Predicting Multi-Gene Mutation Based on Lung Cancer CT Images and Mut-SeResNet
Abstract
:1. Introduction
2. Related Work
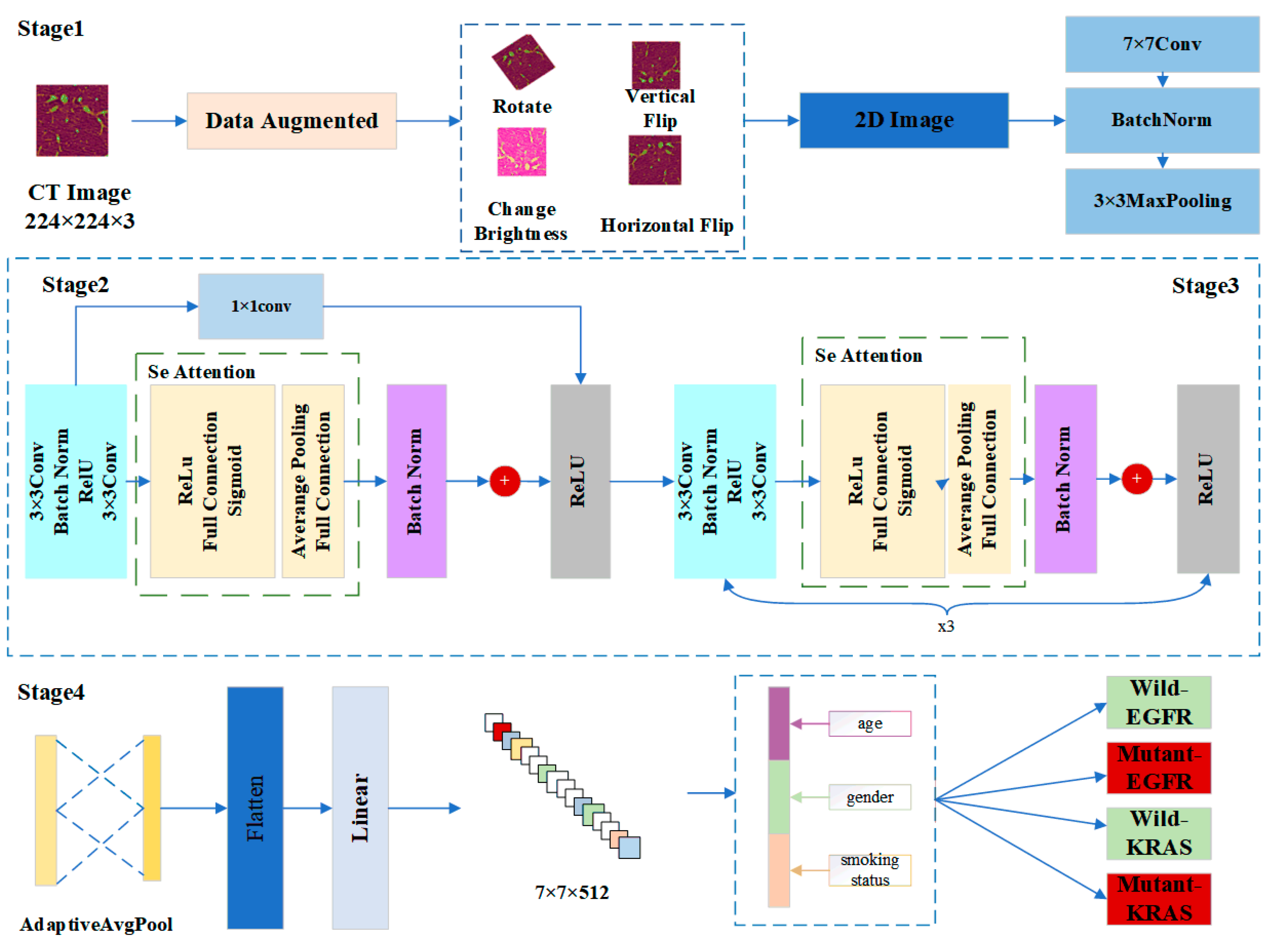
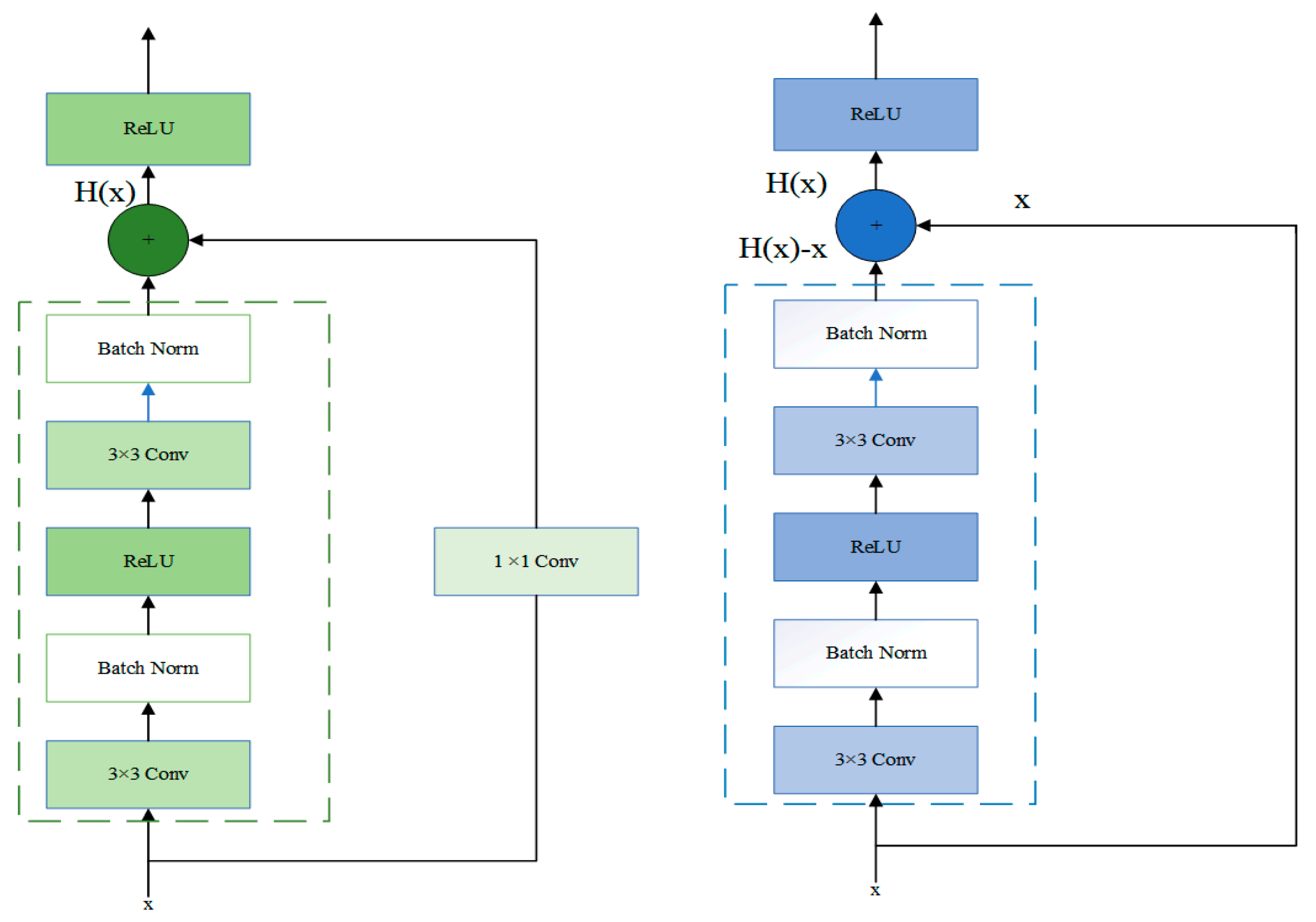
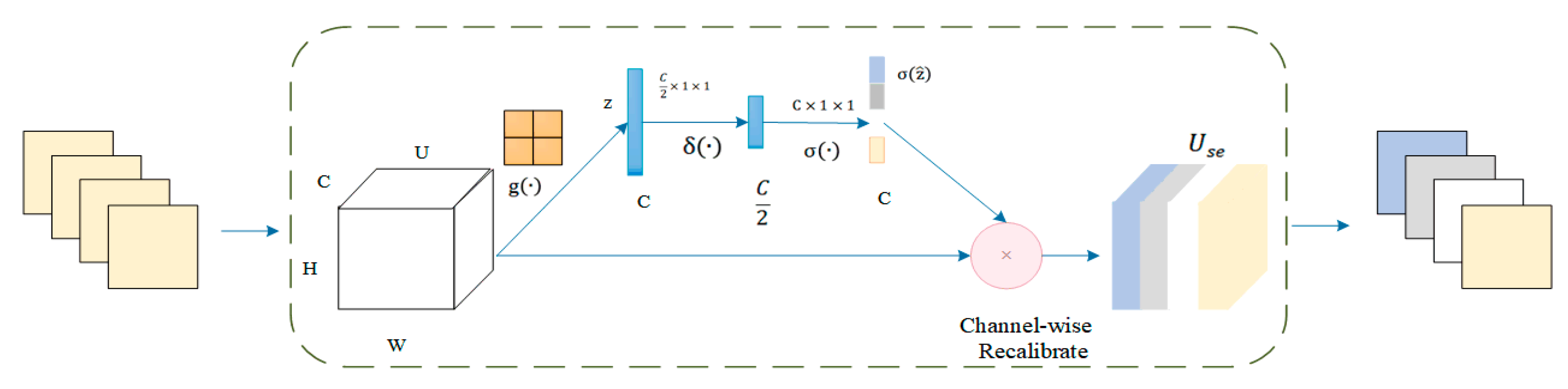
3. Materials and Methods
4. Results
4.1. Dataset Preparation
4.2. Experiment Result
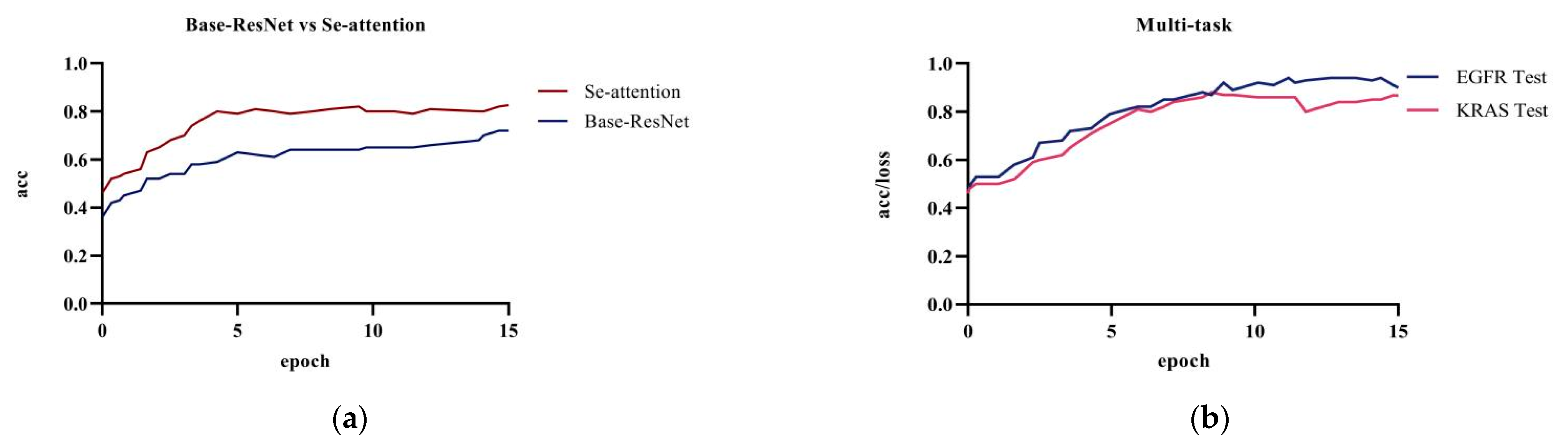
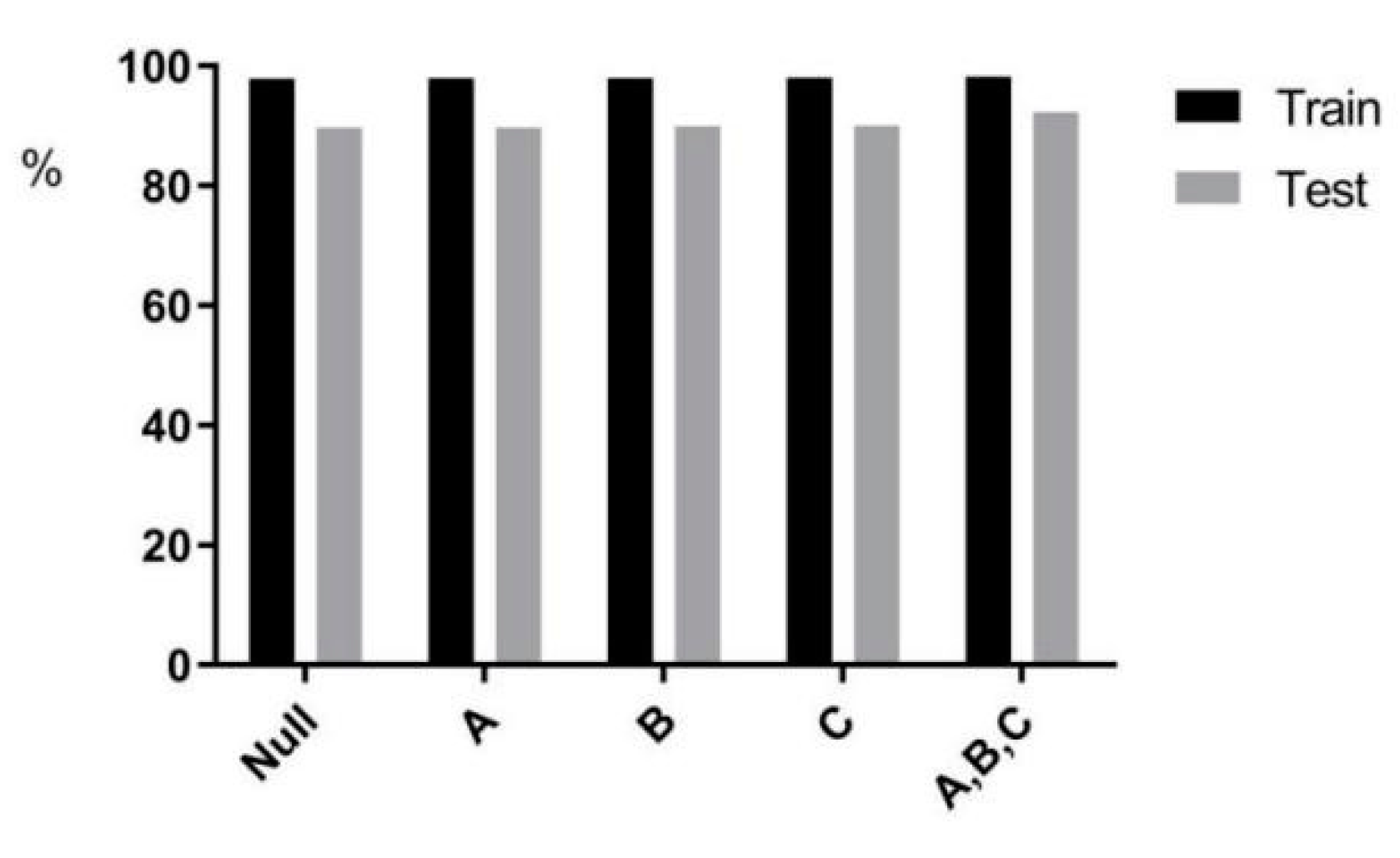
4.3. Ablation Experiment
4.4. Visualization of the Models
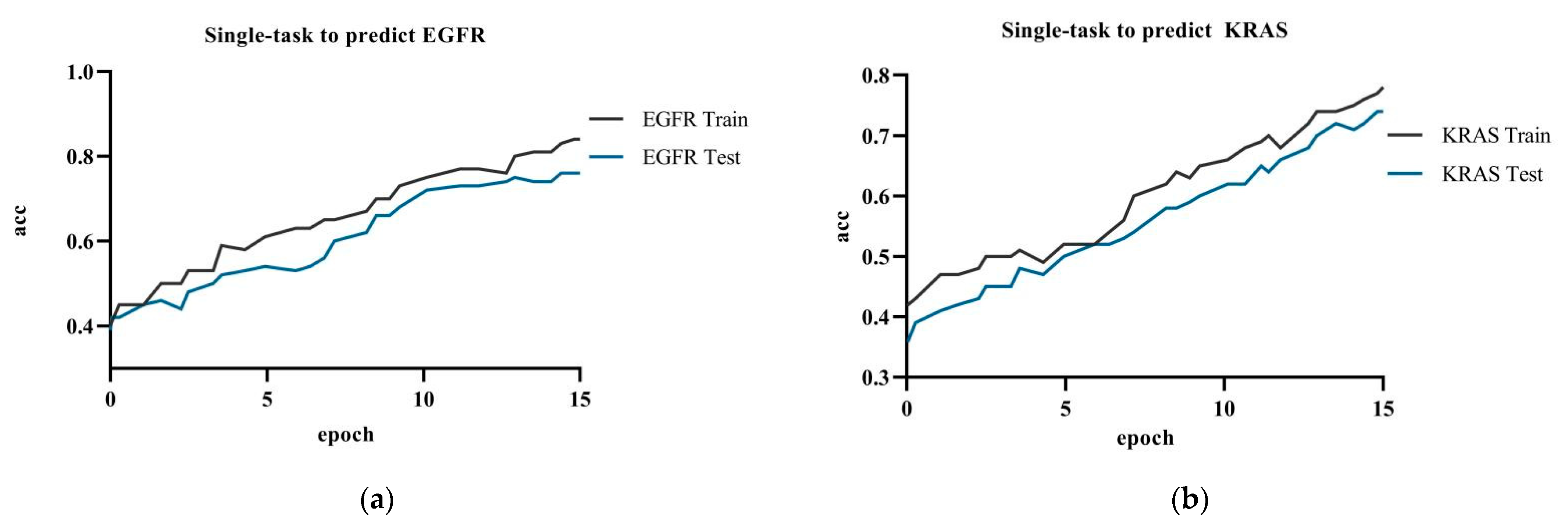
4.4.1. Single-Task versus Multi-Task Attention
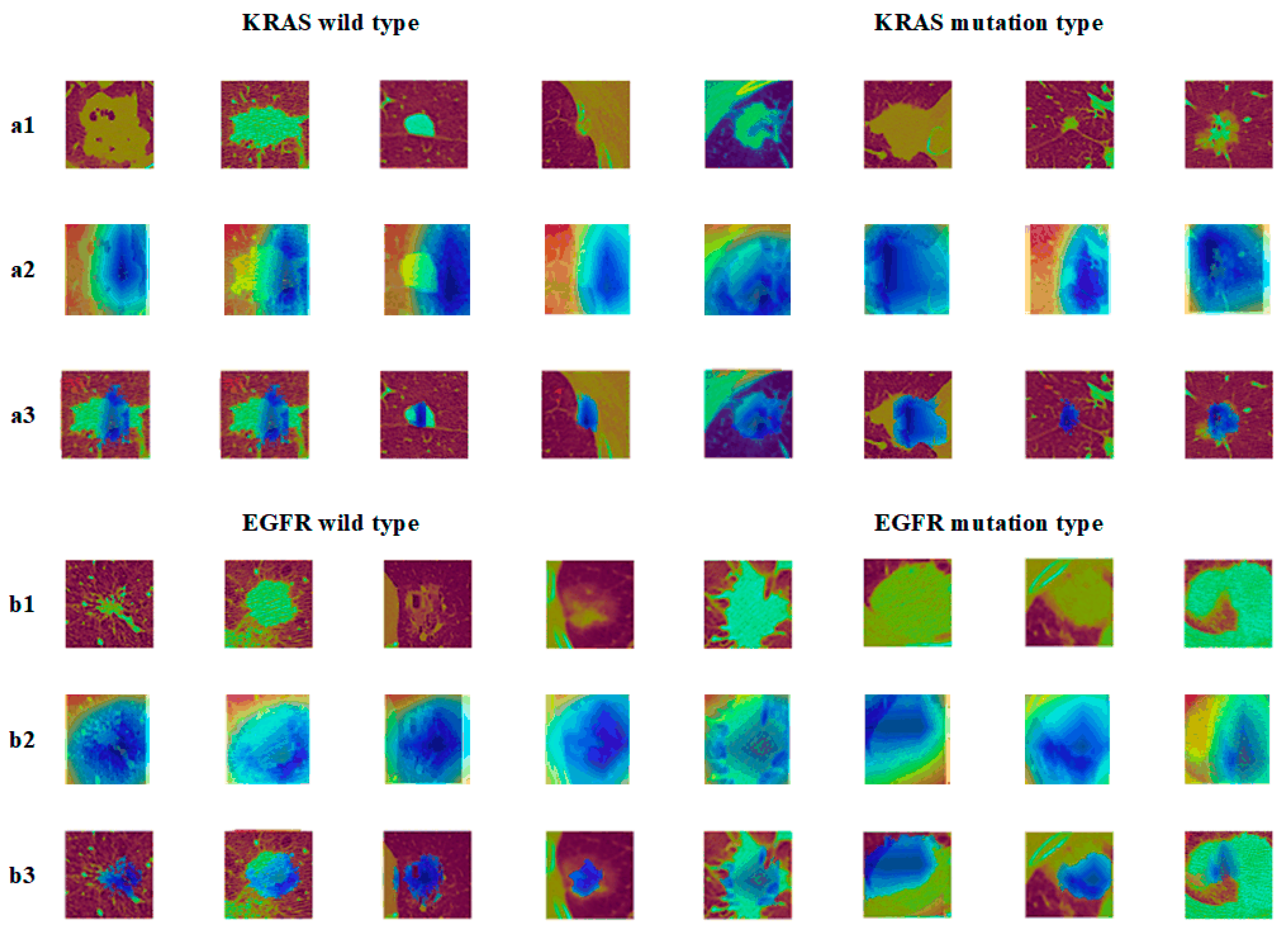
4.4.2. Nodules under Different Gene Mutation Types
5. Discussion
5.1. Impact of PatientPpersonal Information on Results
- It has the potential to support treatment decisions when surgical resection and puncture biopsy are difficult;
- It can be applied to the entire therapeutic EGFR mutation status, as CT images are repeatedly captured in clinical practice;
- CNN tools analyze the entire tumor, not just the tissue sample. They may be able to overcome the problem of heterogeneity;
- The use of CNN is a fast, easy, and low-cost method.
5.2. Comparison with Deep Methods
5.3. Comparison with Recent Studies and Methods
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Cent. 2022, 2, 1–9. [Google Scholar] [CrossRef]
- Liu, F.; Chen, Z.; Sun, P. Detection and segmentation of pulmonary nodules based on improved 3D VNet algorithm. SPIE 2022, 12176, 51–59. [Google Scholar]
- Goldstraw, P.; Ball, D.; Jett, J.R.; Le Chevalier, T.; Lim, E.; Nicholson, A.G.; Shepherd, F.A. Non-small-cell lung cancer. Nat. Rev. Dis. Prim. 2015, 1, 15048. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyo, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Motono, N.; Funasaki, A.; Sekimura, A.; Usuda, K.; Uramoto, H. Prognostic value of epidermal growth factor receptor mutations and histologic subtypes with lung adenocarcinoma. Med. Oncol. 2018, 35, 22. [Google Scholar] [CrossRef]
- Murphy, K.; van Ginneken, B.; Schilham, A.M.R.; de Hoop, B.J.; Gietema, H.A.; Prokop, M. A large scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classifification. Med. Image Anal. 2009, 13, 757–770. [Google Scholar] [CrossRef]
- Messay, T.; Hardie, R.C.; Rogers, S.K. A new computationally effificient CAD system for pulmonary nodule detection in CT imagery. Med. Image Anal. 2010, 14, 390–406. [Google Scholar] [CrossRef]
- Jacobs, C.; van Rikxoort, E.; Twellmann, T.; Scholten, E.; de Jong, P.; Kuhnigk, J.; Oudkerk, M.; de Koning, H.; Prokop, M.; Schaefer-Prokop, C.; et al. Automatic detection of subsolid pulmonary nodules in thoracic computed tomography images. Med. Image Anal. 2014, 18, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Ma, L.; Liu, X.; Fei, B. Computer-aided classification of lung nodules on CT images with expert knowledge. Proc. SPIE Int. Soc. Opt. Eng. 2021, 11598, 115982K. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.K.; Yang, E.; Chin, A.Y.; Park, M.; Moon, J.W.; Kim, J.H. Can deep learning model undergo the same process as a human radiologist when determining malignancy of pulmonary nodules? SPIE 2022, 12033, 844–848. [Google Scholar]
- Mahdy, A.M.S. A numerical method for solving the nonlinear equations of Emden-Fowler models. J. Ocean. Eng. Sci. 2022. [Google Scholar] [CrossRef]
- Mahdy, A.M.S.; Lotfy, K.; El-Bary, A.A. Use of optimal control in studying the dynamical behaviors of fractional financial awareness models. Soft Comput. 2022, 26, 3401–3409. [Google Scholar] [CrossRef]
- Prasoon, A.; Petersen, K.; Igel, C.; Lauze, F.; Dam, E.; Nielsen, M. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013: 16th International Conference, Nagoya, Japan, 22–26 September 2013; pp. 246–253. [Google Scholar]
- Cruz-Roa, A.A.; Arevalo Ovalle, J.E.; Madabhushi, A.; González Osorio, F.A. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2013: 16th International Conference, Nagoya, Japan, 22–26 September 2013; pp. 403–410. [Google Scholar]
- Li, R.; Zhang, W.; Suk, H.I.; Wang, L.; Li, J.; Shen, D.; Ji, S. Deep learning based imaging data completion for improved brain disease diagnosis. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014: 17th International Conference, Boston, MA, USA, 14–18 September 2014; pp. 305–312. [Google Scholar]
- Roth, H.R.; Lu, L.; Seff, A.; Cherry, K.M.; Hoffman, J.; Wang, S.; Liu, J.; Turkbey, E.; Summers, R.M. A new 2.5 D representation for lymph node detection using random sets of deep convolutional neural network observations. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014: 17th International Conference, Boston, MA, USA, 14–18 September 2014; pp. 520–527. [Google Scholar]
- Brosch, T.; Yoo, Y.; Li, D.K.B.; Traboulsee, A.; Tam, R. Modeling the variability in brain morphology and lesion distribution in multiple sclerosis by deep learning. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2014: 17th International Conference, Boston, MA, USA, 14–18 September 2014; Volume 8674, pp. 462–469. [Google Scholar]
- Shen, W.; Zhou, M.; Yang, F. Mutil-scale convolutional neural networks for lung nodule classification. In Proceedings of the International Conference on Information Processing in Medical Imaging, Isle of Skye, UK, 28 June–3 July 2015; pp. 588–599. [Google Scholar]
- Hussein, S.; Cao, K.; Song, Q. Risk stratification of lung nodules using 3d cnn-based multi-task learning. In Proceedings of the International Conference on Information Processing in Medical Imaging, Boone, NC, USA, 25–30 June 2017; pp. 249–260. [Google Scholar]
- Zhu, W.; Liu, C.; Fan, W.; Xie, X.H. Deeplung: 3d deep convolutional nets for automated pulmonary nodule detection and classfication. arXiv 2017, arXiv:1709.05538. [Google Scholar]
- Chen, Y.; Li, J.; Xiao, H.; Jin, X.; Yan, S.; Feng, J. Dual path networks. arXiv 2017, arXiv:1707.01629. [Google Scholar]
- Nibali, A.; He, Z.; Wollersheim, D. Pulmonary nodule classification with deep residual networks. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1799–1808. [Google Scholar]
- Ettinger, D.S.; Wood, D.; Akerley, W.; Bazhenova, L.; Borghaei, H.; Camidge, D.R.; Cheney, R.T.; Chirieac, L.R.; Damico, T.A.; Dilling, T.J. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 4.2016. J. Natl. Compr. Cancer Netw. 2016, 14, 255–264. [Google Scholar] [CrossRef]
- Nakatani, K.; Yamaoka, T.; Ohba, M.; Fujita, K.; Arata, S.; Kusumoto, S.; Takitakemoto, I.; Kamei, D.; Iwai, S.; Tsurutani, J. KRAS and EGFR Amplifications Mediate Resistance to Rociletinib and Osimertinib in Acquired Afatinib-Resistant NSCLC Harboring Exon 19 Deletion/T790M in EGFR. Mol. Cancer Ther. 2019, 18, 112–126. [Google Scholar] [CrossRef]
- Ye, Z. Genetic Mutations in Causing Non-small Cell Lung Cancers and Related Drugs’ Efficiency. In 2022 International Conference on Social Sciences and Humanities and Arts (SSHA 2022); Atlantis Press: Amsterdam, The Netherlands, 2022; pp. 1141–1145. [Google Scholar]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Zhang, J.; Zhao, X.; Zhao, Y.; Zhang, J.; Zhang, Z.; Wang, J.; Wang, Y.; Dai, M.; Han, J. Value of pre-therapy 18 F-FDG PET/CT radiomics in predicting EGFR mutation status in patients with non-small cell lung cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1137–1146. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excition networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Guan, Q.J.; Huang, Y.P. Multi-label chest X-ray image classification via category-wise residual attention learning. Pattern Recognit. Lett. 2020, 130, 259–266. [Google Scholar] [CrossRef]
- Velazquez, E.R.; Liu, Y.; Parmar, C.; Narayan, V.; Gillies, R.; Aerts, H. MO-DE-207B-08: Radiomic CT Features Complement Semantic Annotations to Predict EGFR Mutations in Lung Adenocarcinomas. Med. Phys. 2016, 43, 3706. [Google Scholar] [CrossRef]
- Ying, L.; Kim, J.; Balagurunathan, Y.; Qian, L.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated With EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441. [Google Scholar]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y. Predicting EGFR Mutation Status in Lung Adenocarcinoma on CT Image Using Deep Learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef]
- Rios Velazquez, E.; Parmar, C.; Liu, Y.; Coroller, T.P.; Cruz, G.; Stringfield, O.; Ye, Z.; Makrigiorgos, M.; Fennessy, F.; Mak, R.H.; et al. Somatic Mutations Drive Distinct Imaging Phenotypes in Lung Cancer. Cancer Res. 2017, 77, 3922–3930. [Google Scholar] [CrossRef]
- Gwerder, M.; Khan, A.; Neppl, C.; Zlobec, I. Detection of lung cancer metastases in lymph nodes using a multiple instance learning approach. SPIE 2022, 12039, 332–337. [Google Scholar]
- Xiong, J.F.; Jia, T.Y.; Li, X.Y.; Yu, W.; Xu, Z.Y.; Cai, X.W.; Fu, L.; Zhang, J.; Qin, B.J.; Fu, X.L.; et al. Identifying epidermal growth factor receptor mutation status in patients with lung adenocarcinoma by three-dimensional convolutional neural networks. Br. J. Radiol. 2018, 91, 20180334. [Google Scholar] [CrossRef]
- Yuchen, G.; Jialong, Z. Lung cancer diagnosis system based on 3D CNN. SPIE 2022, 12179, 255–263. [Google Scholar]
- Diao, Y.; Shi, Y.; Hou, Y.; Gao, Y.; Yu, H. Lung Cancer Detection with 3D Ensemble Convolution Neural Network. In Proceedings of the 2019 3rd International Conference on Computer Science and Artificial Intelligence, Normal, IL, USA, 6–8 December 2019; pp. 64–70. [Google Scholar]
- Sharma, M.; Bhatt, J.S.; Joshi, M.V. Early detection of lung cancer from CT images: Nodule segmentation and classification using deep learning. SPIE 2018, 10696, 226–233. [Google Scholar]


| Train Dataset (n = 363) | |||
|---|---|---|---|
| Sex (n/%) | EGFR (n/%) | ||
| Male | 156 (43%) | Mutant type | 164 (45.2%) |
| Female | 207 (57%) | Wild type | 199 (54.8%) |
| Smoking status (n/%) | Unknown or not collected | 0 | |
| Yes | 236 (65%) | KRAS (n/%) | |
| No | 127 (65%) | Mutant type | 84 (23.1%) |
| Age (n/%) | Wild type | 279 (76.9%) | |
| Minimum | 43 | Unknown or not collected | 0 (0%) |
| Maximum | 80 | EGFR&KRAS (n/%) | |
| Median | 62 | Mutant type | 0 |
| Wild type | 363 | ||
| Setting | Acc | |
|---|---|---|
| a | ResNet | 72.1% |
| b | ResNet+Attention mechanism | 82.6% |
| c | ResNet+Single-task | EGFR 76.1% KRAS 74.0% |
| d | ResNet+Multi-task | EGFR 87.8% KRAS 86.7% |
| Null | A | B | C | A, B, C | |
|---|---|---|---|---|---|
| Train | 97.90% | 97.92% | 97.99% | 98% | 98.2% |
| Test | 89.70% | 89.73% | 89.99% | 90% | 92.29% |
| Model | Data | EGFR (Single-Task) | KRAS (Single-Task) | EGFR (Mut-Task) | KRAS (Mut-Task) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc | Sen | Spe | Acc | Sen | Spe | Acc | Sen | Spe | Acc | Sen | Spe | ||
| (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | (%) | ||
| VGG16 | Train | 72.1 | 67.8 | 79.2 | 67.1 | 32.3 | 70.8 | 74.1 | 68.5 | 78.4 | 74.6 | 69.9 | 75.3 |
| Test | 70.3 | 65.0 | 78.4 | 65.8 | 60.1 | 68.4 | 72.9 | 67.27 | 76.9 | 73.3 | 67.8 | 76.8 | |
| InceptionV3 | Train | 63.7 | 60.5 | 67.1 | 62.4 | 61.0 | 64.9 | 65.18 | 64.82 | 66.2 | 64.1 | 63.2 | 65.6 |
| Test | 60.2 | 58.6 | 64.6 | 59.8 | 58.5 | 61.9 | 62.34 | 62.13 | 62.5 | 60.9 | 59.02 | 62.4 | |
| Inception-ResNet-v2 | Train | 71.0 | 69.2 | 71.4 | 67.5 | 66.2 | 69.0 | 71.29 | 68.56 | 73.4 | 69.2 | 67.1 | 71.1 |
| Test | 67.9 | 67.0 | 68.1 | 64.4 | 63.5 | 66.1 | 68.03 | 65.07 | 70.0 | 66.3 | 64.2 | 67.74 | |
| Mut-SeResnet | Train | 84.9 | 72.0 | 75.4 | 78.2 | 75.2 | 76.5 | 96.7 | 78.2 | 81.3 | 95.8 | 71.5 | 74.2 |
| Test | 76.1 | 68.9 | 71.8 | 74.0 | 70.3 | 73.3 | 89.7 | 71.28 | 76.6 | 88.3 | 68.2 | 75.2 | |
| Literature | Number of Patients | Images | Extracted Features | AUC | Acc (%) | Sen (%) | Spe (%) | |
|---|---|---|---|---|---|---|---|---|
| A | Wang [36] | 14926 | CT | Clinical features | 0.85 | - | - | - |
| B | Eur Respir J [37] | 285 | CT | Radiomics features | EGFR (0.82) KRAS (0.67) | - | - | |
| C | Gwerder Mauro et al [38] | 668 | CT | Radiomics features | 0.903 | 84 | - | - |
| D | Xiong [39] | 503 | CT | Deep features, clinical features | 0.838 | 77.20 | 75.80% | 79.10% |
| E | Yu chen Ge et al [40] | DSB17 | CT | Deep features | - | 64 | - | - |
| F | Yongdong Diao et al [41] | - | CT | Clinical features | - | 82.89 | - | - |
| G | Manu Sharma et al [42] | 6306 | CT | Deep features | - | 84.13 | 91.69% | 73.16% |
| Mut-SeResNet | 363 | CT | Deep features, clinical features | EGFR (89.7%) KRAS (88.3%) | EGFR (71.28) KRAS (68.2) | EGFR (76.6) KRAS (75.2) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Dong, Y.; Xu, S.; Feng, X.; Fan, X. Predicting Multi-Gene Mutation Based on Lung Cancer CT Images and Mut-SeResNet. Appl. Sci. 2023, 13, 1921. https://doi.org/10.3390/app13031921
Sun L, Dong Y, Xu S, Feng X, Fan X. Predicting Multi-Gene Mutation Based on Lung Cancer CT Images and Mut-SeResNet. Applied Sciences. 2023; 13(3):1921. https://doi.org/10.3390/app13031921
Chicago/Turabian StyleSun, Lichao, Yunyun Dong, Shuang Xu, Xiufang Feng, and Xiaole Fan. 2023. "Predicting Multi-Gene Mutation Based on Lung Cancer CT Images and Mut-SeResNet" Applied Sciences 13, no. 3: 1921. https://doi.org/10.3390/app13031921
APA StyleSun, L., Dong, Y., Xu, S., Feng, X., & Fan, X. (2023). Predicting Multi-Gene Mutation Based on Lung Cancer CT Images and Mut-SeResNet. Applied Sciences, 13(3), 1921. https://doi.org/10.3390/app13031921





