Featured Application
Convolutional neural networks can accurately identify the Inferior Alveolar Canal, rapidly generating precise 3D data. The datasets and source code used in this paper are publicly available, allowing the reproducibility of the experiments performed.
Abstract
Introduction: The need of accurate three-dimensional data of anatomical structures is increasing in the surgical field. The development of convolutional neural networks (CNNs) has been helping to fill this gap by trying to provide efficient tools to clinicians. Nonetheless, the lack of a fully accessible datasets and open-source algorithms is slowing the improvements in this field. In this paper, we focus on the fully automatic segmentation of the Inferior Alveolar Canal (IAC), which is of immense interest in the dental and maxillo-facial surgeries. Conventionally, only a bidimensional annotation of the IAC is used in common clinical practice. A reliable convolutional neural network (CNNs) might be timesaving in daily practice and improve the quality of assistance. Materials and methods: Cone Beam Computed Tomography (CBCT) volumes obtained from a single radiological center using the same machine were gathered and annotated. The course of the IAC was annotated on the CBCT volumes. A secondary dataset with sparse annotations and a primary dataset with both dense and sparse annotations were generated. Three separate experiments were conducted in order to evaluate the CNN. The IoU and Dice scores of every experiment were recorded as the primary endpoint, while the time needed to achieve the annotation was assessed as the secondary end-point. Results: A total of 347 CBCT volumes were collected, then divided into primary and secondary datasets. Among the three experiments, an IoU score of 0.64 and a Dice score of 0.79 were obtained thanks to the pre-training of the CNN on the secondary dataset and the creation of a novel deep label propagation model, followed by proper training on the primary dataset. To the best of our knowledge, these results are the best ever published in the segmentation of the IAC. The datasets is publicly available and algorithm is published as open-source software. On average, the CNN could produce a 3D annotation of the IAC in 6.33 s, compared to 87.3 s needed by the radiology technician to produce a bidimensional annotation. Conclusions: To resume, the following achievements have been reached. A new state of the art in terms of Dice score was achieved, overcoming the threshold commonly considered of 0.75 for the use in clinical practice. The CNN could fully automatically produce accurate three-dimensional segmentation of the IAC in a rapid setting, compared to the bidimensional annotations commonly used in the clinical practice and generated in a time-consuming manner. We introduced our innovative deep label propagation method to optimize the performance of the CNN in the segmentation of the IAC. For the first time in this field, the datasets and the source codes used were publicly released, granting reproducibility of the experiments and helping in the improvement of IAC segmentation.
1. Introduction
The very recent development of new technologies, particularly in the field of artificial intelligence, is leading to important innovations in the healthcare sector [1,2]. They range from surgical outcomes prediction to anatomical studies, the correlation between clinical and radiological findings, and even to genetic stratification and oncology [3,4,5,6,7,8]. Specifically, the so-called “image-based analysis” is currently highly investigated as the role of the human operator in the setting and reporting of radiological examinations could greatly benefit from technological support [9,10,11].
The preservation of noble anatomical structures is critical in surgery, and, in particular, in the head and neck district. Given the important number of surgical interventions daily performed on the mandible, we decided to focus our work on the segmentation of the inferior alveolar canal (IAC). To date, only a small number of studies regarding the automatic segmentation of the inferior alveolar canal have been conducted. Limited results in terms of accuracy have been achieved, with the exception of the recent paper published by Lahoud et al. [12]. Moreover, the need to obtain accurate data in a quick and automatic manner is a major concern in modern radiology.
Finally, the issue of free access to source codes and datasets in the deep learning field has received considerable critical attention. Reproducibility of the experiments performed cannot be assured if these data are kept private. Previous studies in the field of segmentation of the IAC have not dealt with the open-source sharing of the dataset and algorithm used.
The specific objective of this study was to develop a CNN which is able to overcome the results achieved so far and described in the literature, in order to quickly output a reliable and three-dimensional segmentation of the IAC. This research also sought to optimize the results obtained and to release them publicly together with the dataset and source-codes used to achieve them.
2. Literature Review
The following section of this paper will briefly examine the clinical importance of the IAC, and of its radiological investigation and the results obtained thus far in its segmentation with deep learning techniques.
2.1. The Inferior Alveolar Canal: Clinical Insights
In the field of mandibular surgery (trauma, dental, or reconstructive), the inferior alveolar bundle must be carefully identified and preserved as a noble structure [13]. Consisting of the inferior alveolar nerve and the homonymous vessels (artery and vein), it runs in a bony canal inside the mandible known as the “inferior alveolar canal” (IAC), or improperly called mandibular canal [14] (Figure 1). This canal begins at the level of the lingual aspect of the mandibular ramus (just above the Spix spine) and runs through the ramus, the angle and the mandibular body up to the mental foramen (usually located between the first and second premolar) [15,16]. Its identification during radiological examinations is a key moment in surgical planning [13].
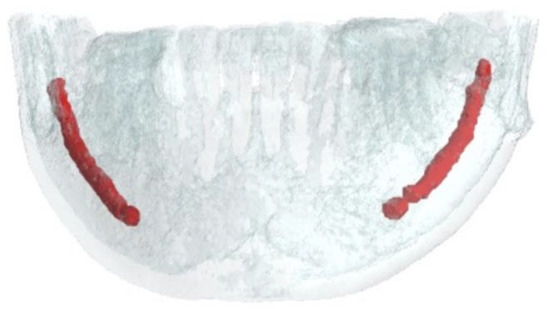
Figure 1.
Position of the Inferior Alveolar Canal within the mandible, 3D annotated on a CBCT volume.
Accurate preoperative identification of the inferior alveolar nerve (IAN) course is crucial in various maxillofacial surgical procedures, such as dental anesthesia, extraction of impacted third molars, placement of endosseous implants, mandibular osteotomies for orthognathic purposes, and fracture plating of the mandibular ramus or angle [17,18,19]. Even though permanent damage to the inferior alveolar bundle is rare, these surgeries can lead to temporary damage to the nerve function. Extraction of impacted teeth, in particular, is the most frequent cause of temporary damage (40.8%), which occurs in up to 7% of cases [20,21,22]. This is the relevant data, considering that dental extraction is one of the most common surgical interventions.
Endosseous dental implant is one of the most frequent dental procedures. However, the risk of neurosensory disorders following implant positioning in the mandible is quite high, and has been estimated at around 13.50/100 person-years (confidence interval (CI): 10.98–16.03) [23]. The persistence of the neurosensory deficit can occur as a result of incorrect positioning of the implant, and can be disabling for the patient [24]. An accurate pre-operative assessment of the three-dimensional position of the IAC within the mandible is recommended, thus allowing for proper planning of implant procedures.
Generally speaking, an injury of the IAN would result in a partial or total loss of lower lip sensitivity and ipsilateral lower dental arch, or painful dysesthesia, together with a proprioceptive alteration of the lower lip. Although not disabling in terms of mobility, neurological damage would certainly cause discomfort to the patient and should, therefore, be avoided if possible [25,26]. In addition, many patients who require elective surgery near the IAN are young and healthy [20]. Hence, it is essential to set up a pre-operative plan as accurately as possible to reduce post-operative complications.
2.2. Cone Beam Computed Tomography and Conventional Radiology
In maxillofacial and dental diagnostics, an important difference exists between two-dimensional and three-dimensional imaging. Orthopantomogram (OPG) radiograph is a two-dimensional radiological investigation, which may be adequate as a first level examination in some clinical conditions, such as mandibular apical-radicular dental lesions. As demonstrated by Vinayahalingam [27], deep learning techniques can also be applied to OPG to identify the IAC with excellent results. However, a three-dimensional study is required in case of more delicate circumstances, such as traumatology, extractions of impacted teeth, or implant surgery [28]. In such cases, the computed tomography (CT) is the main radiological examination. Both the conventional Multi-Detector CT (MDCT) and the more recent Cone Beam CT (CBCT) can be used in dental and maxillofacial surgical planning [29]. Compared to the first, CBCT guarantees an equal or superior image quality in the face with a reduced radiation exposure [30,31]. It should be emphasized that conventional CT is not the first choice for some diagnostic purposes in dentistry and oral surgery, such as the evaluation of impacted teeth and periapical lesions [32] due to the better spatial resolution, CBCT should be preferred in these situations. The lower cost compared to medical CT, the wide availability in out-of-hospital healthcare facilities and greater patient comfort during examination are other noticeable characteristics of CBCT [33]. Given these considerable advantages, the CBCT has therefore established itself as an important diagnostic tool in the oral and maxillofacial region [34,35,36,37]. Furthermore, CBCT application and usefulness in anatomical studies of mandibular structures have been widely described [38,39,40,41].
2.3. CBCT Image Processing
After the acquisition, the CBCT needs some technical adjustments before exporting the final images. First, the best axial slice (spline or panoramic base curve) for the annotations of the canal has to be identified. A curve is drawn on the selected axial slice, and it is usually placed at the equidistant point between the lingual and buccal cortices of the mandible. Since the mesial component of the IAC is usually clinically more relevant due to the presence of teeth, this curve is often shorter than the actual length of the IAC, which instead reaches up to the mandibular ramus. Once the annotation is complete, a coronal image similar to an OPG is generated. It is based on the axial slice selected and almost the entire 2D course of the IAC can be annotated on it. From here, the 2D annotation performed on this slice can be back projected to the original 3D volume, thus providing a 2D sparse annotation of the IAC on the original CBCT scan. Unfortunately, all these annotation steps are performed manually by a dedicated radiology technician, which is a waste of time and resources. The possibility of a machine masking these adjustments with great accuracy is therefore intriguing.
Moreover, 2D annotations fail to identify a considerable amount of inner information about the IAC position and the bone structure, i.e., the accurate contour, position, and diameters of the canal. An incomplete detection of the nerve positioning is often sufficient to facilitate a positive surgical outcome, but it is not an accurate anatomical representation of the IAC, and as such, it may lead to failure. On the other hand, manually producing a 3D annotation of the IAC would be practically unfeasible in the daily medical routine. In fact, this experimental operation requires almost one hour for every patient, and therefore, it is not applicable in the clinical practice. Automatic algorithms could certainly exceed this limitation.
2.4. Automatic Segmentation of the Inferior Alveolar Canal
Recently, great strides have been made in the field of deep learning and, in particular, the refinement of convolutional neural networks (CNN) has determined a strong boost in medical applications.
CNNs are artificial neural networks functionally inspired by the visual cortex. In fact, the animal brain system analyses stimuli in the visual field and interacts according to a layered scheme of increasing complexity. Likewise, convolution filters, constituting the CNNs, are responsible for learning features from the input image (s). A dot product between the filter itself and the image pixel values forms the convolution layer. Such a mathematical model has been successfully applied to both 2D and 3D segmentation, alongside several more computer vision tasks [3,42,43,44,45]. A CNN designed for segmentation purposes would be able to identify the three-dimensional IAC structure correctly without the need for manual adjustments. Unfortunately, the capabilities of convolutional neural networks are strongly limited by the lack of data carefully annotated, which instead are mandatory to train deep learning models. Indeed, despite the significant amount of raw data available in the field of dentistry/maxillofacial surgery, the CNN learning paradigm requires dense 3D annotations to reach its full potential. As previously mentioned, such data are extremely expensive to acquire.
In order to develop a CNN, a dataset to train the network is needed, together with a dataset for its validation. The validation set allows the tuning of the training parameter for the selected model on the specific use case. Finally, a testing dataset is required to evaluate the performance of the model obtained on a real case scenario. (Figure 2).
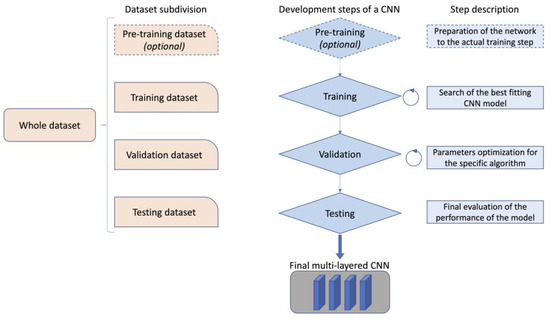
Figure 2.
Example of the functioning and development of a convolutional neural network.
Although investigated by some authors, there are still few works regarding the use of deep learning to identify the IAC. Classical computer vision algorithms for the automatic segmentation of the IAC in CBCT scans have been recently outclassed by modern machine learning and deep learning techniques [1,46,47,48,49,50,51]. In particular, the work from Jaskari et al. is certainly worth of mention [51]. Their CNN’s algorithm was developed upon 2D annotations of the IAC. These annotations have been extended to 3D using a circle expansion technique. This virtual reconstruction assumes that the IAC is regularly and geometrically tubular. The results obtained were certainly interesting, but they were based on an approximation of the real shape of the canal; consequently, the anatomical variants cannot be considered in this model type. Data related to a novel CNN recently described by Lahoud et al. established a new state of the art regarding the IAC segmentation [12]. Their results were developed on a 3D annotated dataset, but neither the dataset nor the algorithm used is publicly accessible, making impossible to verify and reproduce them, and making a proper comparison.
3. Materials and Methods
An experienced team of maxillofacial surgeons, engineers, radiologists, and radiology technicians have joined forces to carry out this scientific work. The research was approved by the local ethical committee, which also gave permission for the publication of the datasets and the source code used (protocol number: 1374/2020/OSS/AOUMO). The study was conducted according to the guidelines of the Declaration of Helsinki.
3.1. Dataset Generation and Annotation
All CBCTs were obtained using a NewTom/NTVGiMK4 machine (3 mA, 110 kV, 0.3 mm cubic voxels) (QR, Verona, Italy). The Affidea center in Modena (Italy) took care of the acquisition and export of the images in DICOM format. The scanned images were anonymized before being exported in DICOM format. Therefore, the only variables accessible to the investigators were the sex and the age of the patients. All images were captured between 2019 and 2020. All the samples are saved as 3D NumPy arrays with a minimum shape of [148, 265, 312] and a maximum shape of [178, 423, 463]. Hounsfield scale has been used to measure the radiodensity of CBCT volumes, and it ranges from −1000 to 4191. The IAC occupies only a small fraction of the whole volume, which is on average 0.1%.
Inclusion criteria were the following:
- Both male and female patients older than 12 years old;
- Patients whose identification of the IAC on CBCT images was performed either by a radiology technician or by a radiologist.
Exclusion criteria were the following:
- Unclear or unreadable CBCT images;
- Presence of gross anatomical mandible anomalies, including those related to previous oncological or respective surgery.
The images were then annotated to identify the course of the IAC.
Sparse annotations routinely performed by the radiology technician in daily clinical practice are present in all CBCTs. The steps of the sparse annotations performed were the following:
- Selection of the axial slice on which to base the subsequent extraction of the simil-OPG coronal slice (Figure 3a);
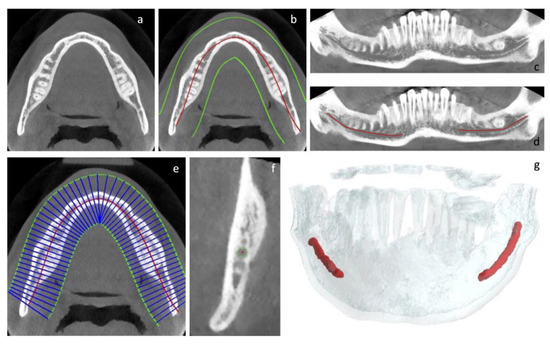 Figure 3. (a–g): Graphical description of the steps required to create the sparse and dense annotations of the CBCT volumes.
Figure 3. (a–g): Graphical description of the steps required to create the sparse and dense annotations of the CBCT volumes. - Drawing of the curve describing the course (base curve) of the canal in the context of the body of the mandible. This curve is a spline automatically identified using the algorithm described in our previous paper and manually adjusted by adding/moving control points when needed [52] (Figure 3b);
- Generation of the coronal slice, similar to the OPG. This is the curved plane perpendicular to the axial plane and containing the base curve (Figure 3c);
- Bidimensional annotation of the IAC course on the coronal plane (Figure 3d).
Different from above, a team of three maxillofacial surgeons devoted themselves to densely annotate CBCT volumes using our previously described tool [52]. Hence, a further dataset of CBCT volumes with both sparse and dense 3D annotations was obtained. The steps to realize it were as follows:
- Selection of the axial slice, on which to base the subsequent extraction of the simil-OPG coronal slice (Figure 3a);
- Generation of the coronal slice, similar to the OPG. This is the curved plane perpendicular to the axial plane and containing the base curve (Figure 3c);
- Bidimensional annotation of the IAC course on the coronal plane (Figure 3d);
- Automatic generation of Cross-Sectional Lines (CSLs), i.e., lines perpendicular to the base curve and always lying on the axial plane (Figure 3e);
- Generation and annotation of the Cross-Sectional Views (CSVs), obtained on the basis of the CLSs already described (Figure 3f). These views are planes containing the CSLs and perpendicular to the direction of the canal, which is derived from the bidimensional annotation previously obtained (Figure 3e);
- Generation of 3D volumes (Figure 3g).
The reader has probably noticed that steps from 1 to 4 of the two aforementioned annotation procedures overlap; nonetheless, the accurate annotations of CSVs in the latter make the difference to achieve a densely annotated dataset. Each step was double-checked by all members of the medical team. If the canal was not unequivocally visible in the CVSs, it was not marked in order to avoid erroneous annotations that could mislead the CNN.
To resume, the whole CBCT dataset obtained with the approach described has been divided in two main groups:
- A secondary dataset: it presents only sparse annotations, thus only showing the descriptive curve of the IAC course on the axial slice and the 2D canal identification on a coronal slice, similar to the OPG;
- A primary dataset: it has both sparse and dense annotations. The latter are all the 3D annotations of the canal, including those performed on the CSVs.
3.2. Model Description and Primary End-Point Measurement
The neural network employed for the experiments was built from scratch, following the description provided in the paper from Jaskari et al. [51]. This 3D segmentation method is based on the U-Net 3D fully convolutional neural network [42]. As classical CNNs, U-Net 3D contains a contraction and an expansion path. The former, also called the encoder, is used to capture the context in the image. The latter, the decoder, is a symmetric expanding path whose goal is to enable precise localization using transposed convolutions. In the encoder path, we employed three down sampling achieved by means of two convolution set, ReLU, and batch normalization, followed by a maxpooling layer. On the other hand, the decoder path is composed by three up sampling phases, each one composed by a convolution, ReLU, and batch normalization, followed by a transposed convolution. Skip connecting are employed among latent space with the same spatial resolution as in the standard UNet. All the layers are detailed in Appendix A. In this specific context, the decoder produces a probability map assigning to each voxel of the input volume a score between 0 and 1. This output score has been later assessed by the threshold to distinguish voxels inside and outside the volume.
The PyTorch-based [34] implementation of this model can be found on https://ditto.ing.unimore.it/maxillo/ (accessed on 28 February 2023).
The primary endpoint has been the measurement of the accuracy of the annotations created by the CNN, compared to the ground-truth volume generated by the medical team. Sørensen-Dice similarity (Dice) score and Intersection over union (IoU) measurements have been performed for every test. These allow us to calculate the similarity and the amount of overlapping between two objects. They both range between 0 and 1; the closer the value is to 1, the greatest is the accuracy of the model.
IoU is the Intersection over Union, and in this study, serves to measure the quality of model prediction. More specifically, we name P and G the set of foreground voxels in the predicted segmentation and ground-truth volume, respectively. The IoU is then calculated as:
where # is the cardinality of the set. If the prediction is equal to the ground-truth, the score is 1. If the predicted voxels have no overlap with the ground-truth, the score is 0.
The Dice score, also known as Sørensen–Dice index, is given by the following formula:
In order to ensure comparison with other papers, both Intersection over Union and Sørensen–Dice similarity (Dice score) are presented in the experimental Results section, even though their values are very strictly correlated. Their values have been calculated on the test set.
Three experiments with specific aims have been conducted. They are schematized in Figure 4 and listed below:
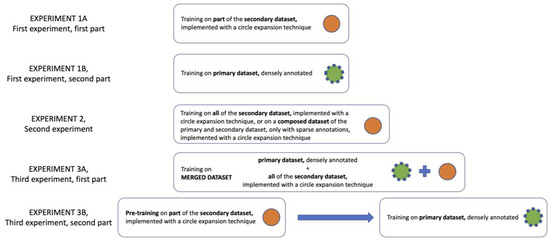
Figure 4.
Synthetic and schematic description of the experiments performed.
- In the first experiment, the CNN was trained twice on the primary dataset. First, only using the sparse annotations extended with the circular expansion technique (experiment 1A). Second, the CNN was trained on the same number of volumes, but this time using dense annotations (experiment 1B). The results in terms of IoU and Dice score were then compared.
- In the second experiment, the results of the CNN obtained with two different techniques were compared (experiment 2). In one case, the CNN was trained using only the secondary dataset with circular expansion. In the second case, the CNN was trained with a dataset composed of the whole secondary dataset and of the sparse annotations on the primary dataset, both undergoing a circular expansion.
- In the third experiment, two attempts were made. In the first case, the CNN was trained on a cumulative dataset which included both the primary (with only dense annotations) and secondary datasets (experiment 3A). In the second case, however, a pre-training of CNN was performed using the secondary dataset, and subsequently, the pre-trained CNN was actually trained on the primary dataset with 3D dense annotations only (experiment 3B).
For what concerns the pre-training, we have performed a total of 28 epochs, while the training phase counts a total of 100 epochs. In both cases, we have used Stochastic Gradient Descent (SGD) as an optimizer with an initial learning rate of 0.1 that would be halved if no improvements on the evaluation loss are detected for 7 successive epochs, a batch size of 6 and a patch shape of 80 × 80 × 80 pixels.
As for the circular expansion technique, it can be briefly described in the following passages:
- For each point in the sparse annotation, the direction of the canal is first determined using the coordinates of the next point.
- A 1.6 voxel-long radius is computed to be orthogonal to the direction of the canal in that point, and a circle is drawn.
- The radius length is set to ensure a circle diameter of 3 mm in real-world measurements. This unit can differ due to the diverse voxel spacing specified in the patient DICOM files (0.3 mm for each dimension, in our data).
- The previous step produces a hollow pipe-shaped 3D structure, that is finally filled with traditional computer vision algorithms.
A more detailed description of this technique can be found in the paper by Jaskari et al. [51].
3.3. Secondary End Point
A secondary endpoint has been established measuring the effective time needed by the radiology technician to annotate every CBCT during his normal work routine. Recently, this time has been compared with the time needed by the CNN to perform its annotations. The time has been measured in seconds and a simple t-test has been conducted (CI 95%, p value < 0.05).
4. Results
4.1. Demographic Data Collected
As already explained above, the volumes exported have been completely anonymized. The only demographic data collected were the patients’ age and sex. The total number of enrolled patients was 347, including 205 female patients and 142 male patients. The mean age of the patients was 50 years, with a median of 52 years, and a standard deviation of 19.78. The most represented age groups were those aged between 60 and 70 years, and those between 20 and 30 years. The youngest patient was 13 years old, while the oldest patient was 96 years old.
4.2. Radiographic Data and Subdivision of the Datasets
In order to train, validate, and test the CNN, the overall dataset was split into a primary and a secondary dataset. A total of 256 CBCT volumes composed the secondary dataset, containing only the sparse annotations already described.
A sample of 91 CBCTs (randomly selected) were densely annotated by the medical team, forming the primary dataset. This primary dataset was then divided in three sub-sections:
- A primary dataset for the training, made of 68 CBCTs;
- A primary dataset for the validation, made of 8 CBCTs;
- A primary dataset for the testing, made of 15 CBCTs.
The overall dataset, obtained from the sum of the primary and secondary datasets, is made of 347 CBCT volumes.
A graphic representation of the subdivision can be seen in Figure 5.
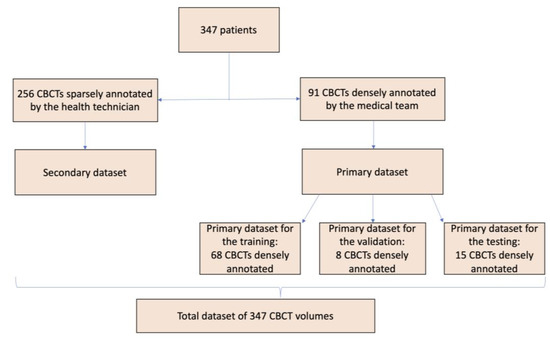
Figure 5.
Synthetic and schematic description of the division of the CBCT volumes to generate the different datasets used.
All datasets are available online, upon registration on the site, at https://ditto.ing.unimore.it/maxillo/ (accessed on 28 February 2023).
4.3. Results of the Experiments
4.3.1. Primary Endpoints
In the first experiment, the CNN was trained in two distinct ways. In the former case, it was trained on a dataset of 68 CBCTs with sparse annotations, subject to circular expansion. In the latter case, however, the CNN was trained using the same number of CBCTs, but this time using dense annotations. The IoU values obtained in the respective experiments were 0.39 and 0.56, while the Dice score values were 0.52 and 0.67, respectively.
In the second experiment, the CNN was trained with two different techniques. In the former scenario, the CNN was trained using only the secondary dataset with circular expansion, obtaining an IoU score of 0.42 and a Dice score of 0.60. In the latter scenario, the CNN was trained with a dataset composed of the whole secondary dataset and of the sparse annotations on the primary dataset, both undergoing a circular expansion, obtaining IoU and Dice score values of 0.45 and 0.62, respectively.
In the third and final experiment, we tried to implement the CNN in two other ways. In the former case, it was trained on all of the primary and secondary datasets, obtaining an IoU and Dice score of 0.45 and 0.62, respectively. In the latter case, it was decided to perform a pre-training on the secondary dataset and the pre-trained CNN was then actually trained on the primary dataset. The IoU obtained was 0.54, together with a Dice score of 0.69. Both values are higher than those obtained in the first experiment. In particular, we obtained a 3% improvement in the Dice score and a 4% improvement in the IoU score.
4.3.2. Optimization of the Algorithm
After the experiment 3B, we tried to further optimize the results obtained. In order to do that, we developed a novel label propagation technique. The deep expansion model is based on the same network used for segmentation, the main difference regards the input layer, which has been changed in order to accept a concatenation of both the raw volume data and the sparse annotations, rendered as a binary channel. This way, the network is allowed to exploit the information about the sparse 2D annotations to produce a 3D dense canal map, which is expected to be better than just using the raw volume as input. By applying this technique to the secondary dataset, we are able to generate a 3D dense annotation for all the patients that previously had only a sparse annotation, thus having a more informative dataset that can be used as a pretraining for the final task (i.e., segmenting the IAC by using as input only the raw volume). Moreover, a positional embedding patch was introduced in the algorithm. Since our sub-volumes have been extracted from the original scan following a fixed grid, we exploited positional information derived from the location of the top-left and bottom-right corners of the sub-volume. Exploiting positional information ensures two extremely important benefits:
- During training, the CNN is fed with implicit information about areas close to the edges of the scan where the IAN is very unlikely to be present.
- Information about cut positions helps the network to better shape the output: sub-volumes located close to the mental foramen generally present a much thinner canal than those located in the mandibular foramen.
These two adjustments granted more efficacy in the pre-training step and determined a further improvement of the results. The IoU score obtained was 0.64, while the Dice score reached 0.79.
To summarize, the training of our network was divided in two phases: Deep Label Propagation, and IAC segmentation. They both used a modified version of UNet3D, where the only difference was the number of input channels. During the Deep Label Propagation, our model took the original 3D volume and the sparse annotation as input, and it was trained to output a 3D segmentation. This process allowed to generate a dense segmentation for all the volumes in our dataset that only have a sparse segmentation. We refer to this data as synthetic (or generated) data. By using exclusively this data (i.e., 3D annotations obtained from 2D ones), we trained the same model to accept only the plain CBCT volume (with no IAC-related information) as input and to output the dense segmentation. As a final step we fine-tuned this latter model with the ground-truth 3D labels.
During both phases, we used a patch-based training. For example, sub-patches were extracted from the whole input, fed into the network, and aggregated back together. During the Deep Label Propagation, patches have a size of 120 × 120 × 120 and a batch size of 2. On the other hand, during the segmentation task, a patch size of 80 × 80 × 80 and batch size of 6 are employed. In both phases a learning rate of 0.1 with SGD as optimizer have been used for 100 epochs. The learning rate was decreased if no improvements in the evaluation were obtained.
The results of the three experiments are summarized in Table 1.

Table 1.
Summary of the results of all experiments. The closer to 1 is the value of the IoU or of the Dice score, the greatest is the accuracy of the model.
4.3.3. Secondary Endpoint
The secondary endpoint was based on the measurements of the timing necessary for the development of the annotated volumes. On average, the time spent by the radiology technician in daily clinical practice to annotate the volumes was 87.3 s to obtain a bidimensional annotation of the mandibular canal. On an NVIDIA Quadro RTX 5000, the inference process takes, on average, 6.33 s to produce a 3D dense annotation of the IAC.
5. Discussion
The inferior alveolar canal has always been a key topic in oral and maxillofacial surgery and several anatomical studies attempted to describe its course [16] accurately. The available literature analyses both the vertical and the horizontal canal path since the three-dimensional relationship with the surrounding structures is crucial in clinical practice [53,54,55]. Most of these topographic anatomical studies were performed on cadaver models [41]. However, improved imaging modalities have recently promoted radiological research. The growing diffusion of CBCT in common clinical practice explains the choice of this method for the present work. The experimental model we have applied might appear complex, but the data have been exploited in a different informative way. In fact, each test performed is independent and pursues specific objectives. We have proceeded to discuss the results one by one as reported below.
As shown in Figure 4, the first experiment was composed of two different parts.
In the first experiment, the CNN was trained in two distinct ways. In the former case, it was trained on a dataset of 68 CBCTs with sparse annotations subject to circular expansion. In the latter case, dense annotations based on the same number of CBCT volumes were used for CNN training.
The IoU values obtained were 0.39 (sparse mode) and 0.56 (dense mode), while the Dice score values were 0.52 (sparse mode) and 0.67 (dense mode). Thus, an improvement of the scores of 44% and 29% were found, respectively.
The substantial amelioration (IoU 0.39 vs. 0.52, and Dice Score 0.56 vs. 0.67) has to be related to the methodology applied. In fact, while in the experiment, 1A, a bidimensional annotation of the circularly expanded canal, was carried out; in the experiment 1B, a 3D voxel-based annotation system was used. The method with dense annotations ensures the elimination of IAC shape approximation biases, which instead characterizes experiments based on circular expansion. This is anatomically correct because the mandibular canal does not maintain a constant tubular shape and diameter, but it is rather subject to inter- and intra-individual variations along its path [16,54,56]. Exactly the same number of CBCT volumes were used in experiments 1A and 1B, and this must also be emphasized; the only differences relied on the type of annotations used. To conclude, the first experiment proves that dense-annotation based dataset is more accurate than the one based on the circular expansion technique only.
In the second experiment, the training of the CNN was performed on two circularly expanded datasets. These differed only in the number of CBCT volumes: in the former case, only the secondary dataset was used (256 CBCT volumes), while in the latter test, the secondary dataset was combined with the primary training dataset provided with sparse annotations only (total of 324 CBCT volumes). Despite a substantial rise in the CNN training volumes used in the second scenario (26.6% enhancement), the results were actually pretty similar. In the first case, the IoU and Dice scores were 0.42 and 0.60, while values of 0.45 and 0.62 were obtained by combining the datasets, respectively. Although they represent a good starting point for researchers, the data produced using the circular expansion technique appear to be limited and unsatisfactory according to these results.
The third and last experiment was divided in two different parts. In the former stage, the primary and secondary datasets were used together to train CNN resulting in an IoU score of 0.45 and a Dice score of 0.62. These results were not very satisfactory considering that these values are lower than those obtained in the first experiment. Moreover, in the experiment 1B, the overall dataset was smaller because only the primary dataset was used. We hypothesize a performance decrease due to the heterogeneity of the CNN training annotations used in experiment 3A. In fact, the CNN was trained at the same time on a dataset implemented by the circular expansion technique and on a dataset with voxel-based annotations.
Therefore, we sought to maximize the primary and secondary datasets in experiment 3B to overcome this limit. In particular, using the secondary dataset with sparse annotations and the circular expansion method, we effectively pre-trained the CNN. The use of pre-training has been shown to perform a role in improving CNNs performance [57]. Indeed, the pre-trained CNN was then properly trained with the primary dataset. The IoU score obtained was 0.54 and the Dice score was 0.69. Regarding both parameters, these were certainly the best overall results obtained.
The main difference between the experiments 1B and 3B consisted of the pre-training performed on the secondary dataset, which could have performed a key-role in providing the CNN a raw understanding of the IAC anatomy. This uptake was then refined with the primary dataset. Therefore, these results demonstrated the usefulness of the circular expansion technique mainly in the pre-training phase of developing a CNN.
The aim of this paper was to create a reliable CNN to three-dimensionally identify the mandibular canal and provide a public voxel-level annotations dataset of the Inferior Alveolar Canal. However, some considerations about the numerical results obtained are mandatory.
Comparing our results to the previously published ones, two main papers are to be considered [12,51]. When the results of this paper were collected, the previous best results were obtained by Jaskari et al. based on 637 CBCT volumes sparsely annotated. Using a circular expansion technique, they obtained a Dice score value of 0.58. It has to be noted that the data produced in this experiment are bidimensional. In the experiment 3B, we were able to overcome this amazing result, achieving a Dice score of 0.69 and an IoU score of 0.54. As explained, this was achieved by pre-training the CNN on a sparse annotated dataset and applying a circular expansion technique; subsequently, a voxel-level annotated dataset of the IAC was employed. Thus, our findings are superior both in terms of accuracy and amount of data compared to Jaskari’s, since the IAC is annotated on a 3D-base.
In a similar way, Lahoud et al. used 3D annotations of the IAC, obtaining a Dice score of 0.77 and an IoU score of 0.64 [12]. Similarly to our technique, Lahoud et al. also centered the annotation protocol on a three-dimensional basis. Nonetheless, this result was acquired using a much wider densely annotated dataset (235 CBCT volumes with 3D annotations) compared with the present study (91 CBCT volumes densely annotated).
Interestingly, in both cases (the present paper and Lahoud’s one), the number of volumes needed to overcome Jaskari’s results was significantly lower: Jaskari needed 637 CBCT volumes compared to Lahoud’s 235 and our 91 CBCT volumes [12,51]. These findings prove that the voxel-based annotation technique is much more informative and efficient than the circular expansion one.
Finally, we committed ourselves to overcome the result presented by Lahoud et al., who recorded a Dice score of 0.77 and an IoU score of 0.64 [12]. So, a new expansion technique of the sparsely-annotated dataset was added. In fact, we were able to transform the 2D-annotations of the IAC into 3D-voxel level information and we also introduced the positional concept in our algorithm (as explained in the subsection “4.3.2. Optimization of the algorithm”). We called this novel method “deep label propagation method” and further technical insights were described in a previous paper [58]. A Dice score of 0.79 and IoU score of 0.64 were recorded. While this achievement has an enormous room for improvement, it has to be said that the Dice score threshold of 0.75 is commonly considered an acceptable score for the use in clinical practice [12]. To the best of our knowledge, our result of 0.79 in terms of Dice score was the best ever recorded, setting a new state of the art in the field of the segmentation of the IAC. Once again, it is also interesting to notice that the number of CBCT volumes we used (91 volumes) was much lower than Lahoud’s one (235 volumes). Together with the better results achieved, this demonstrates the successfulness of the optimized algorithm, which could make the dataset much more informative even with a lower number of CBCTs.
It Is also interesting to observe the improvement of the results in the secondary end-point, i.e., the time required for the development of the annotated volumes. Cost-effectiveness in medicine, and especially in radiology, is becoming a critical topic. In recent years, there has been an increasing interest in extending the amount of radiologic data produced, while reducing the quantity of hours dedicated to their extraction. Therefore, we focused on time as a secondary end-point, in order to measure the effectiveness of our algorithm also in terms of processing speed. As previously stated, there has been a critical reduction in the amount of time required to segmentate the IAC: the CNN is able to accurately annotate it in less than 7.5% of the time needed by the radiology technician (6.33 s vs. 87.3 s). In addition, only the data produced by the present CNN are voxel-based, while the data composed by the radiology technician in the clinical practice are bidimensional.
So, the main advantages reached in these investigations are two: waste of time reduction and improvement in the quality of the information obtained. In fact, a few clicks are enough to obtain a reliable annotation of the mandibular canal using our CNN. During the daily work, it takes more than a minute for the radiology technicians to identify that structure. The time saved can allow the operator to devote his/her time to other tasks while the software performs the annotations independently. Moreover, the annotations conventionally obtained are bidimensional. On the contrary, a properly trained CNN can provide tridimensional identification of the mandibular canal, thus allowing for example a better pre-operative planning of a surgical intervention. The integration of our CNN in a multidisciplinary clinical practice can enable better cost-effectiveness. The beneficiaries will be many:
- The surgeon has access to a three-dimensional annotation of the IAC, thus being able to better visualize the data and better plan the surgical procedures (i.e., positioning of a dental implant or approaching to an impacted tooth).
- The radiologist can examine the CBCT volume and describe the IAC course more detailly.
- The standard of care would be improved, providing the patient a safer and more predictable morphological diagnosis.
- The waste of time for the radiology technician is minimized, while maximizing the amount of information provided to the clinicians.
- The radiology center would increase the cost-effectiveness of the CBCT exam.
A final consideration is to be made regarding the possibility to access the full anonymized dataset, which can be consulted at the following address: at https://ditto.ing.unimore.it/maxillo/ (accessed on 28 February 2023). Along with it, the source code was also made accessible to all. To the best of our knowledge, this is the first time in the medical scientific literature. Yet, we firmly believe in a common effort towards science development, and we hope this choice will encourage more researchers to do the same.
The main limitation of this paper is the low number of densely annotated CBCT volumes. This aspect will soon be improved by enhancing the number of voxel-based annotations of the mandibular canal in new CBCT volumes. Another possible improvement action concerns the study of anatomical variants due to the variability of the IAC shape, bifurcated or trifurcated forms are occasionally encounterable [59]. Furthermore, IAC characteristics differ according to age, sex, and especially oncologic pediatric mandibular surgery can be very delicate [16,60,61].
Other issues to be addressed are the identification of the canal in poor-resolution CBCTs and in cases of pathologies involving the IAC. Introducing these variables into the algorithm could further improve the CNN, together with an expansion of the available datasets. It is our intention to work further in this direction.
6. Conclusions
This paper aimed at developing a CNN capable of automatically and accurately identifying the IAC, starting from an analysis based on CBCT data. To achieve this goal, a new densely annotated 3D dataset has been collected for the first time. The annotations were performed in terms of voxels, thus guaranteeing 3D data and not virtual reconstructions of the IAC.
This preliminary study allowed to improve significantly the manual work of the radiologist/radiology technician. Therefore, the facilitation obtained can be the first step towards the subsequent implementation of the software, with the aim of obtaining a substantial integration between the contribution of the neural network and the human labor. In fact, in the context of modern surgery, obtaining 3D data regarding the anatomy of specific noble structures (e.g., the IAC) is increasingly fundamental. This work sets a new full stop in the segmentation of the inferior alveolar canal on CBCT volumes, recording on of the highest Dice scores currently existing. To summarize, we have demonstrated the following experimental results:
- EXP1A-B: a dataset with dense annotations is more accurate than a dataset which uses a circular expansion technique only;
- EXP2: the circle expansion technique can have limited results;
- EXP3A-B: a sparsely annotated dataset implemented with circle expansion technique can be helpful to pre-train a CNN. The algorithm was further improved thanks to our innovative deep label propagation method applied to the secondary dataset, to enhance the pre-training of the CNN. This crucial step allowed to achieve the best-ever Dice score recorded in the segmentation of the IAC. The results were also obtained with a considerably lower number of CBCT volumes compared to previously published papers in this field.
In addition, for the first time it was provided the free access to the volumes and source code used, so as to be able to work more and more towards the implementation of deep learning in surgical programming. They can be found upon registration at https://ditto.ing.unimore.it/maxillo/ (accessed on 28 February 2023). To the best of our knowledge, this is the first public maxillofacial dataset with voxel-level annotations of the Inferior Alveolar Canal.
Author Contributions
Conceptualization, M.D.B., A.P., F.B., C.G. and A.A.; methodology, S.N., P.M., R.N. and G.C.; software, L.L., M.C., S.A. and F.P.; formal analysis, M.D.B., A.P., F.B., L.L., F.P., R.N. and G.C.; investigation, M.C., S.N., L.C., C.G. and A.A.; resources, P.M., L.C., C.G. and A.A.; writing—original draft preparation, M.D.B., A.P., F.B., M.C., P.M. and R.N.; writing—review and editing, L.L., S.N., S.A., F.P., G.C., L.C., C.G. and A.A.; visualization, M.C., S.N. and F.P.; supervision, M.D.B., A.P., F.B., C.G. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of University Hospital of Modena (1374/2020/OSS/AOUMO approved on the 10 June 2021).
Informed Consent Statement
Patients consent was waived due to the retrospective nature of the study and the completely anonymization of the CBCT volumes dataset used.
Data Availability Statement
We grant the possibility of free access to both the source code and the datasets used to train and test the CNN. They can be found upon registration at https://ditto.ing.unimore.it/maxillo/ (accessed on 28 February 2023).
Acknowledgments
We want to acknowledge Marco Buzzolani for his commendable and precious work.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Internal structure of the network.
Table A1.
Internal structure of the network.
| Layer | Input Channels | Output Channels | Skip Connections |
|---|---|---|---|
| 3D Conv. Block 0 | 1 | 32 | No |
| 3D Conv. Block 1 + MaxPool | 32 | 64 | Yes |
| 3D Conv. Block 2 | 64 | 64 | No |
| 3D Conv. Block 3 + MaxPool | 64 | 128 | Yes |
| 3D Conv. Block 4 | 128 | 128 | No |
| 3D Conv. Block 5 + MaxPool | 128 | 256 | Yes |
| 3D Conv. Block 6 | 256 | 256 | No |
| 3D Conv. Block 7 + MaxPool | 256 | 512 | No |
| Transpose Conv. 0 | 513 | 512 | No |
| 3D Conv. Block 8 | 512 + 256 | 256 | Yes |
| 3D Conv. Block 9 | 256 | 256 | No |
| Transpose Conv. 1 | 256 | 256 | No |
| 3D Conv. Block 10 | 256 + 128 | 128 | Yes |
| 3D Conv. Block 11 | 128 | 128 | No |
| Transpose Conv. 2 | 128 | 128 | No |
| 3D Conv. Block 12 | 128 + 64 | 64 | Yes |
| 3D Conv. Block 13 | 64 | 64 | No |
| 3D Conv. Block 14 | 64 | 1 | No |
References
- Hwang, J.-J.; Jung, Y.-H.; Cho, B.-H.; Heo, M.-S. An overview of deep learning in the field of dentistry. Imaging Sci. Dent. 2019, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Crowson, M.G.; Ranisau, J.; Eskander, A.; Babier, A.; Xu, B.; Kahmke, R.R.; Chen, J.M.; Chan, T.C.Y. A contemporary review of machine learning in otolaryngology-head and neck surgery. Laryngoscope 2020, 130, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Allegretti, S.; Bolelli, F.; Pollastri, F.; Longhitano, S.; Pellacani, G.; Grana, C. Supporting Skin Lesion Diagnosis with Content-Based Image Retrieval. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021. [Google Scholar]
- Alkhadar, H.; Macluskey, M.; White, S.; Ellis, I.; Gardner, A. Comparison of machine learning algorithms for the prediction of five-year survival in oral squamous cell carcinoma. J. Oral Pathol. Med. 2021, 50, 378–384. [Google Scholar] [CrossRef]
- Almangush, A.; Alabi, R.O.; Mäkitie, A.A.; Leivo, I. Machine learning in head and neck cancer: Importance of a web-based prognostic tool for improved decision making. Oral Oncol. 2021, 124, 105452. [Google Scholar] [CrossRef]
- Chinnery, T.; Arifin, A.; Tay, K.Y.; Leung, A.; Nichols, A.C.; Palma, D.A.; Mattonen, S.A.; Lang, P. Utilizing Artificial Intelligence for Head and Neck Cancer Outcomes Prediction From Imaging. Can. Assoc. Radiol. J. 2021, 72, 73–85. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Hsueh, P.-C.; Wu, C.-C.; Tseng, Y.-J. Machine Learning Based Risk Prediction Models for Oral Squamous Cell Carcinoma Using Salivary Biomarkers. Stud. Health Technol. Inform. 2021, 281, 498–499. [Google Scholar] [CrossRef]
- Ariji, Y.; Yanashita, Y.; Kutsuna, S.; Muramatsu, C.; Fukuda, M.; Kise, Y.; Nozawa, M.; Kuwada, C.; Fujita, H.; Katsumata, A.; et al. Automatic detection and classification of radiolucent lesions in the mandible on panoramic radiographs using a deep learning object detection technique. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Brosset, S.; Dumont, M.; Bianchi, J.; Ruellas, A.; Cevidanes, L.; Yatabe, M.; Goncalves, J.; Benavides, E.; Soki, F.; Paniagua, B.; et al. 3D Auto-Segmentation of Mandibular Condyles. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; Volume 2020, pp. 1270–1273. [Google Scholar] [CrossRef]
- Qiu, B.; van der Wel, H.; Kraeima, J.; Glas, H.H.; Guo, J.; Borra, R.J.H.; Witjes, M.J.H.; van Ooijen, P.M.A. Automatic Segmentation of Mandible from Conventional Methods to Deep Learning-A Review. J. Pers. Med. 2021, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Lahoud, P.; Diels, S.; Niclaes, L.; Van Aelst, S.; Willems, H.; Van Gerven, A.; Quirynen, M.; Jacobs, R. Development and validation of a novel artificial intelligence driven tool for accurate mandibular canal segmentation on CBCT. J. Dent. 2021, 116, 103891. [Google Scholar] [CrossRef]
- Agbaje, J.O.; de Casteele, E.V.; Salem, A.S.; Anumendem, D.; Lambrichts, I.; Politis, C. Tracking of the inferior alveolar nerve: Its implication in surgical planning. Clin. Oral Investig. 2017, 21, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Iwanaga, J.; Matsushita, Y.; Decater, T.; Ibaragi, S.; Tubbs, R.S. Mandibular canal vs. inferior alveolar canal: Evidence-based terminology analysis. Clin. Anat. 2021, 34, 209–217. [Google Scholar] [CrossRef]
- Ennes, J.P.; Medeiros, R.M. de Localization of Mandibular Foramen and Clinical Implications. Int. J. Morphol. 2009, 27, 1305–1311. [Google Scholar] [CrossRef]
- Juodzbalys, G.; Wang, H.-L.; Sabalys, G. Anatomy of mandibular vital structures. Part I: Mandibular canal and inferior alveolar neurovascular bundle in relation with dental implantology. J. Oral Maxillofac. Res. 2010, 1, e2. [Google Scholar] [CrossRef]
- Komal, A.; Bedi, R.S.; Wadhwani, P.; Aurora, J.K.; Chauhan, H. Study of Normal Anatomy of Mandibular Canal and its Variations in Indian Population Using CBCT. J. Maxillofac. Oral Surg. 2020, 19, 98–105. [Google Scholar] [CrossRef]
- Oliveira-Santos, C.; Capelozza, A.L.Á.; Dezzoti, M.S.G.; Fischer, C.M.; Poleti, M.L.; Rubira-Bullen, I.R.F. Visibility of the mandibular canal on CBCT cross-sectional images. J. Appl. Oral Sci. 2011, 19, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Lofthag-Hansen, S.; Gröndahl, K.; Ekestubbe, A. Cone-beam CT for preoperative implant planning in the posterior mandible: Visibility of anatomic landmarks. Clin. Implant Dent. Relat. Res. 2009, 11, 246–255. [Google Scholar] [CrossRef]
- Weckx, A.; Agbaje, J.O.; Sun, Y.; Jacobs, R.; Politis, C. Visualization techniques of the inferior alveolar nerve (IAN): A narrative review. Surg. Radiol. Anat. 2016, 38, 55–63. [Google Scholar] [CrossRef]
- Libersa, P.; Savignat, M.; Tonnel, A. Neurosensory disturbances of the inferior alveolar nerve: A retrospective study of complaints in a 10-year period. J. Oral Maxillofac. Surg. 2007, 65, 1486–1489. [Google Scholar] [CrossRef]
- Monaco, G.; Montevecchi, M.; Bonetti, G.A.; Gatto, M.R.A.; Checchi, L. Reliability of panoramic radiography in evaluating the topographic relationship between the mandibular canal and impacted third molars. J. Am. Dent. Assoc. 2004, 135, 312–318. [Google Scholar] [CrossRef]
- Padmanabhan, H.; Kumar, A.V.; Shivashankar, K. Incidence of neurosensory disturbance in mandibular implant surgery—A meta-analysis. J. Indian Prosthodont. Soc. 2020, 20, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Al-Sabbagh, M.; Okeson, J.P.; Khalaf, M.W.; Bhavsar, I. Persistent pain and neurosensory disturbance after dental implant surgery: Pathophysiology’s etiology, and diagnosis. Dent. Clin. N. Am. 2015, 59, 131–142. [Google Scholar] [CrossRef]
- Leung, Y.Y.; Cheung, L.K. Risk factors of neurosensory deficits in lower third molar surgery: A literature review of prospective studies. Int. J. Oral Maxillofac. Surg. 2011, 40, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Van der Cruyssen, F.; Peeters, F.; Gill, T.; De Laat, A.; Jacobs, R.; Politis, C.; Renton, T. Signs and symptoms, quality of life and psychosocial data in 1331 post-traumatic trigeminal neuropathy patients seen in two tertiary referral centres in two countries. J. Oral Rehabil. 2020, 47, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Vinayahalingam, S.; Xi, T.; Bergé, S.; Maal, T.; de Jong, G. Automated detection of third molars and mandibular nerve by deep learning. Sci. Rep. 2019, 9, 9007. [Google Scholar] [CrossRef]
- Ueda, M.; Nakamori, K.; Shiratori, K.; Igarashi, T.; Sasaki, T.; Anbo, N.; Kaneko, T.; Suzuki, N.; Dehari, H.; Sonoda, T.; et al. Clinical Significance of Computed Tomographic Assessment and Anatomic Features of the Inferior Alveolar Canal as Risk Factors for Injury of the Inferior Alveolar Nerve at Third Molar Surgery. J. Oral Maxillofac. Surg. 2012, 70, 514–520. [Google Scholar] [CrossRef]
- Jacobs, R. Dental cone beam ct and its justified use in oral health care. J. Belg. Soc. Radiol. 2011, 94, 254. [Google Scholar] [CrossRef]
- Ludlow, J.B.; Ivanovic, M. Comparative dosimetry of dental CBCT devices and 64-slice CT for oral and maxillofacial radiology. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2008, 106, 106–114. [Google Scholar] [CrossRef]
- Signorelli, L.; Patcas, R.; Peltomäki, T.; Schätzle, M. Radiation dose of cone-beam computed tomography compared to conventional radiographs in orthodontics. J. Orofac. Orthop. 2016, 77, 9–15. [Google Scholar] [CrossRef]
- Saati, S.; Kaveh, F.; Yarmohammadi, S. Comparison of Cone Beam Computed Tomography and Multi Slice Computed Tomography Image Quality of Human Dried Mandible using 10 Anatomical Landmarks. J. Clin. Diagn. Res. 2017, 11, ZC13–ZC16. [Google Scholar] [CrossRef]
- Nasseh, I.; Al-Rawi, W. Cone Beam Computed Tomography. Dent. Clin. N. Am. 2018, 62, 361–391. [Google Scholar] [CrossRef]
- Schramm, A.; Rücker, M.; Sakkas, N.; Schön, R.; Düker, J.; Gellrich, N.-C. The use of cone beam CT in cranio-maxillofacial surgery. Int. Congr. Ser. 2005, 1281, 1200–1204. [Google Scholar] [CrossRef]
- Scarfe, W.C.; Farman, A.G.; Sukovic, P. Clinical applications of cone-beam computed tomography in dental practice. J. Can. Dent. Assoc. 2006, 72, 75–80. [Google Scholar] [PubMed]
- Anesi, A.; Di Bartolomeo, M.; Pellacani, A.; Ferretti, M.; Cavani, F.; Salvatori, R.; Nocini, R.; Palumbo, C.; Chiarini, L. Bone Healing Evaluation Following Different Osteotomic Techniques in Animal Models: A Suitable Method for Clinical Insights. Appl. Sci. 2020, 10, 7165. [Google Scholar] [CrossRef]
- Negrello, S.; Pellacani, A.; di Bartolomeo, M.; Bernardelli, G.; Nocini, R.; Pinelli, M.; Chiarini, L.; Anesi, A. Primary Intraosseous Squamous Cell Carcinoma of the Anterior Mandible Arising in an Odontogenic Cyst in 34-Year-Old Male. Rep. Med. Cases Images Videos 2020, 3, 12. [Google Scholar] [CrossRef]
- Matherne, R.P.; Angelopoulos, C.; Kulild, J.C.; Tira, D. Use of cone-beam computed tomography to identify root canal systems in vitro. J. Endod. 2008, 34, 87–89. [Google Scholar] [CrossRef]
- Kamburoğlu, K.; Kiliç, C.; Ozen, T.; Yüksel, S.P. Measurements of mandibular canal region obtained by cone-beam computed tomography: A cadaveric study. OFral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2009, 107, e34–e42. [Google Scholar] [CrossRef]
- Chen, J.C.-H.; Lin, L.-M.; Geist, J.R.; Chen, J.-Y.; Chen, C.-H.; Chen, Y.-K. A retrospective comparison of the location and diameter of the inferior alveolar canal at the mental foramen and length of the anterior loop between American and Taiwanese cohorts using CBCT. Surg. Radiol. Anat. 2013, 35, 11–18. [Google Scholar] [CrossRef]
- Kim, S.T.; Hu, K.-S.; Song, W.-C.; Kang, M.-K.; Park, H.-D.; Kim, H.-J. Location of the mandibular canal and the topography of its neurovascular structures. J. Craniofacial Surg. 2009, 20, 936–939. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation; Spring: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Bolelli, F.; Baraldi, L.; Pollastri, F.; Grana, C. A Hierarchical Quasi-Recurrent approach to Video Captioning. In Proceedings of the 2018 IEEE International Conference on Image Processing, Applications and Systems (IPAS), Sophia Antipolis, France, 12–14 December 2018; pp. 162–167. [Google Scholar]
- Pollastri, F.; Maroñas, J.; Bolelli, F.; Ligabue, G.; Paredes, R.; Magistroni, R.; Grana, C. Confidence Calibration for Deep Renal Biopsy Immunofluorescence Image Classification. In Proceedings of the 25th International Conference on Pattern Recognition, Milan, Italy, 10–15 January 2021; pp. 1298–1305. [Google Scholar]
- Kainmueller, D.; Lamecker, H.; Seim, H.; Zinser, M.; Zachow, S. Automatic extraction of mandibular nerve and bone from cone-beam CT data. Med. Image Comput. Comput. Assist. Interv. 2009, 12, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Moris, B.; Claesen, L.; Yi, S.; Politis, C. Automated tracking of the mandibular canal in CBCT images using matching and multiple hypotheses methods. In Proceedings of the 2012 Fourth International Conference on Communications and Electronics (ICCE), Hue, Vietnam, 1–3 August 2012; pp. 327–332. [Google Scholar]
- Abdolali, F.; Zoroofi, R.A.; Abdolali, M.; Yokota, F.; Otake, Y.; Sato, Y. Automatic segmentation of mandibular canal in cone beam CT images using conditional statistical shape model and fast marching. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, Y. Inferior alveolar canal segmentation based on cone-beam computed tomography. Med. Phys. 2021, 48, 7074–7088. [Google Scholar] [CrossRef]
- Kwak, G.H.; Kwak, E.-J.; Song, J.M.; Park, H.R.; Jung, Y.-H.; Cho, B.-H.; Hui, P.; Hwang, J.J. Automatic mandibular canal detection using a deep convolutional neural network. Sci. Rep. 2020, 10, 5711. [Google Scholar] [CrossRef] [PubMed]
- Jaskari, J.; Sahlsten, J.; Järnstedt, J.; Mehtonen, H.; Karhu, K.; Sundqvist, O.; Hietanen, A.; Varjonen, V.; Mattila, V.; Kaski, K. Deep Learning Method for Mandibular Canal Segmentation in Dental Cone Beam Computed Tomography Volumes. Sci. Rep. 2020, 10, 5842. [Google Scholar] [CrossRef]
- Mercadante, C.; Cipriano, M.; Bolelli, F.; Pollastri, F.; Di Bartolomeo, M.; Anesi, A.; Grana, C. A Cone Beam Computed Tomography Annotation Tool for Automatic Detection of the Inferior Alveolar Nerve Canal. In Proceedings of the 16th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications-Volume 4: VISAPP, Online, 8–10 February 2021; Volume 4, pp. 724–731. [Google Scholar]
- Heasman, P.A. Variation in the position of the inferior dental canal and its significance to restorative dentistry. J. Dent. 1988, 16, 36–39. [Google Scholar] [CrossRef]
- Rajchel, J.; Ellis, E.; Fonseca, R.J. The anatomical location of the mandibular canal: Its relationship to the sagittal ramus osteotomy. Int. J. Adult Orthod. Orthognath. Surg. 1986, 1, 37–47. [Google Scholar]
- Levine, M.H.; Goddard, A.L.; Dodson, T.B. Inferior alveolar nerve canal position: A clinical and radiographic study. J. Oral Maxillofac. Surg. 2007, 65, 470–474. [Google Scholar] [CrossRef]
- Sato, I.; Ueno, R.; Kawai, T.; Yosue, T. Rare courses of the mandibular canal in the molar regions of the human mandible: A cadaveric study. Okajimas Folia Anat. Jpn. 2005, 82, 95–101. [Google Scholar] [CrossRef]
- Clancy, K.; Aboutalib, S.; Mohamed, A.; Sumkin, J.; Wu, S. Deep Learning Pre-training Strategy for Mammogram Image Classification: An Evaluation Study. J. Digit. Imaging 2020, 33, 1257–1265. [Google Scholar] [CrossRef]
- Cipriano, M.; Allegretti, S.; Bolelli, F.; Pollastri, F.; Grana, C. Improving Segmentation of the Inferior Alveolar Nerve through Deep Label Propagation. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 19–20 June 2022; pp. 21105–21114. [Google Scholar]
- Valenzuela-Fuenzalida, J.J.; Cariseo, C.; Gold, M.; Díaz, D.; Orellana, M.; Iwanaga, J. Anatomical variations of the mandibular canal and their clinical implications in dental practice: A literature review. Surg. Radiol. Anat. 2021, 43, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Angel, J.S.; Mincer, H.H.; Chaudhry, J.; Scarbecz, M. Cone-beam computed tomography for analyzing variations in inferior alveolar canal location in adults in relation to age and sex. J. Forensic Sci. 2011, 56, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Pellacani, A.; Negrello, S.; Buchignani, M.; Nocini, R.; Di Massa, G.; Gianotti, G.; Pollastri, G.; Colletti, G.; Chiarini, L.; et al. Ameloblastoma in a three-year-old child with Hurler Syndrome (Mucopolysaccharidosis Type I). Reports 2022, 5, 10. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).