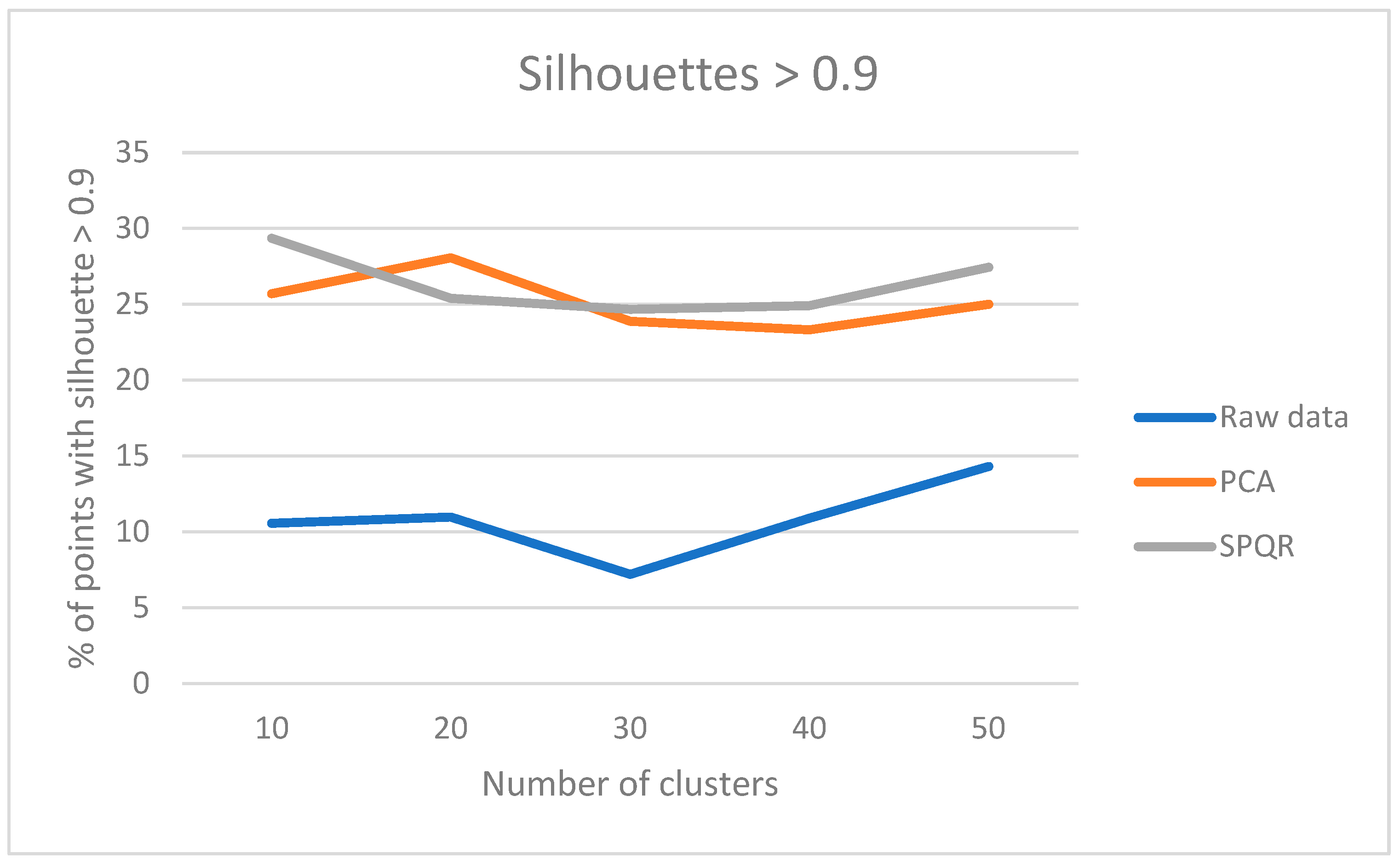
Figure 7.
Results of the “multicompare” procedure.
Figure 7.
Results of the “multicompare” procedure.
Table 1.
Clustering performance in terms of silhouettes using original data.
Table 1.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 43.95 | 37.44 | 43.28 | 50.46 | 60.81 |
| 0.8 | 21.60 | 16.29 | 29.31 | 39.40 | 51.45 |
| 0.9 | 0 | 0.40 | 16.75 | 35.92 | 37.340 |
Table 2.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 2.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 43.95 | 37.44 | 43.28 | 50.46 | 60.81 |
| 0.8 | 21.60 | 16.29 | 29.31 | 39.40 | 51.45 |
| 0.9 | 0 | 0.40 | 16.75 | 35.92 | 37.340 |
Table 3.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 3.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 92.54 | 76.06 | 78.93 | 82.49 | 87.80 |
| 0.8 | 89.55 | 69.64 | 69.59 | 75.41 | 74.40 |
| 0.9 | 78.59 | 47.83 | 57.753 | 46.103 | 48.463 |
Table 4.
Clustering performance in terms of silhouettes using normalized data.
Table 4.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0.06 | 0.08 | 0.17 | 0.08 | 0.23 |
| 0.8 | 0.06 | 0.08 | 0.10 | 0.08 | 0.15 |
| 0.9 | 0.06 | 0.08 | 0.06 | 0.06 | 0.15 |
Table 5.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 5.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0 | 1.92 | 1.96 | 0.02 | 0.17 |
| 0.8 | 0 | 0.04 | 0 | 0.02 | 0.17 |
| 0.9 | 0 | 0.04 | 0 | 0.02 | 0.17 |
Table 6.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 6.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 8.81 | 3.37 | 2.61 | 2.40 | 1.85 |
| 0.8 | 0 | 1.60 | 0 | 2.21 | 1.01 |
| 0.9 | 0 | 0 | 0 | 0.46 | 0.63 |
Table 7.
Clustering performance in terms of silhouettes using original data.
Table 7.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 61.85 | 68.70 | 59.29 | 67.41 | 56.31 |
| 0.8 | 55.32 | 49.05 | 31.20 | 39.98 | 46.01 |
| 0.9 | 50.29 | 34.87 | 5.56 | 9.22 | 22.68 |
Table 8.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 8.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 67.64 | 68.19 | 61.77 | 61.23 | 56.06 |
| 0.8 | 58.49 | 47.50 | 43.93 | 44.03 | 45.42 |
| 0.9 | 48.98 | 34.78 | 7.918 | 6.74 | 16.00 |
Table 9.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 9.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 63.34 | 54.01 | 62.71 | 62.11 | 57.87 |
| 0.8 | 46.83 | 35.16 | 46.13 | 26.06 | 40.93 |
| 0.9 | 4.68 | 13.31 | 2.59 | 3.29 | 19.58 |
Table 10.
Clustering performance in terms of silhouettes using normalized data.
Table 10.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 15.03 | 20.39 | 13.74 | 19.47 | 23.40 |
| 0.8 | 6.17 | 5.67 | 4.04 | 8.10 | 8.65 |
| 0.9 | 0 | 0.01 | 0.02 | 1.03 | 0.03 |
Table 11.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 11.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 51.43 | 41.16 | 38.56 | 37.08 | 32.20 |
| 0.8 | 32.08 | 21.46 | 21.27 | 17.01 | 14.96 |
| 0.9 | 4.34 | 0.01 | 1.58 | 0.83 | 3.14 |
Table 12.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 12.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 53.34 | 56.47 | 55.40 | 61.69 | 62.98 |
| 0.8 | 37.88 | 35.16 | 34.88 | 40.51 | 40.34 |
| 0.9 | 13.51 | 19.07 | 10.68 | 9.81 | 17.37 |
Table 13.
Clustering performance in terms of silhouettes using original data.
Table 13.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 48.91 | 25.48 | 21.60 | 16.59 | 15.48 |
| 0.8 | 27.31 | 14.24 | 8.83 | 8.06 | 7.062 |
| 0.9 | 0 | 5.59 | 3.35 | 2.24 | 0.82 |
Table 14.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 14.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.45 | 64.04 | 66.27 | 64.92 | 67.10 |
| 0.8 | 52.44 | 50.85 | 52.62 | 51.68 | 54.21 |
| 0.9 | 23.19 | 24.48 | 25.37 | 23.90 | 27.78 |
Table 15.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 15.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 63.39 | 61.86 | 59.98 | 59.86 | 59.74 |
| 0.8 | 50.68 | 47.38 | 46.44 | 46.08 | 47.14 |
| 0.9 | 22.31 | 19.36 | 18.07 | 17.83 | 17.83 |
Table 16.
Clustering performance in terms of silhouettes using normalized data.
Table 16.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 27.49 | 11.42 | 0.12 | 2.77 | 3.12 |
| 0.8 | 17.66 | 0 | 0.12 | 0 | 0 |
| 0.9 | 0 | 0 | 0.12 | 0 | 0 |
Table 17.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 17.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 25.60 | 17.89 | 10.24 | 12.48 | 5.59 |
| 0.8 | 11.48 | 14.12 | 9.71 | 4.00 | 2.77 |
| 0.9 | 0 | 0 | 0 | 0 | 0 |
Table 18.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 18.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 27.37 | 15.30 | 9.24 | 0.06 | 3.06 |
| 0.8 | 14.42 | 10.12 | 0.18 | 0.06 | 0.06 |
| 0.9 | 0 | 0 | 0 | 0.06 | 0.06 |
Table 19.
Clustering performance in terms of silhouettes using original data.
Table 19.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 38.64 | 36.29 | 36.33 | 12.09 | 36.51 |
| 0.8 | 38.17 | 21.03 | 21.16 | 11.91 | 21.34 |
| 0.9 | 17.74 | 17.74 | 17.78 | 0.17 | 17.96 |
Table 20.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 20.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.27 | 52.95 | 52.01 | 63.44 | 61.53 |
| 0.8 | 45.16 | 48.29 | 46.93 | 58.51 | 58.19 |
| 0.9 | 38.64 | 43.27 | 31.09 | 55.51 | 53.22 |
Table 21.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 21.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 48.47 | 48.81 | 48.76 | 48.56 | 52.93 |
| 0.8 | 37.09 | 40.36 | 40.32 | 40.12 | 48.31 |
| 0.9 | 21.84 | 29.05 | 29.00 | 40.12 | 39.8 |
Table 22.
Clustering performance in terms of silhouettes using normalized data.
Table 22.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 36.82 | 30.39 | 36.37 | 30.10 | 30.04 |
| 0.8 | 21.07 | 29.92 | 21.21 | 29.63 | 29.56 |
| 0.9 | 0.04 | 18.18 | 17.83 | 17.89 | 17.83 |
Table 23.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 23.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.27 | 58.06 | 58.71 | 58.75 | 58.51 |
| 0.8 | 45.16 | 52.86 | 53.67 | 54.45 | 54.83 |
| 0.9 | 38.64 | 34.83 | 45.43 | 49.84 | 50.33 |
Table 24.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 24.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 48.16 | 48.90 | 48.76 | 49.66 | 52.91 |
| 0.8 | 28.33 | 40.45 | 40.32 | 41.21 | 44.46 |
| 0.9 | 28.33 | 29.14 | 29.01 | 29.90 | 39.85 |
Table 25.
Clustering performance in terms of silhouettes using original data.
Table 25.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.92 | 57.86 | 42.13 | 40.45 | 42.13 |
| 0.8 | 45.50 | 38.20 | 25.28 | 24.15 | 28.65 |
| 0.9 | 10.67 | 7.86 | 6.18 | 10.11 | 14.61 |
Table 26.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 26.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 70.22 | 70.79 | 73.59 | 74.16 | 68.54 |
| 0.8 | 58.43 | 59.55 | 58.99 | 58.99 | 57.30 |
| 0.9 | 28.65 | 26.40 | 31.46 | 31.46 | 39.89 |
Table 27.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 27.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 65.73 | 71.35 | 75.28 | 74.16 | 74.16 |
| 0.8 | 52.81 | 60.11 | 61.80 | 57.30 | 60.11 |
| 0.9 | 21.91 | 31.46 | 34.83 | 29.77 | 41.57 |
Table 28.
Clustering performance in terms of silhouettes using normalized data.
Table 28.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0.56 | 1.12 | 1.68 | 1.68 | 1.123 |
| 0.8 | 0.56 | 0.56 | 1.12 | 1.68 | 0.56 |
| 0.9 | 0.56 | 0.56 | 1.12 | 1.68 | 0.56 |
Table 29.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 29.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 5.62 | 8.99 | 8.99 | 10.11 | 12.92 |
| 0.8 | 0 | 1.68 | 4.49 | 3.93 | 8.42 |
| 0.9 | 0 | 1.123 | 0 | 0 | 5.62 |
Table 30.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 30.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 14.61 | 14.61 | 10.67 | 19.10 | 14.61 |
| 0.8 | 3.93 | 4.49 | 3.93 | 8.99 | 8.43 |
| 0.9 | 0 | 0 | 3.37 | 2.81 | 2.81 |
Table 31.
Clustering performance in terms of silhouettes using original data.
Table 31.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 22.40 | 2.40 | 0.70 | 0.35 | 0.70 |
| 0.8 | 15.95 | 0 | 0 | 0 | 0 |
| 0.9 | 0 | 0 | 0 | 0 | 0 |
Table 32.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 32.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 56.55 | 32.30 | 24.95 | 24.10 | 23.45 |
| 0.8 | 38.65 | 10.90 | 7.10 | 8.50 | 6.400 |
| 0.9 | 15.65 | 0 | 0.35 | 0.35 | 0 |
Table 33.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 33.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 65.35 | 55.10 | 48.60 | 44.85 | 44.95 |
| 0.8 | 51.60 | 35.80 | 28.75 | 25.85 | 25.50 |
| 0.9 | 22.45 | 8.85 | 3.65 | 3.55 | 2.10 |
Table 34.
Clustering performance in terms of silhouettes using normalized data.
Table 34.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0 | 0.20 | 0.10 | 0.15 | 0 |
| 0.8 | 0 | 0.20 | 0.10 | 0.15 | 0 |
| 0.9 | 0 | 0.20 | 0.10 | 0.15 | 0 |
Table 35.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 35.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 21.05 | 8.70 | 6.90 | 2 | 1.40 |
| 0.8 | 4.15 | 0.40 | 0.65 | 0 | 0 |
| 0.9 | 0 | 0 | 0 | 0 | 0 |
Table 36.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 36.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 73.70 | 74.85 | 69.25 | 67.75 | 72.35 |
| 0.8 | 67.70 | 64.60 | 57.20 | 58.50 | 61.75 |
| 0.9 | 46.45 | 45.45 | 29.80 | 23.35 | 40.60 |
Table 37.
Clustering performance in terms of silhouettes using original data.
Table 37.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.15 | 59.83 | 60.83 | 60.52 | 59.70 |
| 0.8 | 48.18 | 45.64 | 46.20 | 45.95 | 45.34 |
| 0.9 | 17.24 | 16.09 | 16.72 | 15.99 | 14.38 |
Table 38.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 38.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.20 | 61.43 | 61.27 | 60.45 | 61.16 |
| 0.8 | 48.29 | 46.52 | 46.95 | 46.86 | 46.85 |
| 0.9 | 17.46 | 17.48 | 18.41 | 18.37 | 19.23 |
Table 39.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 39.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 62.26 | 61.13 | 61.31 | 61.02 | 60.66 |
| 0.8 | 48.14 | 47.12 | 47.61 | 46.80 | 46.85 |
| 0.9 | 17.40 | 17.43 | 18.61 | 18.40 | 18.11 |
Table 40.
Clustering performance in terms of silhouettes using normalized data.
Table 40.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 13.16 | 4.39 | 3.22 | 0.55 | 0.53 |
| 0.8 | 3.11 | 2.36 | 2.31 | 0 | 0 |
| 0.9 | 0.15 | 0.037 | 0.01 | 0 | 0 |
Table 41.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 41.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 30.07 | 22.94 | 19.43 | 17.23 | 17.13 |
| 0.8 | 10.10 | 6.74 | 5.55 | 3.91 | 4.74 |
| 0.9 | 0 | 0 | 0 | 0 | 0 |
Table 42.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 42.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 30.55 | 21.61 | 20.99 | 19.95 | 21.64 |
| 0.8 | 11.39 | 4.94 | 4.33 | 4.63 | 5.99 |
| 0.9 | 0 | 0 | 0 | 0 | 0 |
Table 43.
Clustering performance in terms of silhouettes using original data.
Table 43.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 11.94 | 37.07 | 29.13 | 45.43 | 45.31 |
| 0.8 | 11.31 | 32.07 | 20.36 | 39.45 | 39.34 |
| 0.9 | 9.70 | 27.02 | 5.54 | 35.07 | 34.99 |
Table 44.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 44.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 68.40 | 74.05 | 78.86 | 75.34 | 77.41 |
| 0.8 | 58.14 | 65.12 | 70.83 | 66.66 | 68.81 |
| 0.9 | 41.40 | 41.38 | 51.89 | 46.52 | 50.42 |
Table 45.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 45.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 90.66 | 85.51 | 81.28 | 84.09 | 79.63 |
| 0.8 | 85.30 | 80.79 | 76.01 | 78.15 | 72.96 |
| 0.9 | 81.17 | 65.34 | 59.48 | 63.08 | 56.15 |
Table 46.
Clustering performance in terms of silhouettes using normalized data.
Table 46.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 34.28 | 20.97 | 18.43 | 9.97 | 11.9 |
| 0.8 | 9.65 | 13.52 | 15.20 | 2.99 | 8.56 |
| 0.9 | 7.08 | 5.12 | 5.42 | 1.97 | 0.40 |
Table 47.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 47.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 48.17 | 25.93 | 21.18 | 16.80 | 12.46 |
| 0.8 | 18.63 | 15.13 | 14.07 | 8.93 | 4.05 |
| 0.9 | 8.01 | 2.48 | 1.94 | 0 | 1.16 |
Table 48.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 48.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 28.40 | 27.88 | 14.55 | 12.74 | 12.79 |
| 0.8 | 15.10 | 11.13 | 9.59 | 6.58 | 7.49 |
| 0.9 | 6.88 | 3.37 | 4.38 | 2.75 | 3.95 |
Table 49.
Clustering performance in terms of silhouettes using original data.
Table 49.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0.002 | 0.002 | 0.004 | 0.013 | 0.019 |
| 0.8 | 0.002 | 0.002 | 0.004 | 0.013 | 0.019 |
| 0.9 | 0.002 | 0.002 | 0.004 | 0.013 | 0.019 |
Table 50.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 50.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 10.06 | 7.16 | 4.99 | 6.56 | 3.79 |
| 0.8 | 0.82 | 2.91 | 2.88 | 2.858 | 0.68 |
| 0.9 | 0 | 1.54 | 1.42 | 1.19 | 0 |
Table 51.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 51.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 23.61 | 25.91 | 15.64 | 20.30 | 37.95 |
| 0.8 | 8.39 | 5.75 | 0.01 | 15.531 | 21.03 |
| 0.9 | 5.38 | 0.01 | 0.01 | 5.47 | 8.31 |
Table 52.
Clustering performance in terms of silhouettes using normalized data.
Table 52.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0.006 | 0.007 | 0.011 | 0.015 | 0.020 |
| 0.8 | 0.006 | 0.007 | 0.011 | 0.015 | 0.020 |
| 0.9 | 0.006 | 0.007 | 0.011 | 0.015 | 0.020 |
Table 53.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 53.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 13.45 | 10.93 | 5.20 | 2.15 | 1.46 |
| 0.8 | 1.46 | 4.32 | 3.18 | 0 | 0 |
| 0.9 | 0 | 1.40 | 1.46 | 0 | 0 |
Table 54.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 54.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 53.43 | 82.86 | 90.65 | 88.22 | 85.94 |
| 0.8 | 25.01 | 79.76 | 86.64 | 83.62 | 81.28 |
| 0.9 | 0 | 57.74 | 79.67 | 70.08 | 67.80 |
Table 55.
Clustering performance in terms of silhouettes using original data.
Table 55.
Clustering performance in terms of silhouettes using original data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0 | 0.18 | 0.09 | 0.28 | 0.37 |
| 0.8 | 0 | 0.18 | 0.09 | 0.28 | 0.37 |
| 0.9 | 0 | 0.18 | 0.09 | 0.28 | 0.37 |
Table 56.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
Table 56.
Clustering performance in terms of silhouettes using the reduced dataset with PCA.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 63.42 | 62.41 | 63.70 | 63.70 | 64.63 |
| 0.8 | 50.09 | 49.07 | 48.61 | 50.92 | 50.65 |
| 0.9 | 23.05 | 19.35 | 21.85 | 21.76 | 19.07 |
Table 57.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
Table 57.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 58.15 | 61.85 | 64.35 | 63.15 | 63.15 |
| 0.8 | 47.59 | 50.92 | 49.44 | 49.91 | 49.72 |
| 0.9 | 17.87 | 21.30 | 22.68 | 21.39 | 22.41 |
Table 58.
Clustering performance in terms of silhouettes using normalized data.
Table 58.
Clustering performance in terms of silhouettes using normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 0 | 0 | 0 | 0.09 | 0.18 |
| 0.8 | 0 | 0 | 0 | 0.09 | 0.18 |
| 0.9 | 0 | 0 | 0 | 0.09 | 0.18 |
Table 59.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
Table 59.
Clustering performance in terms of silhouettes using the reduced dataset with PCA applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 60.09 | 61.85 | 62.41 | 62.68 | 64.44 |
| 0.8 | 46.30 | 49.44 | 48.70 | 48.98 | 50.65 |
| 0.9 | 19.26 | 20.74 | 20.46 | 20.37 | 19.81 |
Table 60.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
Table 60.
Clustering performance in terms of silhouettes using the reduced dataset with SPQR applied to normalized data.
| Silhouette\N. Clusters | 10 | 20 | 30 | 40 | 50 |
|---|
| 0.7 | 61.11 | 62.78 | 59.91 | 61.941 | 61.851 |
| 0.8 | 48.981 | 50.181 | 45.551 | 48.051 | 48.421 |
| 0.9 | 19.26 | 20.83 | 17.78 | 18.80 | 20.37 |