy of periapical lesion classification.
2.3.1. Accuracy of PRA Detector
Average Precision (AP) is commonly used to measure the accuracy of object detection models. We used an IoU of 0.5 to calculate AP, which is also known as AP50. Average precision considers both the precision and recall detection performance. A detection is considered successful using the IoU by checking whether a bounding box overlaps with the corresponding groundtruth box by a minimum of 50%.
2.3.2. Accuracy of Periapical Lesion Classifier
We used accuracy, sensitivity, and specificity to measure the performance of the periapical lesion classifier. These classification metrics were calculated as below:
where TP is True Positive, TN is True Negative, FP is False Positive, and FN is False Negative.
3. Results
Our proposed solution consisted of two main CNNs: PRA detector and lesion classifier. First, the entire panoramic radiograph was used as an input to the presented system. Then the detection model (i.e., detector) was used to process the panoramic radiograph to localized and define the boundaries of each PRA by a bounding box. Finally, the PRAs extracted by the system, were fed them to the PL Classifier. The PL Classifier in turn classified each cropped PRA as Periapical Lesion (PL) or Healthy (H). To automate the process of periapical lesion localization and classification, our proposed solution combined the detection and classification models as shown in
Figure 1. The system final output was an image showing the periapical lesion on the OPG.
Our proposed detector identified the PRAs using a custom function based on the model aster R-RCNN. We tested two strategies to train our detector. In the first strategy (method 1) we only included unhealthy PRA (PL) for the training, while for the second strategy (method 2) we included both healthy (H) and unhealthy (PL). We trained for 1000 iterations and evaluated the detection performance of both methods based on Average Precision (AP) calculated at intersection-over-union (IOU) =0.5 on a test set of 143 images. Method 1 achieved AP50 of 38.5% while method 2 achieved AP50 of 74.95% at iteration 1000. We observed that training the detector on PRAs including H and PL classes achieved faster convergence and higher detection rates. We therefore adopted this approach in our further experiments. The proposed system then cropped and normalized all predicted bounding boxes that were then submitted to the classifier model.
Figure 2 shows one example of the output of the detection model. The detector was able to adapt to the size and extension of the periapical region automatically depending on the size and extension of the roots and the lesions. This was demonstrated by the high AP50 of the algorithm. Each periapical area was labeled independently regardless of whether it was found on single rooted or multi rooted teeth.
The periapical lesion classifier was responsible to label the bounding box of each detected PRAs as PL or H using a fine-tuned CNN. We trained and validated three classification models (VGG16, Inception v3, and Xception) on a balanced subset of the original dataset consisted of 3249 PRAs. As a baseline, all models were initialized from the ImageNet pre-trained weights and trained for 50 epochs. We calculated the accuracy, sensitivity, and specificity on a test set of 707 PRA to assess the performance of these classifiers at epoch 50 as shown in
Table 2. Our findings showed that Inception v3 outperformed both VGG16 and Xception in all metrics. Therefore, we selected Inception v3 as the classification model in the proposed architecture. We resumed training until convergence.
Figure 3 shows Inception v3’s learning curves and error rates, where it shows that the model converged on epoch 100. The final performance results of the Inception model are shown in
Table 3. In this test evaluating the performance of the final model, a total of 312 PRAs were successfully classified as H, and 282 PRAs were successfully classified as PL. While 48 PRAs were incorrectly classified as PL and 65 PRAs were incorrectly labelled as H. The classifier accuracy, sensitivity, and specificity were 84, 81, and 86, respectively.
The detection and classification models were combined in order to produce our overall solution that was able to automatically identify the periapical lesions in the OPGs with red bounding boxes corresponding to Periapical Lesions. These boxes included a confidence score that represented a percentage calculated by the AI indicating how sure the AI was the output is correct. The overall solution was tested on a test set of 299 PL on 143 panoramic radiographs. The accuracy, sensitivity, and specify of the overall solution were 84.6%, 72.2%, and 85.6%, respectively.
Figure 4 shows representative examples of the final output generated by the proposed method. This final tool was designed to detect periapical areas and ignore anything else in the radiograph, thus it was not affected by the presence of distractors and artifacts such as implants and crowns, also, the size of the detection box was able to adapt to the actual PRA, and it discriminated between different apices on the same tooth. On average, the proposed combined AI system took 2.3 s to detect and classify all PRAs as H or PL on a panoramic radiograph.
4. Discussion
The experimental results of this study showed the effectiveness of the proposed model to detect periapical lesions on panoramic radiographs. This model could be useful in clinical application for quick and easy detection of periapical lesions; on average, the proposed AI system takes 2.3 s to detect and classify all PRAs as H or PL on a panoramic radiograph.
In the present study, we proposed a two-stage deep learning architecture for periapical lesion detection on panoramic radiographs. We employed Faster-RCNN to localize PRAs and Inception v3 to classify PRA as PL or H. Breaking the task of PL detection into two steps, detection and classification, presented a faster learning convergence and higher detection rates (AP50 74.95%) compared to the baseline model in which we trained purely on PL samples (AP50 38.5%) The Inception-v3 classifier achieved the highest performance compared to other models (VGG16, Xception). When merging both detection and classification models, the overall accuracy, sensitivity, and specificity were 84.6%, 72.2%, and 85.6%, respectively.
Few studies addressed the problem of periapical lesions detection on panoramic radiographs using deep learning. Ekert et al. [
5] proposed a classification model using a small dataset (85 panoramic radiographs). The model was not sensitive enough to be used clinically, and was limited in its ability to automate the detection of the region of interest, PRAs, as it was trained on manually cropped patches of the individual teeth.
In our study, we proposed an AI architecture that can automatically detect and classify PL on an entire panoramic radiograph without the need for manual processing. Automatic evaluation of panoramic radiological images was also explored in another study that assessed the reliability of a CNN based automatic software “Diagnocat” [
26]. Upon evaluation of thirty panoramic radiographs, their CNN based automatic protocol showed very high sensitivity with respect to dental fillings, endodontically treated teeth, residual roots, periodontal bone loss, missing teeth, and prosthetic restorations. However, the reliability obtained for caries and periapical lesions assessments was unacceptable (ICC = 0.681 and 0.619, respectively).
Two other studies followed a segmentation approach to localize PL on panoramic radiographs [
27,
28]. Endres et al. [
27] presented a deep learning-based model trained on 2902 de-identified panoramic radiographs. In order to validate the algorithm 24 oral and maxillofacial surgeons assessed the presence or appearance of periapical radiolucencies on a separate set of panoramic radiographs. The findings of this study showed that the developed model outperformed 14 out of 24 surgeons. Their model achieved a precision of 67% and sensitivity of 51% on a test set of 102 radiographs. Bayrakdar et al. [
28] employed a UNet model trained on 470 panoramic radiographs to segment PLs. The model was tested on a small dataset consisting of 63 PL on 47 panoramic radiographs. The sensitivity, precision, and F1-score of UNet were 0.92, 0.84, and 0.88, respectively [
28]. Our study was validated in a much larger sample of 299 PL on 143 panoramic radiographs, and still we achieved high accuracy, sensitivity, and specificity (84.6%, 72.2%, and 85.6%, respectively).
In addition to OPG, other radiographic modalities have also been investigated for automated diagnosis of PL. In one study, CBCT images of 153 periapical lesions were evaluated by deep CNN, which detected 142 periapical lesions, along with the location and volume of lesions [
29]. Another study used using CNN on periapical radiographs showed the possibility of automatically identifying and judging periapical lesions with a success rate of as high as 92.75% [
11].
Diagnosing and documenting pathologies on dental radiographs is time-consuming and even though general and specialist dentists are well trained to do this, they are not exempt of human error. In fact, most complications in dental practice stem from misdiagnosis, which often involves missing out on noticing periapical lesions [
30]. In this sense, the proposed technology could help clinicians fill dental charts, and minimize diagnosis errors in the detection of periapical lesions. In fact, a previous study has already demonstrated that AI could outperform dental specialists in detection of apical lesions [
27].
This study has several limitations. It is based on panoramic radiographs from a single source restricted to patients from Brazil. Even though our model achieved excellent image recognition and detection results, data collection from multiple other sources and sites would be required in the future to increase the robustness of the algorithm and establish generalization of the applicability of the model across different sites. Another limitation of this study was the use of panoramic radiographs of only permanent dentition. Even though our study included panoramic radiographs for patients with a wide range of ages (between 11 and 84 years of age), mixed dentition was excluded, thus future studies would be needed to address an optimized algorithm for both permanent and mixed dentition. Another limitation of our study was that the OPGs used for training were only diagnosed visually by inspection of the radiograph; more accurate diagnosis necessitates the inclusion of clinical data such as percussion, thermal and electric pulp tests, which have not been taken into account here. Thus, future studies could further improve the performance of the algorithms by training them on images with diagnoses confirmed by a wider range of diagnostic techniques.