Artificial Intelligence (AI)-Based Systems for Automatic Skeletal Maturity Assessment through Bone and Teeth Analysis: A Revolution in the Radiological Workflow?
Abstract
:1. Introduction
Introduction to AI Basic Terminology
2. Left Hand and Wrist Bone Age Assessment
2.1. Traditional Approaches
2.2. AI-Based Approaches
3. Dental Age Assessment
3.1. Traditional Approach
3.2. AI-Based Approach
4. Other Methods
4.1. Traditional Approaches
4.2. AI-Based Approaches
5. Challenges and Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mughal, A.M.; Hassan, N.; Ahmed, A. Bone age assessment methods: A critical review. Pak. J. Med. Sci. 2014, 30, 211–215. [Google Scholar] [CrossRef]
- Satoh, M. Bone age: Assessment methods and clinical applications. Clin. Pediatr. Endocrinol. 2015, 24, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Creo, A.L.; Schwenk, W.F. Bone Age: A Handy Tool for Pediatric Providers. Pediatrics 2017, 140, e20171486. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.D.; Wit, J.M.; Hochberg, Z.; Sävendahl, L.; van Rijn, R.R.; Fricke, O.; Cameron, N.; Caliebe, J.; Hertel, T.; Kiepe, D.; et al. The Use of Bone Age in Clinical Practice—Part 1. Horm. Res. Paediatr. 2011, 76, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, S.M. Prediction of adult height by Tanner-Whitehouse method in young Caucasian male athletes. QJM Int. J. Med. 2012, 106, 341–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.J.; Kwon, A.; Jung, M.K.; Kim, K.E.; Suh, J.; Chae, H.W.; Kim, D.H.; Ha, S.; Seo, G.H.; Kim, H.-S. Incidence and Prevalence of Central Precocious Puberty in Korea: An Epidemiologic Study Based on a National Database. J. Pediatr. 2019, 208, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Lee, Y.S.; Yu, J. Assessment of Bone Age in Prepubertal Healthy Korean Children: Comparison among the Korean Standard Bone Age Chart, Greulich-Pyle Method, and Tanner-Whitehouse Method. Korean J. Radiol. 2015, 16, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Karami, M.; Rabiei, M.; Riahinezhad, M. Evaluation of the pelvic apophysis with multi-detector computed tomography for legal age estimation in living individuals. J. Res. Med. Sci. 2015, 20, 209. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4468222/ (accessed on 17 January 2023).
- Greulich, W.W.; Pyle, S.I. Radiographic Atlas of Skeletal Development of the Hand and Wrist Professor of Anatomy; Stanford University School of Medicine: Stanford, CA, USA, 1959. [Google Scholar]
- Gilsanz, V.; Ratib, O. Hand Bone Age: A Digital Atlas of Skeletal Maturity; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–96. [Google Scholar] [CrossRef]
- Serinelli, S.; Panetta, V.; Pasqualetti, P.; Marchetti, D. Accuracy of three age determination X-ray methods on the left hand-wrist: A systematic review and meta-analysis. Leg. Med. 2011, 13, 120–133. [Google Scholar] [CrossRef]
- Kim, S.; Oh, Y.; Shin, J.; Rhie, Y.; Lee, K. Comparison of the Greulich-Pyle and Tanner Whitehouse (TW3) Methods in Bone age Assessment. J. Korean Soc. Pediatr. Endocrinol. 2008, 13, 50–55. [Google Scholar]
- Berst, M.J.; Dolan, L.; Bogdanowicz, M.M.; Stevens, M.A.; Chow, S.; Brandser, E.A. Effect of Knowledge of Chronologic Age on the Variability of Pediatric Bone Age Determined Using the Greulich and Pyle Standards. Am. J. Roentgenol. 2001, 176, 507–510. [Google Scholar] [CrossRef]
- Cunha, E.; Baccino, E.; Martrille, L.; Ramsthaler, F.; Prieto, J.; Schuliar, Y.; Lynnerup, N.; Cattaneo, C. The problem of aging human remains and living individuals: A review. Forensic Sci. Int. 2009, 193, 1–13. [Google Scholar] [CrossRef]
- Bull, R.K.; Edwards, P.D.; Kemp, P.M.; Fry, S.; Hughes, I.A. Bone age assessment: A large scale comparison of the Greulich and Pyle, and Tanner and Whitehouse (TW2) methods. Arch. Dis. Child. 1999, 81, 172–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, B.C.; Beattie, T.J.; Ramage, I.J.; Lucas, P.; Law, C.; Baird, J. Assessment of Skeletal Maturity and Prediction of Adult Height (TW3 Method), 3rd ed.; W.B. Saunders: London, UK, 2001. [Google Scholar]
- Lee, B.-D.; Lee, M.S. Automated Bone Age Assessment Using Artificial Intelligence: The Future of Bone Age Assessment. Korean J. Radiol. 2021, 22, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Aja-Fernández, S.; Garcia, R.D.L.; Martín-Fernández, M.; Alberola-Lopez, C. A computational TW3 classifier for skeletal maturity assessment. A Computing with Words approach. J. Biomed. Informatics 2004, 37, 99–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellinghaus, M.; Schulz, R.; Vieth, V.; Schmidt, S.; Schmeling, A. Forensic age estimation in living subjects based on the ossification status of the medial clavicular epiphysis as revealed by thin-slice multidetector computed tomography. Int. J. Leg. Med. 2009, 124, 149–154. [Google Scholar] [CrossRef]
- Soegiharto, B.M.; Moles, D.; Cunningham, S.J. Discriminatory ability of the skeletal maturation index and the cervical vertebrae maturation index in detecting peak pubertal growth in Indonesian and white subjects with receiver operating characteristics analysis. Am. J. Orthod. Dentofac. Orthop. 2008, 134, 227–237. [Google Scholar] [CrossRef]
- DiMéglio, A.; Charles, Y.P.; Daures, J.-P.; De Rosa, V.; Kaboré, B. Accuracy of the Sauvegrain Method in Determining Skeletal Age During Puberty. J. Bone Jt. Surg. 2005, 87, 1689–1696. [Google Scholar] [CrossRef]
- Canavese, F.; Charles, Y.P.; DiMeglio, A. Skeletal age assessment from elbow radiographs. Review of the literature. Chir. Organi Mov. 2008, 92, 1–6. [Google Scholar] [CrossRef]
- Michael, D.; Nelson, A. HANDX: A model-based system for automatic segmentation of bones from digital hand radiographs. IEEE Trans. Med. Imaging 1989, 8, 64–69. [Google Scholar] [CrossRef]
- Coppola, F.; Faggioni, L.; Regge, D.; Giovagnoni, A.; Golfieri, R.; Bibbolino, C.; Miele, V.; Neri, E.; Grassi, R. Artificial intelligence: Radiologists’ expectations and opinions gleaned from a nationwide online survey. La Radiol. Med. 2020, 126, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Nakaura, T.; Higaki, T.; Awai, K.; Ikeda, O.; Yamashita, Y. A primer for understanding radiology articles about machine learning and deep learning. Diagn. Interv. Imaging 2020, 101, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Pirovano, M.; Ciocca, M.; Gibelli, D.; Floridi, C.; Oliva, G. Radiomic analysis of the optic nerve at the first episode of acute optic neuritis: An indicator of optic nerve pathology and a predictor of visual recovery? La Radiol. Med. 2021, 126, 698–706. [Google Scholar] [CrossRef]
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. La Radiol. Med. 2021, 126, 1296–1311. [Google Scholar] [CrossRef] [PubMed]
- Jung, A. Machine Learning; Springer Nature: Singapore, 2022. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Setola, S.V.; Dell’Aversana, F.; Grassi, F.; Belli, A.; Silvestro, L.; Ottaiano, A.; et al. Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. La Radiol. Med. 2022, 127, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.-M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Colombo, E.; Fick, T.; Esposito, G.; Germans, M.; Regli, L.; van Doormaal, T. Segmentation techniques of brain arteriovenous malformations for 3D visualization: A systematic review. La Radiol. Med. 2022, 127, 1333–1341. [Google Scholar] [CrossRef]
- Matsoukas, S.; Scaggiante, J.; Schuldt, B.R.; Smith, C.J.; Chennareddy, S.; Kalagara, R.; Majidi, S.; Bederson, J.B.; Fifi, J.T.; Mocco, J.; et al. Accuracy of artificial intelligence for the detection of intracranial hemorrhage and chronic cerebral microbleeds: A systematic review and pooled analysis. La Radiol. Med. 2022, 127, 1106–1123. [Google Scholar] [CrossRef]
- Chiu, H.-Y.; Chao, H.-S.; Chen, Y.-M. Application of Artificial Intelligence in Lung Cancer. Cancers 2022, 14, 1370. [Google Scholar] [CrossRef]
- Moore, M.M.; Slonimsky, E.; Long, A.D.; Sze, R.W.; Iyer, R.S. Machine learning concepts, concerns and opportunities for a pediatric radiologist. Pediatr. Radiol. 2019, 49, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.J.; Cheor, W.L.; Lim, L.L.; Ab Rahman, K.S.; Bakrin, I.H. Artificial Intelligence (AI) in Breast Imaging: A Scientometric Umbrella Review. Diagnostics 2022, 12, 3111. [Google Scholar] [CrossRef] [PubMed]
- Ullah, N.; Khan, J.A.; Almakdi, S.; Khan, M.S.; Alshehri, M.; Alboaneen, D.; Raza, A. A Novel CovidDetNet Deep Learning Model for Effective COVID-19 Infection Detection Using Chest Radiograph Images. Appl. Sci. 2022, 12, 6269. [Google Scholar] [CrossRef]
- Nakamura, Y.; Higaki, T.; Honda, Y.; Tatsugami, F.; Tani, C.; Fukumoto, W.; Narita, K.; Kondo, S.; Akagi, M.; Awai, K. Advanced CT techniques for assessing hepatocellular carcinoma. La Radiol. Med. 2021, 126, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Lee, J.; Lee, S. Activation Fine-Tuning of Convolutional Neural Networks for Improved Input Attribution Based on Class Activation Maps. Appl. Sci. 2022, 12, 12961. [Google Scholar] [CrossRef]
- Alshehri, A.; AlSaeed, D. Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms. Appl. Sci. 2022, 12, 12922. [Google Scholar] [CrossRef]
- Han, D.; Chen, Y.; Li, X.; Li, W.; Zhang, X.; He, T.; Yu, Y.; Dou, Y.; Duan, H.; Yu, N. Development and validation of a 3D-convolutional neural network model based on chest CT for differentiating active pulmonary tuberculosis from community-acquired pneumonia. La Radiol. Med. 2022, 128, 68–80. [Google Scholar] [CrossRef]
- Parekh, V.S.; Jacobs, M.A. Deep learning and radiomics in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 59–72. [Google Scholar] [CrossRef] [Green Version]
- Mettler, F.A., Jr.; Huda, W.; Yoshizumi, T.T.; Mahesh, M. Effective Doses in Radiology and Diagnostic Nuclear Medicine: A Catalog. Radiology 2008, 248, 254–263. [Google Scholar] [CrossRef]
- Alshamrani, K.; Messina, F.; Offiah, A.C. Is the Greulich and Pyle atlas applicable to all ethnicities? A systematic review and meta-analysis. Eur. Radiol. 2019, 29, 2910–2923. [Google Scholar] [CrossRef] [Green Version]
- Lin, N.-H.; Ranjitkar, S.; Macdonald, R.; Hughes, T.; Taylor, J.A.; Townsend, G. New growth references for assessment of stature and skeletal maturation in Australians. Aust. Orthod. J. 2006, 22, 1–10. [Google Scholar] [PubMed]
- Soudack, M.; Ben-Shlush, A.; Jacobson, J.; Raviv-Zilka, L.; Eshed, I.; Hamiel, O. Bone age in the 21st century: Is Greulich and Pyle’s atlas accurate for Israeli children? Pediatr. Radiol. 2012, 42, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Büken, B.; Şafak, A.A.; Yazıcı, B.; Büken, E.; Mayda, A.S. Is the assessment of bone age by the Greulich–Pyle method reliable at forensic age estimation for Turkish children? Forensic Sci. Int. 2007, 173, 146–153. [Google Scholar] [CrossRef]
- Calfee, R.P.; Sutter, M.; Steffen, J.A.; Goldfarb, C.A. Skeletal and chronological ages in American adolescents: Current findings in skeletal maturation. J. Child. Orthop. 2010, 4, 467–470. [Google Scholar] [CrossRef] [Green Version]
- Kaplowitz, P.; Srinivasan, S.; He, J.; McCarter, R.; Hayeri, M.R.; Sze, R. Comparison of bone age readings by pediatric endocrinologists and pediatric radiologists using two bone age atlases. Pediatr. Radiol. 2010, 41, 690–693. [Google Scholar] [CrossRef]
- Lin, F.-Q.; Zhang, J.; Zhu, Z.; Wu, Y.-M. Comparative study of Gilsanz-Ratib digital atlas and Greulich-Pyle atlas for bone age estimation in a Chinese sample. Ann. Hum. Biol. 2014, 42, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Kahleyss, S.; Hoepffner, W.; Keller, E.; Willgerodt, H. The determination of bone age by the Greulich-Pyle and Tanner-Whitehouse methods as a basis for the growth prognosis of tall-stature girls. Pediatr. Relat. Top. 1990, 29, 137–140. Available online: https://europepmc.org/article/MED/2352741 (accessed on 17 January 2023).
- Ahmed, M.L.; Warner, J.T. TW2 and TW3 bone ages: Time to change? Arch. Dis. Child. 2007, 92, 371–372. [Google Scholar] [CrossRef] [Green Version]
- Gross, G.W.; Boone, J.M.; Bishop, D.M. Pediatric skeletal age: Determination with neural networks. Radiology 1995, 195, 689–695. [Google Scholar] [CrossRef]
- Halabi, S.S.; Prevedello, L.; Kalpathy-Cramer, J.; Mamonov, A.B.; Bilbily, A.; Cicero, M.; Pan, I.; Pereira, L.A.; Sousa, R.; Abdala, N.; et al. The RSNA Pediatric Bone Age Machine Learning Challenge. Radiology 2019, 290, 498–503. [Google Scholar] [CrossRef]
- Mehta, C.; Ayeesha, B.; Sotakanal, A.; Nirmala, S.R.; Desai, S.D.; Suryanarayana, K.V.; Ganguly, A.D.; Shetty, V. Deep Learning Framework for Automatic Bone Age Assessment. In Proceedings of the 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Virtual, 1–5 November 2021; pp. 3093–3096. [Google Scholar]
- Pan, I.; Thodberg, H.H.; Halabi, S.S.; Kalpathy-Cramer, J.; Larson, D.B. Improving Automated Pediatric Bone Age Estimation Using Ensembles of Models from the 2017 RSNA Machine Learning Challenge. Radiol. Artif. Intell. 2019, 1, e190053. [Google Scholar] [CrossRef]
- Beheshtian, E.; Putman, K.; Santomartino, S.M.; Parekh, V.S.; Yi, P.H. Generalizability and Bias in a Deep Learning Pediatric Bone Age Prediction Model Using Hand Radiographs. Radiology 2023, 306, 2. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Shim, W.H.; Yoon, H.M.; Hong, S.H.; Lee, J.S.; Cho, Y.A.; Kim, S. Computerized Bone Age Estimation Using Deep Learning Based Program: Evaluation of the Accuracy and Efficiency. Am. J. Roentgenol. 2017, 209, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Larson, D.B.; Chen, M.C.; Lungren, M.P.; Halabi, S.S.; Stence, N.V.; Langlotz, C.P. Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology 2018, 287, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mutasa, S.; Chang, P.D.; Ruzal-Shapiro, C.; Ayyala, R. MABAL: A Novel Deep-Learning Architecture for Machine-Assisted Bone Age Labeling. J. Digit. Imaging 2018, 31, 513–519. [Google Scholar] [CrossRef]
- Spampinato, C.; Palazzo, S.; Giordano, D.; Aldinucci, M.; Leonardi, R. Deep learning for automated skeletal bone age assessment in X-ray images. Med. Image Anal. 2017, 36, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Tajmir, S.; Lee, J.; Zissen, M.; Yeshiwas, B.A.; Alkasab, T.K.; Choy, G.; Do, S. Fully Automated Deep Learning System for Bone Age Assessment. J. Digit. Imaging 2017, 30, 427–441. [Google Scholar] [CrossRef] [Green Version]
- Tong, C.; Liang, B.; Li, J.; Zheng, Z. A Deep Automated Skeletal Bone Age Assessment Model with Heterogeneous Features Learning. J. Med. Syst. 2018, 42, 249. [Google Scholar] [CrossRef]
- Xu, X.; Xu, H.; Li, Z. Automated Bone Age Assessment: A New Three-Stage Assessment Method from Coarse to Fine. Healthcare 2022, 10, 2170. [Google Scholar] [CrossRef]
- Somkantha, K.; Theera-Umpon, N.; Auephanwiriyakul, S. Bone Age Assessment in Young Children Using Automatic Carpal Bone Feature Extraction and Support Vector Regression. J. Digit. Imaging 2011, 24, 1044–1058. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Gertych, A.; Liu, B.J.; Huang, H.K. Bone age assessment for young children from newborn to 7-year-old using carpal bones. Med. Imaging 2007 PACS Imaging Inform. 2007, 6516, 651618. [Google Scholar] [CrossRef]
- Bai, M.; Gao, L.; Ji, M.; Ge, J.; Huang, L.; Qiao, H.; Xiao, J.; Chen, X.; Yang, B.; Sun, Y.; et al. The uncovered biases and errors in clinical determination of bone age by using deep learning models. Eur. Radiol. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Thodberg, H.H.; Kreiborg, S.; Juul, A.; Pedersen, K.D. The BoneXpert Method for Automated Determination of Skeletal Maturity. IEEE Trans. Med. Imaging 2008, 28, 52–66. [Google Scholar] [CrossRef]
- Zhao, K.; Ma, S.; Sun, Z.; Liu, X.; Zhu, Y.; Xu, Y.; Wang, X. Effect of AI-assisted software on inter- and intra-observer variability for the X-ray bone age assessment of preschool children. BMC Pediatr. 2022, 22, 644. [Google Scholar] [CrossRef] [PubMed]
- Booz, C.; Yel, I.; Wichmann, J.L.; Boettger, S.; Al Kamali, A.; Albrecht, M.H.; Martin, S.S.; Lenga, L.; Huizinga, N.; D’Angelo, T.; et al. Artificial intelligence in bone age assessment: Accuracy and efficiency of a novel fully automated algorithm compared to the Greulich-Pyle method. Eur. Radiol. Exp. 2020, 4, 6–8. [Google Scholar] [CrossRef]
- Zhang, S.-Y.; Liu, G.; Ma, C.-G.; Han, Y.-S.; Shen, X.-Z.; Xu, R.-L.; Thodberg, H.H. Automated Determination of Bone Age in a Modern Chinese Population. ISRN Radiol. 2013, 2013, 874570. [Google Scholar] [CrossRef] [Green Version]
- Thodberg, H.H.; Sävendahl, L. Validation and Reference Values of Automated Bone Age Determination for Four Ethnicities. Acad. Radiol. 2010, 17, 1425–1432. [Google Scholar] [CrossRef]
- Alshamrani, K.; Hewitt, A.; Offiah, A. Applicability of two bone age assessment methods to children from Saudi Arabia. Clin. Radiol. 2019, 75, 156.e1–156.e9. [Google Scholar] [CrossRef]
- Klünder-Klünder, M.; Espinosa-Espindola, M.; Lopez-Gonzalez, D.; Loyo, M.S.-C.; Suárez, P.D.; Miranda-Lora, A.L. Skeletal Maturation in the Current Pediatric Mexican Population. Endocr. Pract. 2020, 26, 1053–1061. [Google Scholar] [CrossRef]
- Thodberg, H.H.; Thodberg, B.; Ahlkvist, J.; Offiah, A.C. Autonomous artificial intelligence in pediatric radiology: The use and perception of BoneXpert for bone age assessment. Pediatr. Radiol. 2022, 52, 1338–1346. [Google Scholar] [CrossRef]
- Schündeln, M.M.; Marschke, L.; Bauer, J.J.; Hauffa, P.K.; Schweiger, B.; Führer-Sakel, D.; Lahner, H.; Poeppel, T.D.; Kiewert, C.; Hauffa, B.P.; et al. A Piece of the Puzzle: The Bone Health Index of the BoneXpert Software Reflects Cortical Bone Mineral Density in Pediatric and Adolescent Patients. PLoS ONE 2016, 11, e0151936. [Google Scholar] [CrossRef]
- Willems, G. A review of the most commonly used dental age estimation techniques. J. Forensic Odonto-Stomatol. 2001, 19, 9–17. [Google Scholar]
- Farhadian, M.; Salemi, F.; Saati, S.; Nafisi, N. Dental age estimation using the pulp-to-tooth ratio in canines by neural networks. Imaging Sci. Dent. 2019, 49, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Schour, I.; Massler, M. Studies in Tooth Development: The Growth Pattern of Human Teeth. J. Am. Dent. Assoc. 1940, 27, 1778–1793. [Google Scholar] [CrossRef]
- Demirjian, A.; Goldstein, H.; Tanner, J.M. A new system of dental age assessment. Hum. Biol. 1973, 45, 211–227. [Google Scholar] [PubMed]
- Khdairi, N.; Halilah, T.; Khandakji, M.N.; Jost-Brinkmann, P.-G.; Bartzela, T. The adaptation of Demirjian’s dental age estimation method on North German children. Forensic Sci. Int. 2019, 303, 109927. [Google Scholar] [CrossRef]
- Magon, P.; Viswanathan, V.K. “Bone Age”, Revision Classes in Pediatrics; Jaypee Brothers Medical Publishers (P) Ltd.: New Delhi, India, 2008; p. 54. [Google Scholar] [CrossRef]
- Aggarwal, A.; Kulkarni, S.; Sheikh, S.; Aggarwal, O.; Mehta, S.; Gupta, D. Correlation between Radiographic evaluation of dental age and chronological age: A study on 6 to 16 years human population of Ambala using Demirjian method. J. Oral Sign 2012, 4, 63–67. [Google Scholar]
- Willems, G.; Van Olmen, A.; Spiessens, B.; Carels, C. Dental Age Estimation in Belgian Children: Demirjian’s Technique Revisited. J. Forensic Sci. 2001, 46, 15064. [Google Scholar] [CrossRef]
- Kvaal, S.I.; Kolltveit, K.M.; Thomsen, I.O.; Solheim, T. Age estimation of adults from dental radiographs. Forensic Sci. Int. 1995, 74, 175–185. [Google Scholar] [CrossRef]
- Kvaal, S.; Solheim, T. A non-destructive dental method for age estimation. J. Forensic Odonto-Stomatol. 1994, 12, 6–11. Available online: https://pubmed.ncbi.nlm.nih.gov/9227083/ (accessed on 15 January 2023).
- Guo, Y.-C.; Han, M.; Chi, Y.; Long, H.; Zhang, D.; Yang, J.; Yang, Y.; Chen, T.; Du, S. Accurate age classification using manual method and deep convolutional neural network based on orthopantomogram images. Int. J. Leg. Med. 2021, 135, 1589–1597. [Google Scholar] [CrossRef]
- Vila-Blanco, N.; Carreira, M.J.; Varas-Quintana, P.; Balsa-Castro, C.; Tomas, I. Deep Neural Networks for Chronological Age Estimation From OPG Images. IEEE Trans. Med. Imaging 2020, 39, 2374–2384. [Google Scholar] [CrossRef]
- Zaborowicz, K.; Biedziak, B.; Olszewska, A.; Zaborowicz, M. Tooth and Bone Parameters in the Assessment of the Chronological Age of Children and Adolescents Using Neural Modelling Methods. Sensors 2021, 21, 6008. [Google Scholar] [CrossRef]
- Zaborowicz, M.; Zaborowicz, K.; Biedziak, B.; Garbowski, T. Deep Learning Neural Modelling as a Precise Method in the Assessment of the Chronological Age of Children and Adolescents Using Tooth and Bone Parameters. Sensors 2022, 22, 637. [Google Scholar] [CrossRef]
- Kim, S.; Lee, Y.-H.; Noh, Y.-K.; Park, F.C.; Auh, Q.-S. Age-group determination of living individuals using first molar images based on artificial intelligence. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Banjšak, L.; Milošević, D.; Subašić, M. Implementation of Artificial Intelligence in Chronological Age Estimation from Orthopantomographic X-ray Images of Archaeological Skull Remains. Bull. Int. Assoc. Paleodont. 2020, 14, 122–129. Available online: www.paleodontology.com (accessed on 24 February 2023).
- Milošević, D.; Vodanović, M.; Galić, I.; Subašić, M. Automated estimation of chronological age from panoramic dental X-ray images using deep learning. Expert Syst. Appl. 2021, 189, 116038. [Google Scholar] [CrossRef]
- Kahaki, S.M.; Nordin, M.J.; Ahmad, N.; Arzoky, M.; Ismail, W. Deep convolutional neural network designed for age as-sessment based on orthopantomography data. Neural Comput. Appl. 2020, 32, 21–22. [Google Scholar] [CrossRef]
- Sauvegrain, J. Etude de la maturation osseuse du coude. Ann. Radiol. 1962, 5, 542–550. Available online: https://cir.nii.ac.jp/crid/1572543025397534592 (accessed on 17 January 2023).
- Hermetet, C.; Saint-Martin, P.; Gambier, A.; Ribier, L.; Sautenet, B.; Rérolle, C. Forensic age estimation using computed tomography of the medial clavicular epiphysis: A systematic review. Int. J. Leg. Med. 2018, 132, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Benito, M.; Muñoz, A.; Beltrán, I.; Labajo, E.; Perea, B.; Sánchez, J.A. Assessment of adulthood in the living Spanish population based on ossification of the medial clavicle epiphysis using ultrasound methods. Forensic Sci. Int. 2018, 284, 161–166. [Google Scholar] [CrossRef]
- Shedge, R.; Kanchan, T.; Warrier, V.; Dixit, S.G.; Krishan, K. Forensic age estimation using conventional radiography of the medial clavicular epiphysis: A systematic review. Med. Sci. Law 2021, 61, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Schmeling, A.; Schulz, R.; Reisinger, W.; Mühler, M.; Wernecke, K.-D.; Geserick, G. Studies on the time frame for ossification of the medial clavicular epiphyseal cartilage in conventional radiography. Int. J. Leg. Med. 2004, 118, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Kreitner, K.-F.; Schweden, F.J.; Riepert, T.; Nafe, B.; Thelen, M. Bone age determination based on the study of the medial extremity of the clavicle. Eur. Radiol. 1998, 8, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Li, D.T.; Cui, J.J.; DeVries, S.; Nicholson, A.D.; Li, E.; Petit, L.; Kahan, J.B.; Sanders, J.O.; Liu, R.W.; Cooperman, D.R.; et al. Humeral Head Ossification Predicts Peak Height Velocity Timing and Percentage of Growth Remaining in Children. J. Pediatr. Orthop. 2018, 38, e546–e550. [Google Scholar] [CrossRef] [PubMed]
- Bitan, F.D.; Veliskakis, K.P.; Campbell, B.C. Differences in the Risser Grading Systems in the United States and France. Clin. Orthop. Relat. Res. 2005, 436, 190–195. [Google Scholar] [CrossRef]
- Lottering, N.; Alston-Knox, C.L.; MacGregor, D.M.; Izatt, M.T.; Grant, C.A.; Adam, C.J.; Gregory, L.S. Apophyseal Ossification of the Iliac Crest in Forensic Age Estimation: Computed Tomography Standards for Modern Australian Subadults. J. Forensic Sci. 2016, 62, 292–307. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.; Schmeling, A.; Zwiesigk, P.; Pfeiffer, H.; Schulz, R. Sonographic evaluation of apophyseal ossification of the iliac crest in forensic age diagnostics in living individuals. Int. J. Leg. Med. 2011, 125, 271–276. [Google Scholar] [CrossRef]
- Rhee, C.H.; Shin, S.M.; Choi, Y.S.; Yamaguchi, T.; Maki, K.; Kim, Y.I.; Kim, S.S.; Park, S.B.; Son, W.S. Application of statistical shape analysis for the estimation of bone and forensic age using the shapes of the 2nd, 3rd, and 4th cervical vertebrae in a young Japanese population. Forensic Sci. Int. 2015, 257, 513.e1–513.e9. [Google Scholar] [CrossRef]
- Lai, E.H.-H.; Liu, J.-P.; Chang, J.Z.-C.; Tsai, S.-J.; Yao, C.-C.J.; Chen, M.-H.; Chen, Y.-J.; Lin, C.-P. Radiographic Assessment of Skeletal Maturation Stages for Orthodontic Patients: Hand-Wrist Bones or Cervical Vertebrae? J. Formos. Med. Assoc. 2008, 107, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Schlégl, T.; O’Sullivan, I.; Varga, P.; Than, P.; Vermes, C. Determination and correlation of lower limb anatomical parameters and bone age during skeletal growth (based on 1005 cases). J. Orthop. Res. 2016, 35, 1431–1441. [Google Scholar] [CrossRef]
- Li, S.Q.; Nicholson, A.D.; Cooperman, D.R.; Liu, R.W. Applicability of the Calcaneal Apophysis Ossification Staging System to the Modern Pediatric Population. J. Pediatr. Orthop. 2019, 39, 46–50. [Google Scholar] [CrossRef]
- O’Connor, J.E.; Bogue, C.; Spence, L.D.; Last, J. A method to establish the relationship between chronological age and stage of union from radiographic assessment of epiphyseal fusion at the knee: An Irish population study. J. Anat. 2008, 212, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Krämer, J.A.; Schmidt, S.; Jürgens, K.-U.; Lentschig, M.; Schmeling, A.; Vieth, V. Forensic age estimation in living individuals using 3.0T MRI of the distal femur. Int. J. Leg. Med. 2014, 128, 509–514. [Google Scholar] [CrossRef]
- Risser, J.C. The Classic: The Iliac Apophysis: An Invaluable Sign in the Management of Scoliosis. Clin. Orthop. Relat. Res. 2009, 468, 646–653. [Google Scholar] [CrossRef] [Green Version]
- Baik, S.B.; Cha, K.G. A Study on Deep Learning Based Sauvegrain Method for Measurement of Puberty Bone Age. arXiv 2018, arXiv:1809.06965. [Google Scholar]
- Der Mauer, M.A.; Well, E.J.-V.; Herrmann, J.; Groth, M.; Morlock, M.M.; Maas, R.; Säring, D. Automated age estimation of young individuals based on 3D knee MRI using deep learning. Int. J. Leg. Med. 2020, 135, 649–663. [Google Scholar] [CrossRef]
- Dallora, A.L.; Berglund, J.S.; Brogren, M.; Kvist, O.; Ruiz, S.D.; Dübbel, A.; Anderberg, P. Age Assessment of Youth and Young Adults Using Magnetic Resonance Imaging of the Knee: A Deep Learning Approach. JMIR Public Health Surveill. 2019, 7, e16291. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.; Payer, C.; Giuliani, N.; Urschler, M. Automatic Age Estimation and Majority Age Classification From Multi-Factorial MRI Data. IEEE J. Biomed. Health Inform. 2018, 23, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Liao, N.; Dai, J.; Tang, Y.; Zhong, Q.; Mo, S. iCVM: An Interpretable Deep Learning Model for CVM Assessment Under Label Uncertainty. IEEE J. Biomed. Health Inform. 2022, 26, 4325–4334. [Google Scholar] [CrossRef]
- Kim, D.; Kim, J.; Kim, T.; Kim, T.; Kim, Y.; Song, I.; Ahn, B.; Choo, J.; Lee, D. Prediction of hand-wrist maturation stages based on cervical vertebrae images using artificial intelligence. Orthod. Craniofacial Res. 2021, 24, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Wan, L.; Wang, M.W.; Li, Z.; Wang, P.; Liu, T.A.; Wang, Y.H.; Zhao, H. Comparison of Three CNN Models Applied in Bone Age Assessment of Pelvic Radiographs of Adolescents. J. Forensic Med. 2020, 36, 622–630. [Google Scholar] [CrossRef]
- Peng, L.-Q.; Guo, Y.-C.; Wan, L.; Liu, T.-A.; Wang, P.; Zhao, H.; Wang, Y.-H. Forensic bone age estimation of adolescent pelvis X-rays based on two-stage convolutional neural network. Int. J. Leg. Med. 2022, 136, 797–810. [Google Scholar] [CrossRef]
- Chen, H.C.; Wu, C.H.; Lin, C.J.; Liu, Y.H.; Sun, Y.N. Automated segmentation for patella from lateral knee X-ray images. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 3553–3556. [Google Scholar] [CrossRef]
- Xia, Y.; Fripp, J.; Chandra, S.S.; Schwarz, R.; Engstrom, C.; Crozier, S. Automated bone segmentation from large field of view 3D MR images of the hip joint. Phys. Med. Biol. 2013, 58, 7375–7390. [Google Scholar] [CrossRef]
- Vicini, S.; Bortolotto, C.; Rengo, M.; Ballerini, D.; Bellini, D.; Carbone, I.; Preda, L.; Laghi, A.; Coppola, F.; Faggioni, L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: Focus on the three most common cancers. La Radiol. Med. 2022, 127, 819–836. [Google Scholar] [CrossRef]
- Langlotz, C.P. Will Artificial Intelligence Replace Radiologists? Radiol. Artif. Intell. 2019, 1, e190058. [Google Scholar] [CrossRef] [PubMed]
- Ontell, F.K.; Ivanovic, M.; Ablin, D.S.; Barlow, T.W. Bone age in children of diverse ethnicity. Am. J. Roentgenol. 1996, 167, 1395–1398. [Google Scholar] [CrossRef] [PubMed]
- Sardanelli, F.; Colarieti, A. Open issues for education in radiological research: Data integrity, study reproducibility, peer-review, levels of evidence, and cross-fertilization with data scientists. La Radiol. Med. 2022, 128, 133–135. [Google Scholar] [CrossRef]
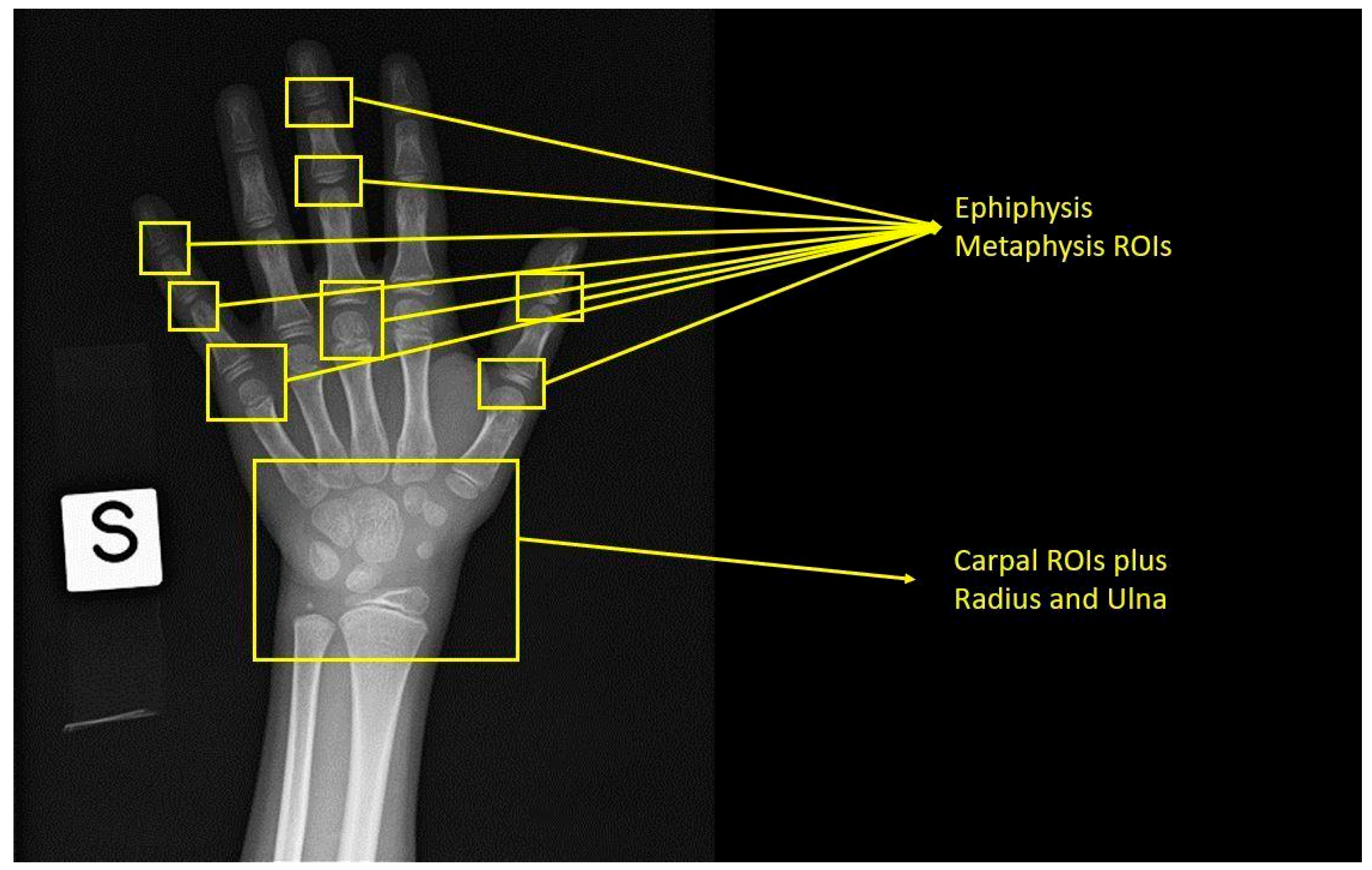

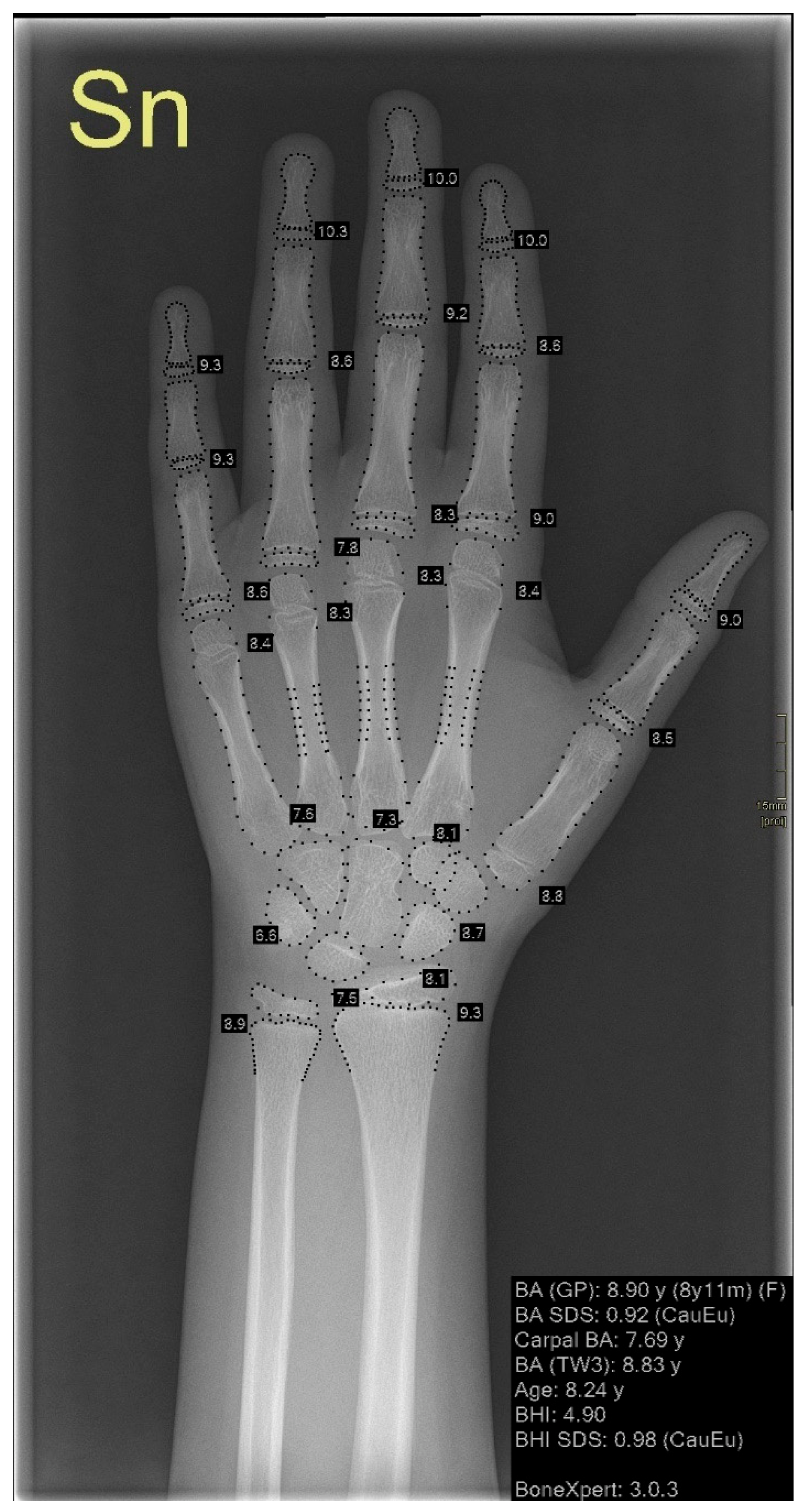
| Authors | Number of Left-Hand Radiographs in the Dataset | Reference Standard Bone Age | AI Technique | RMSE (Years) | MAD (Years) | MAE (Years) |
|---|---|---|---|---|---|---|
| Halabi et al., 2018 [54] | Training set: 12,611 Validation set: 1425 Test set: 200 | Radiology report provided by RSNA | Inception V3 for pixel information, additional dense layers, and multiple high-performing models | 0.35 | ||
| Mehta et al., 2021 [55] | Training set: 12,611 Validation set: 1425 Test set: 200 | Radiology report provided by RSNA | Inception V3 architecture applied on gamma-corrected images | 0.492 | ||
| Pan et al., 2019 [56] | 200 cases test set divided into 1000 validation-test splits | Radiology report provided by RSNA | Combination of 8 models of RSNA Pediatric Bone Age ML Challenge | 0.328 | ||
| Beheshtian et al., 2023 [57] | Internal validation set: 1425 External test set: 1202 | Radiology report | Inception V3 + additional dense layers and multiple high-performing models | 0.567 vs. 0.575 | ||
| Kim et al., 2017 [58] | Training set: 18,940 Test set: 200 | 2 experienced radiologists | VUNO Med-BoneAge (deep learning semiautomatic system, based on GP method) | 0.6 | ||
| Larson et al., 2018 [59] | Training and validation set: 14,036 Test set: 200 | Clinical report and 3 human reviewers | CNN model based on GP method | 0.63 | 0.5 | |
| Mutasa et al., 2018 [60] | Training set: 10,289 Test set: 300 | Radiology report | 14 hidden layer CNN based on GP method | 0.536 | ||
| Lee H. et al., 2017 [62] | Training set: 5828 Test set: 1249 | Radiology report | ImageNet pre-trained, fine-tuned CNN | 0.82–0.93 | ||
| Xu et al., 2022 [64] | Public dataset: 12,600 Clinical Training set: 2014 Clinical Test set: 504 | Radiology report | CNN based on TW3 method | 0.64; 0.54 | ||
| Bai et al., 2022 [67] | Training set: 9607 + 11,226 Test set: 1246 | 10 senior radiologists | 3 deep learning models | 0.42 | ||
| Thodberg et al., 2009 [68] | Training set: 1559 Validation set: 122 | Radiologists applying GP method | BoneXpert 2.1 (3 layers deep learning model based on GP and TW3 methods) | 0.38 | ||
| Lee et al., 2021 [17] | Training set: 2684 Test set: 660 | Radiology report (TW3) | HH-boneage. io (fully automated system based on TW3) | 0.62 | 0.46 | |
| Zhao et al., 2022 [69] | Test set: 54 | 3 expert radiologists | Deep learning software by Deep Wise Artificial Intelligence Lab based on TW3 modified for Chinese people | TW3-RUS: 0.501 TW3-Carpal: 0.323 | TW3-RUS: 0.379 TW3-Carpal: 0.229 |
| Traditional Method | Procedure | Commercially Available AI-Based Tool |
|---|---|---|
| Greulich and Pyle (GP) | Comparison with reference images contained in the Atlas | BoneXpert; VUNO Med-BoneAge |
| Gilsanz and Ratibin (GR) | Comparison with reference images contained in the Digital Atlas | |
| Tanner Whitehouse (TW) | Scoring the level of maturity of specific regions of interest based on the reference scale | BoneXpert; HH-boneage.io |
| How It Works | Advantages | Disadvantages | ||
|---|---|---|---|---|
| TRADITIONAL APPROACH | Greulich and Pyle (GP) method | Comparison between the patient radiography and reference images included in the Atlas |
|
|
| Gilsanz and Ratibin (GR) Atlas | Comparison between the patient radiography and reference images included in the Digital Atlas |
|
| |
| Tanner Whitehouse (TW3) Method | Age derived from a score calculated from the analysis of 20 ROIs |
|
| |
| AI-BASED APPROACH | AI-based software provides an automatic result |
|
| |
| Authors | Anatomical Region | Imaging Technique | Process Description | Age Range of Subjects | Disadvantages |
|---|---|---|---|---|---|
| Sauvegrain et al. [95] | Elbow | Radiography (AP and LL projections) | Evaluation of 4 elbow ossification centers, basing on a 27-point scoring system | 0–18 | The double projection exposes to a higher radiation dose |
| Schmeling et al. [99] | Medial clavicle epiphysis | Radiography | Evaluation of ossification degree of the cartilage, basing on a 5-stage classification system | 18–22 | Conventional clavicle X-ray can be hindered by overlapping images related to the mediastinal structures, vertebrae, or ribs |
| Li et al. [101] | Proximal humeral physis | Radiography | Evaluation of the humeral head epiphysis and fusion of the external portion of the physis, based on a 5-stage scale | 10–15 | Historical collection of radiographs (dated 1926–1942) |
| Lottering et al. [103] | Iliac crest | CT | Evaluation of the apophyseal ossification of the iliac crest, according to Risser’s sign, a 5-score classification | 7–25 | The ossification of iliac crest apophysis is not uniform, thus it can create some discrepancies |
| Schmidt et al. [104] | Iliac crest | US | Apophyseal ossification of the iliac crest, according to Risser’s sign, a 5-score classification | 11–20 | The ossification of iliac crest apophysis is not uniform, thus it can create some discrepancies |
| Soegiharto et al. [105] | Cervical vertebrae | Radiography (lateral cephalometric) | Evaluation of C2, C3 and C4, basing on a 6- stage maturation scale | 8–17 | The method originally was developed more than 5 decades ago, without a fair description of the classification system, until a few years ago. |
| Li et al. [108] | Calcaneal apophysis | Radiography (lateral foot projection) | Evaluation of the calcaneal apophysis, based on a 5-stage scale | 7–16 | Ethnical differences |
| O’Connor et al. [109] | Knee | Radiography (AP and LL knee projections) | Evaluation of the stage of the epiphyseal union at the knee joint, basing on a 5-stage scale | 9–19 | The study was applied to a highly selected population (Irish), thus the results are difficult to generalize |
| Krämer et al. [110] | Distal femur | 3T MRI | Evaluation of the ossification stage of the distal femoral epiphysis, based on a 5-stage scale | 10–30 | Unbalanced age distribution of subjects, particularly in the lower age groups; only one sectional plane and only one MRI weighting were considered. |
| Authors | Object of Analysis | Dataset | AGE | CNN | MAE | Accuracy (%) | RMSE (Years) |
|---|---|---|---|---|---|---|---|
| Bin Baik et al. [112] | Elbow radiographs | 576 | adolescents | U-Net, RPN+, F-RCNN, VGC16 | 2.8 months | ||
| Der Mauer et al. [113] | Knee MRI | 589 | 13–21 | N4ITK, MAdM, U-Net, AgeNet2D | 0.67 ± 0.49 y | 90.9 | |
| Dallora et al. [114] | Knee MRI | 402 | 14–21 | GoogleNet, ResNet-50, Inception-v3, VGG, AlexNet, DenseNet, U-Net | 0.793–0.988 y | 95–98.1 | |
| Štern et al. [115] | MRI of hands, clavicle, and teeth | 322 | 13–25 | Inception V3 | 1.01 ± 0.74 y | ||
| Kim at al. [117] | Cervical vertebrae in lateral cephalograms | 499 | 6–18 | BayesianRidge, Ridge, LinearRegression, HuberRegressor, SGDRegressor, RandomForestRegressors, TheilSenRegressor, AdaBoostRegressor and LinearSV | 0.9 y | 1.2 | |
| Peng et al. [118] | Pelvic radiographs | 962 | 11–21 | Inception-V3, Inception-ResNet-V2, and VGG19 | 0.82–1.02 y | 1.11–1.29 | |
| Peng et al. [119] | Pelvic radiographs | 2164 | 11–21 | Inception-V3, Inception-ResNet-V2, and VGG19, U-Net | 0.93–1.14 y | 1.22–1.63 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caloro, E.; Cè, M.; Gibelli, D.; Palamenghi, A.; Martinenghi, C.; Oliva, G.; Cellina, M. Artificial Intelligence (AI)-Based Systems for Automatic Skeletal Maturity Assessment through Bone and Teeth Analysis: A Revolution in the Radiological Workflow? Appl. Sci. 2023, 13, 3860. https://doi.org/10.3390/app13063860
Caloro E, Cè M, Gibelli D, Palamenghi A, Martinenghi C, Oliva G, Cellina M. Artificial Intelligence (AI)-Based Systems for Automatic Skeletal Maturity Assessment through Bone and Teeth Analysis: A Revolution in the Radiological Workflow? Applied Sciences. 2023; 13(6):3860. https://doi.org/10.3390/app13063860
Chicago/Turabian StyleCaloro, Elena, Maurizio Cè, Daniele Gibelli, Andrea Palamenghi, Carlo Martinenghi, Giancarlo Oliva, and Michaela Cellina. 2023. "Artificial Intelligence (AI)-Based Systems for Automatic Skeletal Maturity Assessment through Bone and Teeth Analysis: A Revolution in the Radiological Workflow?" Applied Sciences 13, no. 6: 3860. https://doi.org/10.3390/app13063860





