1. Introduction
Carnivore action on bone assemblages is difficult to differentiate due to equifinality. Different carnivore species may produce similar bone damage, impeding, in some cases, an accurate taxonomic characterization of the taphonomic agents intervening in archaeopaleontological sites. Furthermore, archaeopaleontological sites are usually described as palimpsests, which implies that several taphonomic agents (e.g., humans, carnivores, etc.) may have modified those bone assemblages. A suitable description of the taphonomic agents involved in the bone modification of archaeopaleontological sites is crucial to formulating hypotheses on hominin-carnivore interactions, hominin subsistence patterns, or site formation processes [
1,
2,
3,
4,
5,
6,
7]. On this basis, much neotaphonomic research has been conducted in order to find variables that would not only allow the identification of modifying agents in the fossil record (i.e., humans or carnivores), but also the specification of carnivore groups and taxa based on the bone surface modifications they generate [
8,
9,
10,
11].
For instance, tooth mark dimensions have been analysed by several authors to statistically differentiate the pits and scores produced by different carnivore species [
12,
13,
14,
15,
16]. Pits have been described as circular depressions, while tooth scores are elongated marks characterized by U-shaped cross-sections [
1,
17]. Both width and length have been recorded on bones modified by different carnivores (e.g., felids, hyenids, canids, ursids, etc), while considering bone region (e.g., epiphysis versus long bone shaft). For instance, pit and score dimensions have been used to discuss the accumulating agent of bone assemblages from Olduvai Gorge (Tanzania) or Sima de los Huesos (Spain) [
12,
13,
14]. The analyses of tooth mark dimensions from the FLK Zinj site (Olduvai Gorge, Tanzania) showed that both felids and hyenids modified the bone assemblage [
12]. Cut and percussion marks produced by hominins were also recorded on the FLK Zinj bone assemblage [
3,
7]. According to the analyses and results obtained, it has been argued that hyenas were the last modifying agent of the ungulate carcasses from the FLK Zinj [
12]. Thus, either hyenas may have scavenged ungulate carcasses that were first defleshed by hominins and felids, or hyenas may have just consumed the marrow of bones first scavenged by hominins after a primary consumption by felids [
12]. These results imply that early hominins from Olduvai Gorge were not the first taphonomic agents in the consumption change [
12]. Further research has demonstrated that carnivore body mass and the location of the tooth mark, i.e., on the epiphysis or diaphysis, determines tooth mark dimensions [
15], so that tooth marks inflicted on long bone shafts by small and large carnivores are statistically differentiable [
14,
15,
16]. However, both pit and score dimensions overlap among different carnivore taxa. That means that while carnivore size could be inferred from tooth mark dimensions, this same variable cannot be used to determine carnivore taxa. Thus, felid and hyenid species could not be differentiated. These results could not support the three-stage model hypothesis (carnivore-hominin-carnivore) for the FLK Zinj bone assemblage [
14].
On the other hand, tooth mark frequency or the number of tooth marks inflicted by bone have been also explored in order to determine agency in bone assemblages, and thus, overcome the equifinality biases that tooth mark size could not totally solve. The extent of bone modification depends on the type of carnivore involved at the site. It has been argued that felids modify skeletal body parts less intensively than hyenas [
2,
12]. Further research showed that a low tooth mark frequency on long bone shafts may be produced by a primary access of felids to animal carcasses, or by a secondary hyenid access [
18]. Thus, in multi-agent scenarios such as archaeopaleontological sites where several taphonomic agents may modify skeletal parts, tooth mark frequency also shows equifinality problems.
Recently, new methodological approaches combining geometric morphometric (GM) analyses and virtual reconstructions of tooth marks have improved carnivore taxa identification [
19,
20,
21,
22,
23]. The examination of tooth mark morphology along with tooth mark dimensions showed promising results regarding the differentiation of carnivore groups. Despite the valuable methodological progress presented by Yravedra et al. [
19], concerning the morphological registration and study of carnivore tooth scores, we believe that the lack of specificity regarding the selection of the score cross-section (most characteristic U-shape section within the 30–70% mark length) could cause biases during data collection, not only in relation to the observer’s own objectivity, but also to the potential intra-specific morphological variation along the score profile. Therefore, we consider it necessary to assess this method in a more detailed manner. For that purpose, in the present study, we evaluate the original, as well as novel landmark-based models, to explore felid and hyenid tooth score morphology at different points along the length of scores, with the aim of proposing some methodological recommendations for future research in this line.
4. Discussion
Tooth scores are elongated U-shaped marks that are commonly found on the cortical surfaces of skeletons modified by carnivores [
1,
17]. The first attempt to analyse score morphology using GM techniques was based on a seven-landmark model that provided promising results in the differentiation of carnivore groups [
19]. However, methodological laxity regarding the selection of the score profile and the subsequent potential lack of representativeness of such cross-sections required a more detailed study, including the morphological comparison of score profiles at different score lengths. The conduction of thorough methodological research, like the one presented here, has not only allowed the evaluation of intra-specific score morphological variation and cross-sectional representativeness, but also demonstrated the effectiveness of landmark and semilandmark models for the differentiation of carnivore families and taxa based on specific cases.
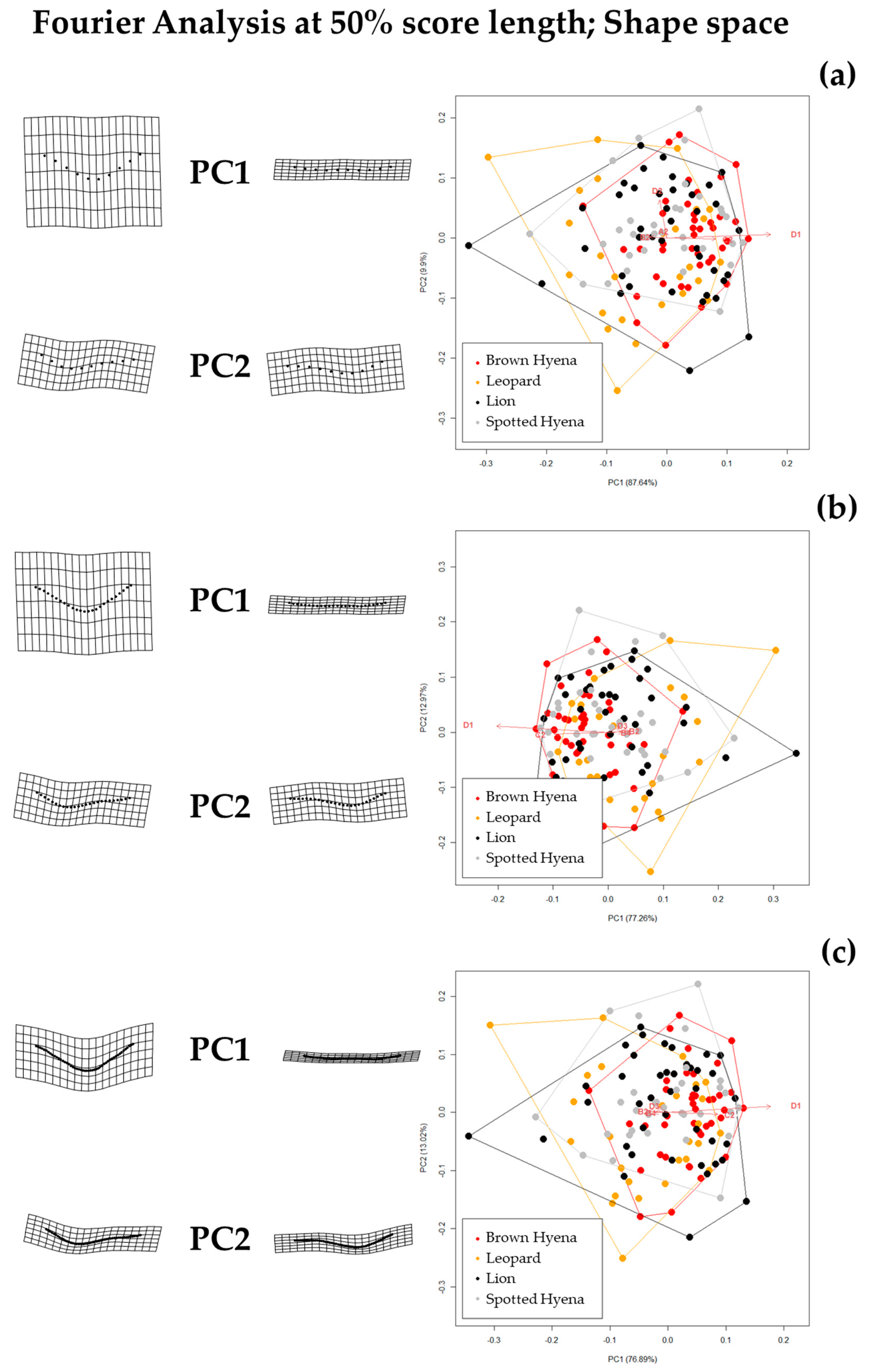
Results show that the selection of the cross-sectional portion does not greatly affect the morphological characterization of the tooth scores produced by lions, leopards, and spotted and brown hyenas, as intra-specific variation does not seem to vary significantly (
Figure S5). However, according to the tests performed here, the discrimination between carnivore groups improves when the sections selected for the present study (e.g., 35/50/65%) are used, instead of the most conspicuous U-shape section [
19] (
Table 2). This is particularly striking in the case of different carnivore families (e.g., hyenids and felids). Thus, a standardization of the model considering specific profiles along the score length might not only help replicate the method, but might also improve discrimination among carnivore groups.
However, when comparing the effect of the selection of different standardized cross-sections, it appears that not all cross-sections are equally valid for carnivore characterization and group separation. For instance, species discrimination slightly improves when the 35% profile is selected (
Table 2). Since 35% and 65% profiles are the result of score orientation and are, thus, randomly grouped as such, it can only be hypothesized that the score ends (e.g., 35% and 65%) may be more suitable for morphological characterization than profiles at half score length (50%). However, analyses considering 65% profiles highlight that lions and leopards cannot be statistically differentiated, neither in shape nor in form space, while brown and spotted hyenas can only be distinguished in form space. These results suggest that score size is important in order to characterize hyenid species. In fact, spotted hyenas present the largest form variance, probably directly associated with the large size range of the scores they produce (
Figure S6). In sum, the seven-landmark model first published by [
19] is able to differentiate between hyenid species in form space, but cannot distinguish between felid species neither in shape nor in form space. Nevertheless, if carnivore species are grouped by family (e.g.,
Felidae and
Hyenidae), groups can be statistically differentiated both in shape and form space.
These results are important for taphonomic studies, since different carnivores may modify and accumulate bones in archaeopaleontological contexts. Additionally, distinguishing the action of felids and hyenids in archaeological sites is also particularly relevant in order to clarify the anthropogenic access order to animal carcasses in those sites where both human and carnivore action has been recognized. It has been hypothesized that felid species may provide ungulate carcasses for human scavenging [
8,
17]. In contrast, hyenid species usually consume their prey entirely [
67,
68]. Thus, if the main carnivore activity at a specific site can be classified as felid-made, there is a possibility that humans may have secondarily intervened, scavenging the skeletal parts previously consumed by felids. On the contrary, if carnivore tooth marks are mainly classified as hyenid-made, this means that the bone assemblage may have been first modified by humans and subsequently scavenged by hyenas.
Furthermore, the methodology tested here is able to differentiate between brown and spotted hyenas based on the form of the scores they generate. This is particularly important because both hyenid species are able to produce bone accumulations. It has been proposed that numerous paleontological sites worked as hyena dens both in Africa and Europe during the Pleistocene. Previous research has explored several taphonomic variables in order to distinguish bone assemblages accumulated by different hyenid species, although not without problems, since hyenids may produce similar taphonomic modifications that impede the discrimination among taxa [
69,
70]. The application of the seven-landmark method can shed light on these equifinality problems and help discern the action of specific hyenid species at paleontological sites. The generalization of this hypothesis, however, requires additional analyses. Striped hyenas, which were not included in the present study, can also produce bone accumulations. Thus, in order to prove useful to the discrimination among all living and skilled bone-accumulator hyenid taxa, the seven-landmark model needs to be tested on spotted, brown, and stripped hyenas.
In addition to the fine-tuning of the already published seven-landmark method [
19], in the present study, we have assessed novel models based on semilandmarks for the characterization and analysis of tooth scores. First, we have explored if the inclusion of a higher number of points provides a better registration of score morphology and, thus, an improvement in carnivore differentiation.
Models including 10, 25, and 50 semilandmarks on a single curve, that is, without specifying the deepest point of the score profile, were evaluated showing similar general morphological trends to the seven-landmark method, but a lower differentiation power between groups (
Table 2,
Table 4 and
Table 5). Conversely to the seven-landmark method, these novel models are not able to distinguish between felids and hyenids, regardless of the methodological approach, namely using GM and EFA approaches. In fact, only hyenid species can be statistically differentiated when shape and size features are considered together, confirming that brown and spotted hyenas are morphologically easier to discriminate than lions and leopards, probably due to the differences in size observed between the former species (
Figure S6).
The second group of models based on semilandmarks (13, 23, and 33) was molded using two curves that end at the score profile lateral ends and the deepest point of the score cross-section. Results show that the differentiation power improves when using the two-curve models in comparison to the one-curve models, which suggests that the deepest point may be morphologically relevant to describe score morphology. Nevertheless, carnivore distinction is still less clear than using the original seven-landmark model, since the two-curve models are not able to consistently separate felids from hyenids based on the tooth scores they generate, and can only statistically distinguish between brown and spotted hyenas in form space, like the rest of the models tested in the present study (
Table 6). Additionally, the EFA approach is also capable of separating the brown hyena sample from the rest of the carnivore groups included in the study when considering only shape variables (
Table 7). However, this is likely due to the number of variables used in the analysis, which would result in an increase of the effects of the curse of dimensionality.
In general, the results obtained here highlight that the use of high numbers of landmarks to describe tooth score cross-sections does not necessarily improve morphological characterization and carnivore differentiation. Thus, the use of semilandmark models is not recommended for the study of archaeopaleontological sites where both felid and hyenid may have acted as taphonomic agents. Anyway, in case of using 2D semilandmark curves to avoid landmarking biases, researchers might want to make sure that the deepest point of the score cross-section is included in the analysis, as its location seems to be relevant for morphological characterization.
Among the carnivores included in the study, hyenids seem to be easier to differentiate based on the tooth score morphology they generate, probably with reference to the effect that size has on the spotted hyena score morphology. Spotted hyenas form, indeed, the most differentiated group (see balanced accuracy in
Table S2), whereas leopards usually present the lowest accuracy rates among the four groups. Thus, while the 3D GMM analysis of carnivore pits has provided useful tools for the separation of carnivores, including the distinction of leopards and lions [
26], as well as wider range of carnivore taxa [
20,
21,
22,
23], the GMM analysis of score morphology might be relevant for the distinction of not only hyenid species, but also different carnivore families in those scenarios, where the tooth pit sample is not large enough to perform statistical analyses due to preservation problems.
Despite the positive results obtained regarding the separation among hyenid species, further analyses are required to prove the validity of the method in a more generalized context that includes a wider range of carnivore taxa. Therefore, works in this line including 2D and 3D landmark data will be conducted in the following years.