Abstract
The recent increase in the prevalence of skin cancer, along with its significant impact on individuals’ lives, has garnered the attention of many researchers in the field of deep learning models, especially following the promising results observed using these models in the medical field. This study aimed to develop a system that can accurately diagnose one of three types of skin cancer: basal cell carcinoma (BCC), melanoma (MEL), and nevi (NV). Additionally, it emphasizes the importance of image quality, as many studies focus on the quantity of images used in deep learning. In this study, transfer learning was employed using the pre-trained VGG-16 model alongside a dataset sourced from Kaggle. Three models were trained while maintaining the same hyperparameters and script to ensure a fair comparison. However, the quantity of data used to train each model was varied to observe specific effects and to hypothesize about the importance of image quality in deep learning models within the medical field. The model with the highest validation score was selected for further testing using a separate test dataset, which the model had not seen before, to evaluate the model’s performance accurately. This work contributes to the existing body of research by demonstrating the critical role of image quality in enhancing diagnostic accuracy, providing a comprehensive evaluation of the VGG-16 model’s performance in skin cancer detection and offering insights that can guide future improvements in the field.
1. Introduction
The incidence of both non-melanoma and melanoma skin cancers has increased in recent decades. Currently, between 2 and 3 million non-melanoma skin cancers and 132,000 melanoma skin cancers occur globally each year, as reported by the World Health Organization (WHO).
The global rise in skin cancer cases underscores the need for effective diagnostic and treatment methods. As indicated by the World Health Organization (WHO), the rising pervasiveness of non-melanoma and malignant melanoma skin growths requests pressing consideration. In the U.S. alone, more than 9500 people find a malignant skin growth day to day, and multiple individuals capitulate to the infection consistently [1,2,3].
One of the key factors that significantly impacts a patient’s life is diagnosis. Correctly diagnosing serious cases can contribute to higher recovery rates by directing patients to early treatment [4]. However, the diagnostic stage faces several challenges that may lead to early misdiagnosis, including difficulties in distinguishing skin lesions with the naked eye, the high costs and unavailability of necessary diagnostic tools and technologies in many healthcare facilities, as well as human errors related to the expertise of specialists and the wide variety of skin lesions, which lack general features for differentiation [5,6,7].
Then again, profound learning, a subset of AI in machine learning, has achieved satisfactory results in the analysis and processing of medical images [8].
Deep learning models have shown significant promise in diagnosing different kinds of skin cancer, including basal cell carcinoma, squamous cell carcinoma, and melanoma. These models can analyze clinical, dermoscopic, and histopathologic images to identify cancerous lesions with high accuracy. By leveraging deep learning as a decision support tool, healthcare professionals can enhance their diagnostic capabilities, leading to earlier detection and improved patient outcomes [9].
The wild development of strange cells in the epidermis, the furthest skin layer, is brought about by unrepaired DNA damage that triggers transformations. These changes lead the skin cells to increase quickly and structure dangerous cancers [10].
The VGG16 model, a convolutional neural network (CNN) architecture developed by the Visual Geometry Group at the University of Oxford, has shown significant promise in skin cancer detection. VGG16 is known for its deep architecture with 16 weight layers, which permits it to catch many-sided subtleties in medical pictures. Studies have demonstrated that VGG16 can effectively classify skin cancer types, such as melanoma and non-melanoma, by analyzing dermoscopic images. Its ability to distinguish subtle differences in skin lesions makes it a powerful tool for early diagnosis. A study published in MDPI’s Diagnostics highlighted that VGG16, when combined with transfer learning, significantly improved the classification performance in differentiating between benign and malignant skin conditions [11]. The application of VGG16 in clinical settings can assist dermatologists in making more exact analyses, eventually prompting better tolerant results and timely treatment [12].
One of the most common types of skin cancer in the non-melanoma category is basal cell carcinoma (BCC). Basal cell carcinoma is the most widely recognized disease in people with a light complexion type and is consistently expanding in frequency [13,14]. In the melanoma category, there is one type, which is melanoma (MEL). Melanoma skin disease is viewed as the most serious of all malignant skin growths [15]. In certain individuals, we can see “moles” called “nevi”. They are non-cancerous skin lesions, but they can develop into skin cancer and manifest in various forms [16]. In a meta-examination on nevi as hazard factors for melanoma, the most noteworthy gamble (around 7-fold) for melanoma was seen in people with an excess of 100 nevi [17].
When it comes to automated diagnosis using machines, some traditional methods heavily rely on manual functions and simple artificial neural networks that require significant effort and time for training, which makes them somewhat limited. Unlike these methods, deep learning has significantly reduced manual intervention, thereby reducing the effort required for model training while allowing for various adjustments to achieve improved results in record time. In addition, profound models have shown fantastic execution in many fields, like clinical picture order, division, sore recognition, and enrollment [18,19,20].
Deep learning primarily relies on deep artificial neural networks that contain more than two hidden layers. A couple of models incorporate convolutional neural organizations [21], profound conviction networks [22], cascaded autoencoders [23], generative ill-disposed networks [24], variational autoencoders [25], flow models [26], recurrent neural networks [27], and consideration-based models [17].
Developing models based on deep learning can enable computers to analyze images and classify them into different categories as needed, contributing to the integration of automation in various fields, particularly in the medical domain [28].
2. The State of the Art
When we generally talk about the classification process, we mean dividing a large and general set of elements that share common properties into subgroups with more specific shared characteristics. For example, we start with a set whose elements share two properties and arrive at subgroups whose elements share more than two properties. The same principle applies to the diagnosis of skin cancer and skin lesions, a task that is somewhat difficult due to the wide variety of lesions and the absence of a general rule to rely on for classification.
In this context, dermatologists utilized the ABCDE criteria for the detection and diagnosis of skin lesions. However, they later realized that these criteria are not always sufficient. The research on the early location of nodular melanoma conducted by Chamberlain et al. [29] showed that nodular melanoma skin lesions do not follow the ABCDE criteria, which proves the limitations of these criteria at times.
Later, other methods such as machine learning emerged, but they also faced several challenges, including manual feature extraction, limited accuracy (as these methods were not effective in extracting complex features), high resource requirements for training, and slow processing times.
Based on previous limitations, efforts converged to further enhance machine learning, resulting in the emergence of deep learning.
Profound learning permits computational models that are made out of various handling layers in view of brain organizations to learn portrayals of information with numerous degrees of reflection [30]. Due to its depth and multiple layers, along with the automation of feature extraction, the ability to learn detailed and complex features which improve model accuracy, and the capability to integrate deep learning models into other applications, deep learning has been able to overcome the obstacles faced by traditional methods.
Following the outcome of PC vision, the primary uses of profound learning regarding clinical information involve picture handling, particularly in the examination of attractive reverberation imaging (X-ray) to foresee Alzheimer’s disease and its varieties [31,32].
The profound structures applied to the medical services area have been, for the most part, founded on convolutional brain organizations (CNNs) [33], intermittent neural networks (RNNs) [34], Restricted Boltzmann Machines (RBMs) [35], and autoencoders (AEs) [36]. The most common one is a CNN.
Lately, Convolutional Neural Networks (CNNs) have arisen as the most prevalent architecture among deep learning models, as highlighted in various comprehensive reviews that delve into their applications, advancements, and performance across different domains. Nazari et al. emphasized this dominance in their review [37], highlighting CNNs’ effectiveness in analyzing clinical images for skin cancer location. Similarly, Dildar et al. [38] pointed to CNNs as a cornerstone of deep learning approaches for skin cancer detection, exploring their success in extracting features to separate dangerous and harmless cancers. The review by Naqvi et al. further underscored this by discussing the dominance of CNN architectures in achieving promising results for skin cancer classification [39]. They delved into the capabilities of CNNs in learning hierarchical features from clinical images, ultimately aiding in cancer detection.
A CNN is the most effective learning algorithm for comprehending picture material [31]. CNNs depend on neighborhood associations and tied loads across units followed by pooling (subsampling) to obtain interpretations of invariant descriptors [40]. Several models have been developed based on CNNs, including DenseNet, ResNet, and VGGNet.
Regarding the use of CNN models, researchers have the option to use pre-trained models and adapt them to their specific needs with fine-tuning or transfer learning, or to build their own convolutional networks.
In the current study, VGG16 was chosen as the CNN backbone to create a custom model by applying transfer learning, and TensorFlow 2 was the selected framework used to circulate computations to more than one computer processor or GPU with a solitary programming interface [41].
3. Methods and Materials
3.1. Dataset
Given the importance of data, a search was conducted for a dataset containing an adequate number of images related to the three types we aim to classify. We chose datasets from Kaggle to train three models. For the first model, 14,454 images were used for training, with 1671 images used for validation. The second model was trained with 13,232 images with 820 for validation, and it was also tested with 4500 images. The third model was trained with 10,232 images and validated with 820 images. In all stages for the second and third models, the preprocessed HAM 10,000 dataset was employed.
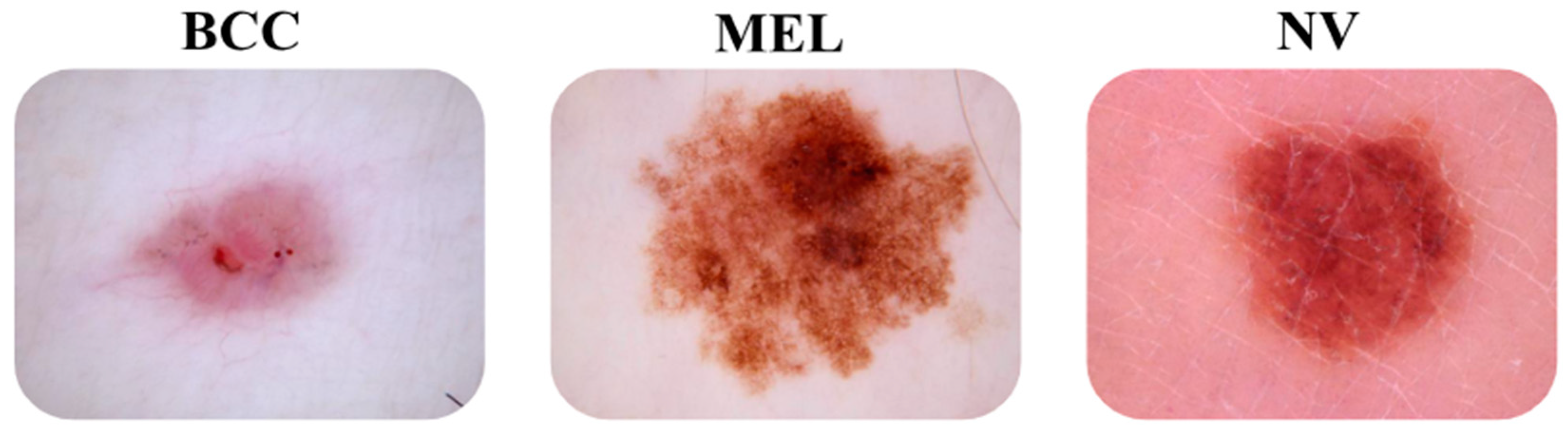
Figure 1 below illustrates some images of the three types of models.
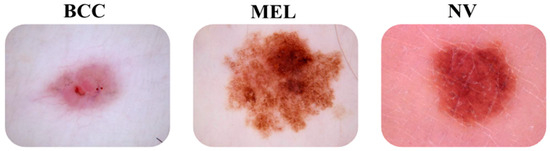
Figure 1.
Images of “bcc” and “mel” skin cancer types and “nv” skin lesions.
3.2. Methodology
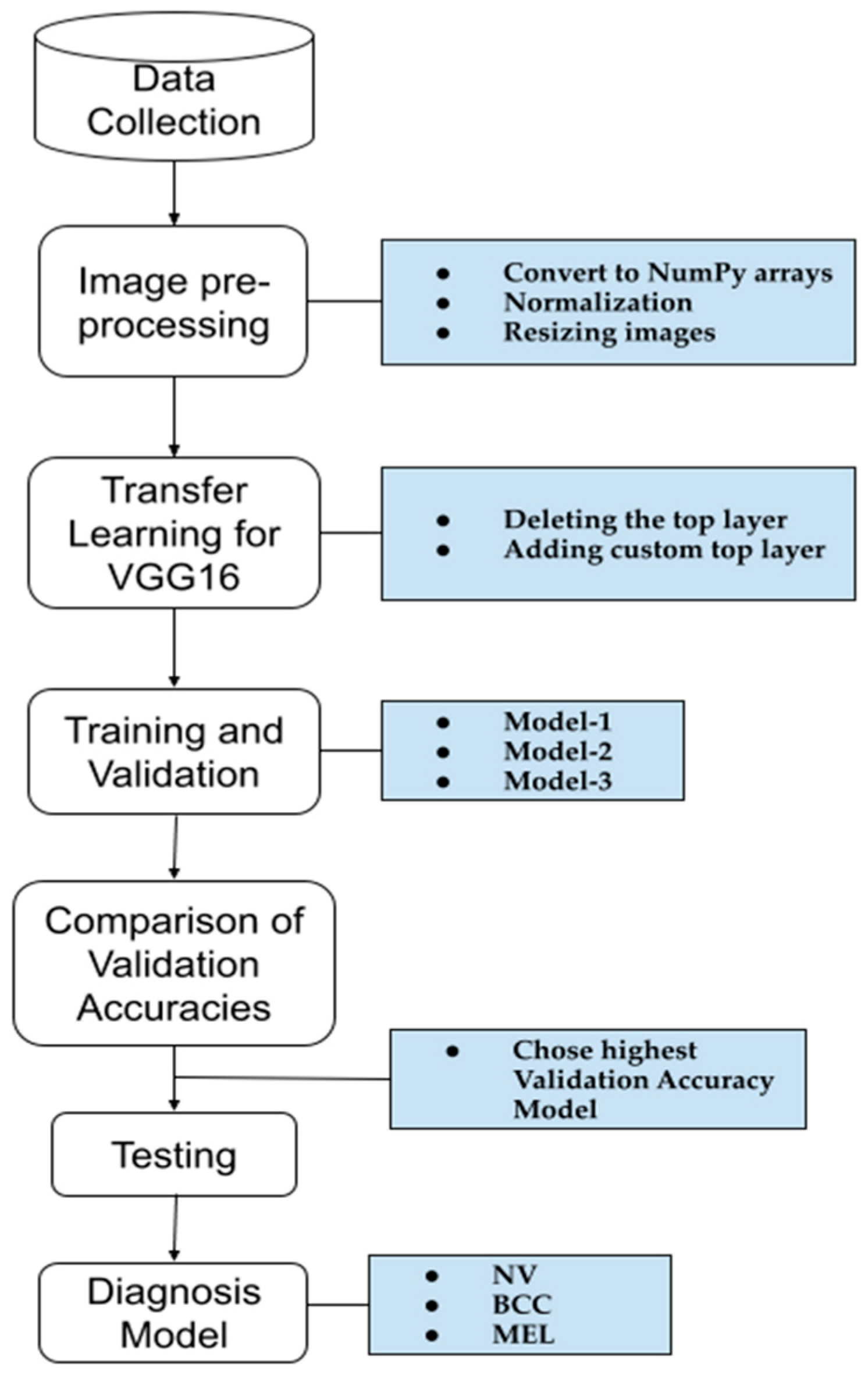
A schematic representation of the methodology used to create our model for skin cancer diagnosis is shown in Figure 2. The first four steps were applied to the three models, each with a different dataset size. The model with the highest accuracy was then tested to confirm its performance. A detailed explanation of each step is provided below.
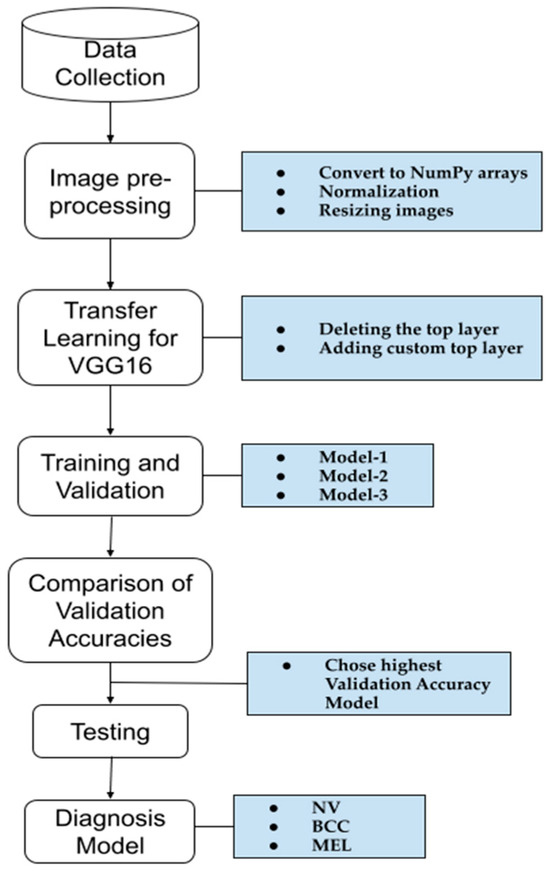
Figure 2.
A schematic representation of the methodology used for the skin cancer diagnosis model.
3.2.1. Data Collection
In deep learning, the most challenging part of building a model is data collection. To achieve high accuracy, we need large datasets, but obtaining real data that precisely fit the problem is often difficult. As mentioned in the “Dataset” Section, Kaggle provides a mid-solution by offering datasets that can potentially fit our study needs while providing an acceptable number of data points for deep learning models. The choice of a different amount of data for each of these three models was intentional as it allowed us to compare the results based on varying data quantities. The rule of data balance for training deep learning models was considered during the collection of the training and test data to avoid model bias towards any of these types: BCC, MEL, or NV.
3.2.2. Image Pre-Processing
After the data collection stage, we moved on to the preprocessing stage where three main concepts were applied to obtain uniform data in terms of size, pixel values, and array dimensions. The following three methods were applied: converting to NumPy arrays, normalization, and resizing images.
- Converting to NumPy arrays
In this step, all of the used images for training, validation, and testing were converted into NumPy arrays, which means each pixel in the image will typically be represented as a NumPy array of shape (3 matrix).
The first framework addresses the force of the red channel. The second addresses the force of the green channel. The third network addresses the power of the blue channel.
Converting data to NumPy arrays enhances performance, simplifies code, and facilitates interoperability with other numerical computing libraries. In detail, they allow for faster operations and utilize less memory, providing a wide range of mathematical functions that operate on entire arrays of data without the need for explicit loops, making code concise and easier to read.
- 2.
- Normalization
In this applied step, the data values (pixels) were adjusted to a common scale by dividing by 255 without distorting differences in the ranges of values. The purpose of this method is to improve convergence and accuracy and avoid numerical instability.
- 3.
- Resizing images
In this study, the data images were resized to 100 × 100 pixels to facilitate easier handling and processing, especially when dealing with large datasets where images might vary in size, as in our case. Additionally, resizing helps reduce the computational load and memory requirements during model training and prediction. While VGG16 typically requires input data of 416 × 416 pixels, resizing to 100 × 100 pixels helps avoid session crashes in Google Collab due to memory limitations.
3.2.3. Transfer Learning for VGG16
Move Learning is frequently utilized to classify skin injuries. Initially, a CNN is prepared on ImageNet; then, at that point, its weighting boundaries are changed to meet the characterization task. Esteva et al. [41] made a significant contribution to the field. Transfer learning was selected for two main reasons: the first was to avoid issues with data scarcity, and the second was to make a comparison with previous work in the same task. In our study, we applied two main actions that fall under the transfer learning method.
- Deleting the Top Layer
In this step, we effectively removed the dense (classification) layers that were originally trained on ImageNet, allowing for the customizing of the top layers of the model according to our classification requirements.
- 2.
- Adding Custom Top Layer
In this step, additional layers (Conv2D, Flatten, Dropout, and Dense) were defined to create a new top layer structure tailored to our specific classification task where the number of classes = 3 (bcc, mel, and nv).
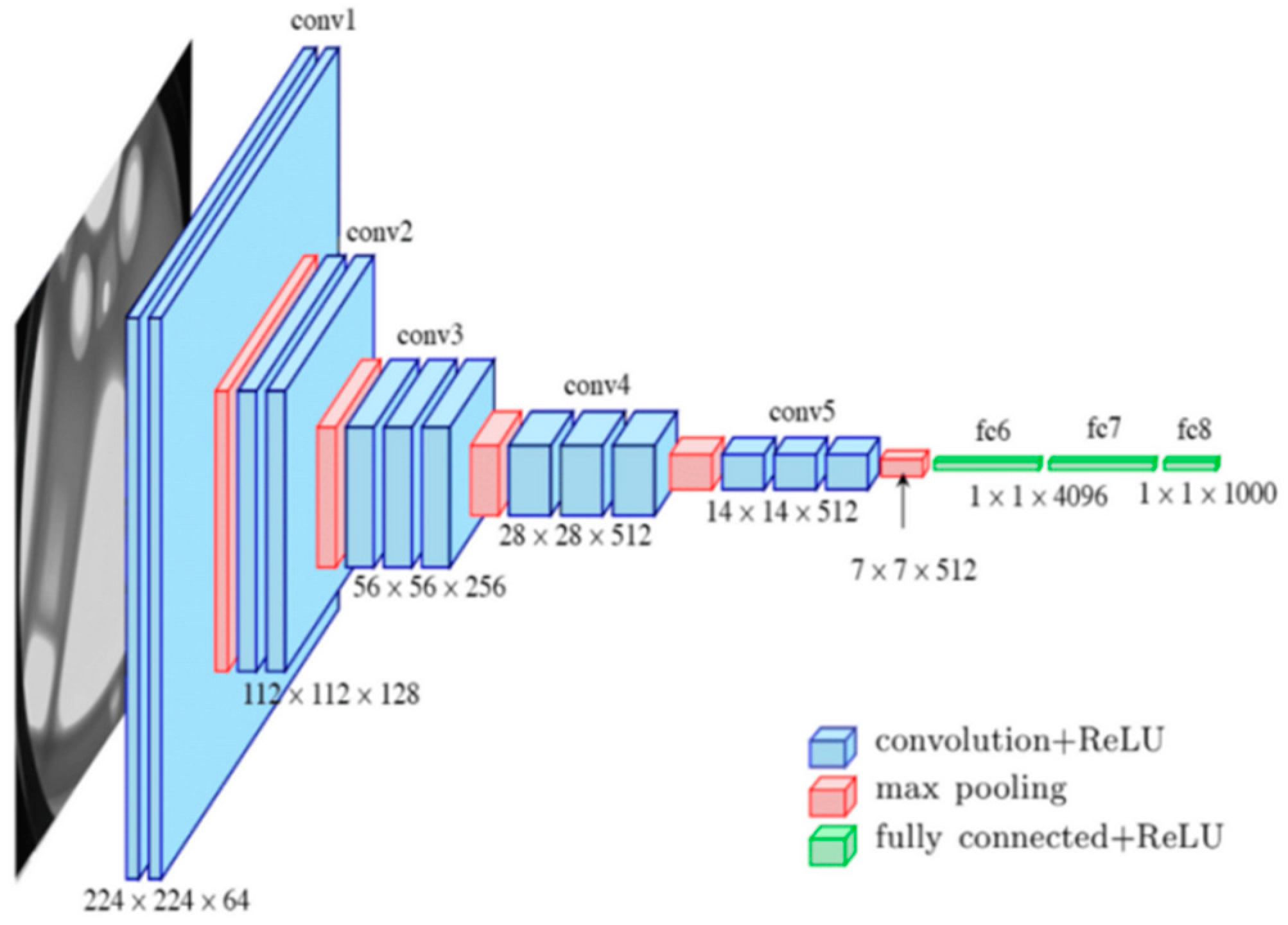
Transfer learning was applied to a pre-trained VGG16 model. For further clarification, the diagram in Figure 3 illustrates the structure of the model before customization.
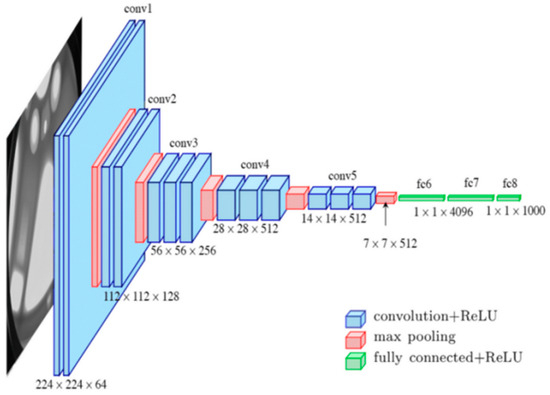
Figure 3.
VGG16’s pre-trained model architecture.
The pre-prepared VGG16 was prepared on a subset of the ImageNet dataset, and an assortment of more than 14 million pictures having a place with 22,000 classifications [42] was selected to create a custom skin anomaly diagnosis model for the following reasons: VGG has more convolution and pooling layers compared to previous architectures. In addition, VGG16 achieved good results with only 7.0% of the top 5 test errors [42]. Our last reason for this selection was to compare it with a previous example that used the same backbone for the same goal.
The main functions of each layer of VGG16 are as follows:
- Convolutional Layers
Convolutional layers extract features from the image, from basic edges to complex patterns.
- Max-Pooling Layers
These layers reduce the spatial dimensions to control overfitting and reduce computational complexity.
- Fully Connected Layers
These layers combine and refine the highlights extricated by the convolutional layers into a last feature vector.
Between the layers that we used in our model, two activation functions were applied:
- ReLU: ReLU acquaints non-linearity with the organization, permitting it to learn complex examples in the information.
- Softmax: This function converts the classification scores into probabilities, providing the final output for classification.
The transfer learning process applied in our current study fundamentally altered the information specific to the first layer of the model according to this study’s needs.
3.2.4. Training and Validation Model 1, Model 2, and Model 3
After customizing the VGG16 model to fit our task, the training and validation steps were applied. We gave specific values to specific parameters needed for the training phase such as:
- Epochs = 40: The number of times the model will iterate over the entire training dataset.
- Batch_size = 32: The number of samples that will be propagated through the network at one time. After processing this batch, the model’s weights will be updated.
- Callbacks: These are special functions that can be called during training at certain points.
- They are used for various purposes, such as saving the model after each epoch or stopping training early if the validation loss stops improving.
- Verbosity mode = 1: This controls the verbosity of the output, and “1” means progress updates will be shown during training.
- Optimizer = Adam: Adam is a popular optimization algorithm used for training machine learning models, especially deep learning models. “Adam” represents a versatile second assessment. It is an augmentation of the Stochastic Slope Plunge (SGD) calculation that determines versatile learning rates for every boundary.
After defining all the needed parameters and their values, training and validation will begin, where the training phase is the period during which the model is exposed to the training dataset. During this phase, the model learns to map input features to output labels. This is achieved by adjusting its weight based on the loss from predictions and true labels. The validation phase involves evaluating the model’s execution on a different approval dataset that the model has not seen during preparation. This helps to monitor how well the model generalizes to new, unseen data.
3.2.5. Comparison of Validation Accuracies
After training the models in the same way and using the same settings, we proceeded to compare their results using the validation accuracy metric. Our goal was to make observations, draw conclusions, or propose hypotheses.
3.2.6. Testing
Once the comparison stage was complete, we proceeded to the testing phase where the model with the highest validation accuracy was selected. Subsequently, the model was evaluated using a set of images (test dataset) that the model had not seen before to obtain a true accuracy measure of the model’s performance. Validation accuracy alone is insufficient to determine the real performance of a model. It was used solely for comparison purposes.
4. Metrics
4.1. Accuracy (ACC)
This metric measures the general rightness of the model’s forecasts. It works out the proportion of accurately anticipated perceptions to the absolute number of perceptions. While exactness is a helpful measurement, it may not be reasonable for imbalanced datasets as it can be biased towards the majority class.
4.2. Loss
This is a proportion of how well the model is performing during preparation. It addresses the contrast between the anticipated qualities and the real qualities. The objective during preparation is to limit this distinction. Normal misfortune capabilities incorporate cross-entropy misfortune for grouping undertakings and mean squared blunder for relapse assignments. Depending on the task, the loss function will be selected; in our case, for three types of classification, the formula of the applied function is as follows:
where
- is the number of samples;
- is the number of classes;
- is a binary indicator (0 or 1) if the class label is the correct classification for the sample ;
- is the predicted probability of the sample being classified as class .
4.3. Precision ()
Precision is estimated as the extent of unequivocally anticipated perceptions to all normal positive perceptions.
4.4. Recall ()
The extent of by and large pertinent outcomes that the calculation appropriately perceives is alluded to as review (Recall):
4.5. F-measure
The F1 score is the mean of the accuracy and recall, respectively. The highest F score is 1, indicating perfect precision and recall scores.
4.6. Learning Curves
Learning curves are visual representations of the model’s performance metrics (such as loss, accuracy, P_curve, R_curve, and PR_curve) on both the training and validation datasets as a function of training epochs or iterations. These curves provide valuable insights into how well the model is learning from the data over time and whether it is overfitting or underfitting. By analyzing learning curves, we can gain insights into the following: model performance, where we assess how well the model is learning from the training data and whether it is generalizing well to unseen data; overfitting and underfitting, where discrepancies between the training and validation curves can indicate overfitting (if the training curve continues to improve while the validation curve stagnates or worsens) or underfitting (if both curves show poor performance); and optimization, where earning curves can guide hyperparameter tuning and optimization efforts. For example, if the model is overfitting, regularization techniques can be applied to mitigate it.
5. Results
5.1. A Comparison of the Three Models Using Validation Accuracy
Table 1 shows the CNN architecture, number of epochs, batch size, optimizer, dataset sizes, types of skin cancers/lesions, number of kernels, kernel size, padding, dropout rates, and activation functions. This thorough comparison highlights the differences and similarities that influence each model’s performance and accuracy.

Table 1.
A comparison of the 3 models during the training and validation steps using some factors.
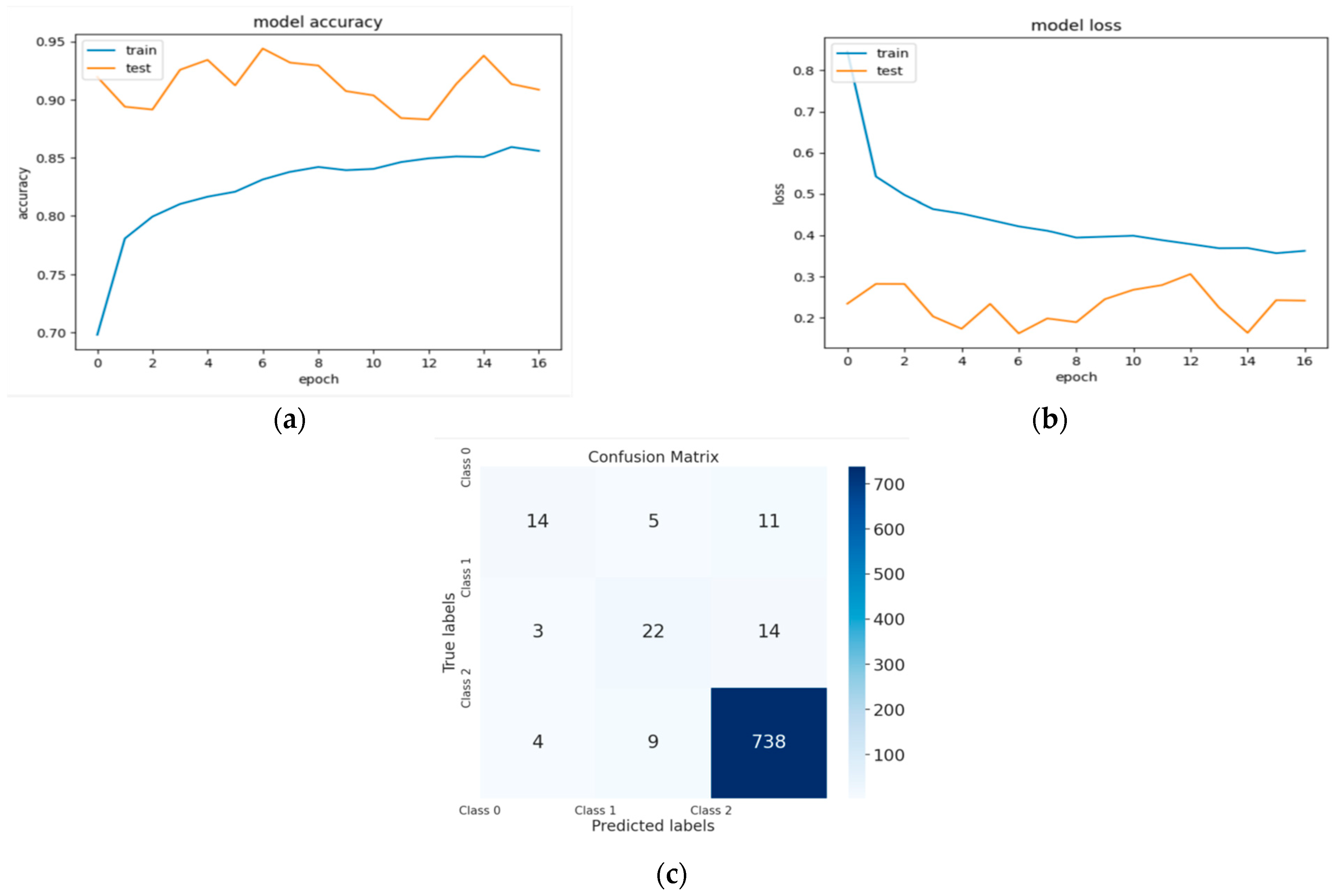
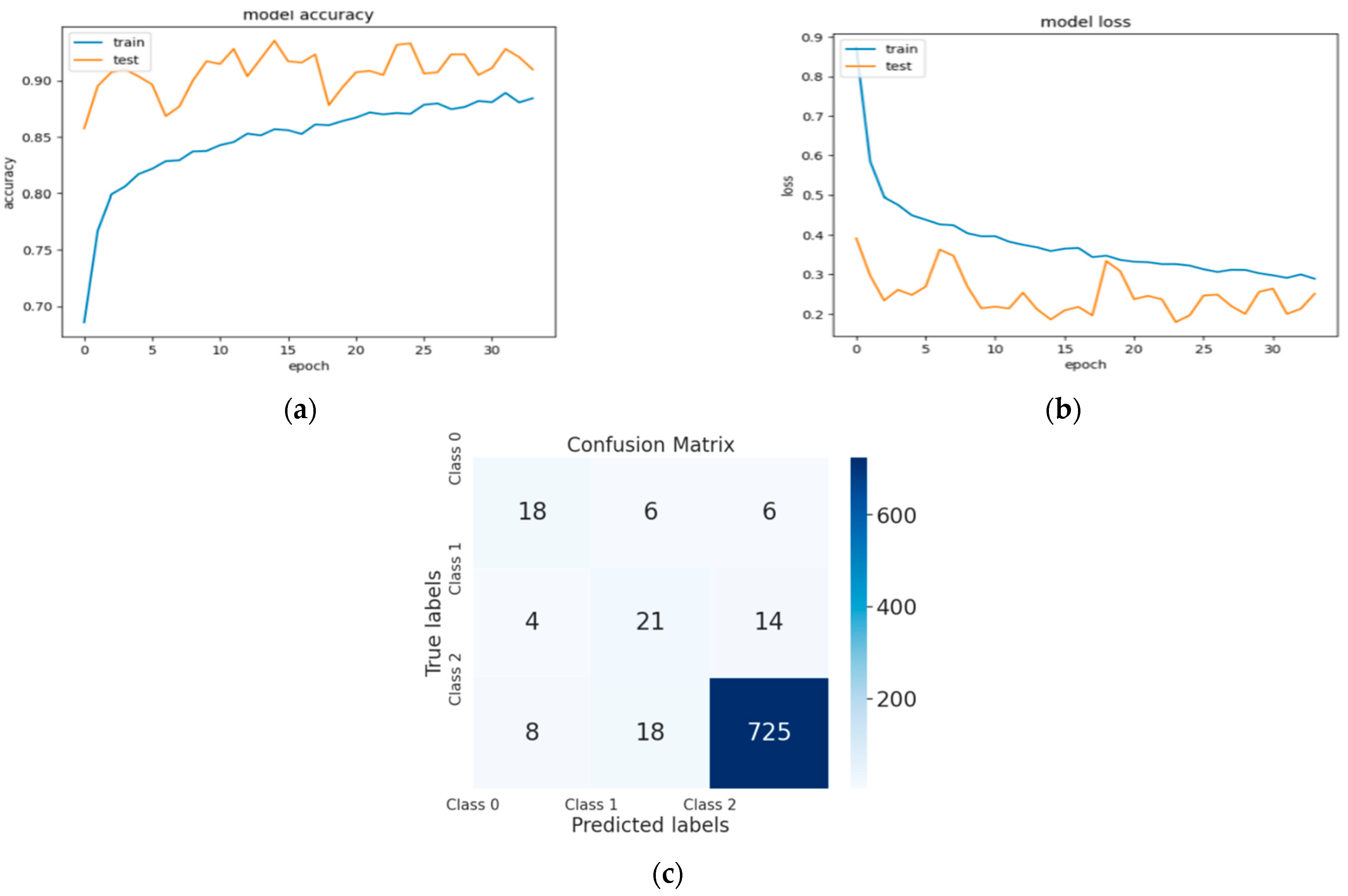
Table 2 summarizes the results of the three models. Model 1, trained on 14,454 images, achieved a validation accuracy of 86%. In contrast, Model 2 and Model 3, trained on smaller datasets of 13,232 images and 10,232 images, respectively, achieved higher validation accuracies of 94% and 93%. For the loss values of these models, Model 2 and Model 3 achieved better results compared to Model 1.

Table 2.
A comparison of the 3 models’ performance.
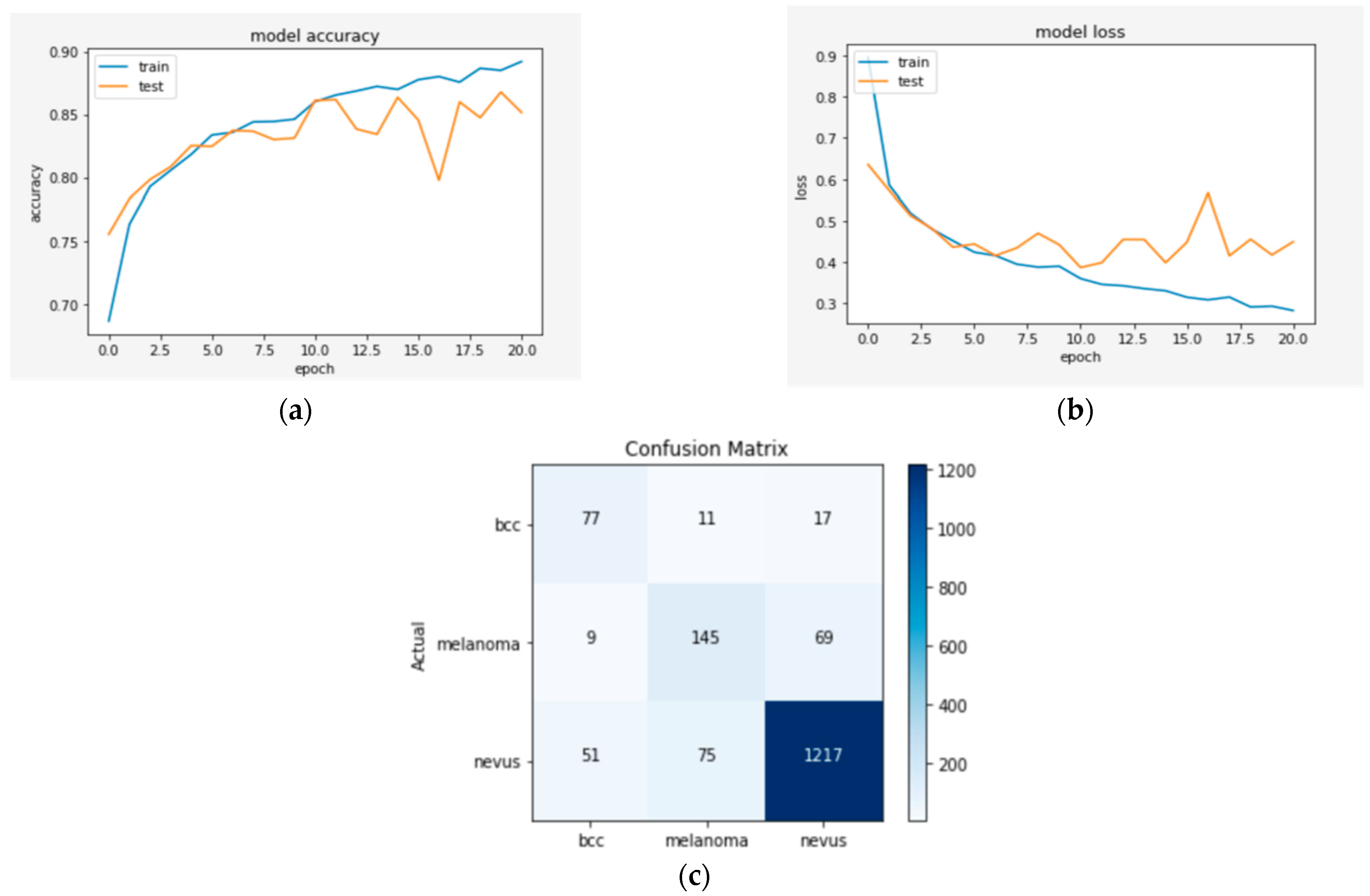
Figure 4, Figure 5 and Figure 6 represent a set of visual representations that help clarify the obtained results further. Each figure includes accuracy learning curves, loss learning curves, and the confusion matrix for each model.
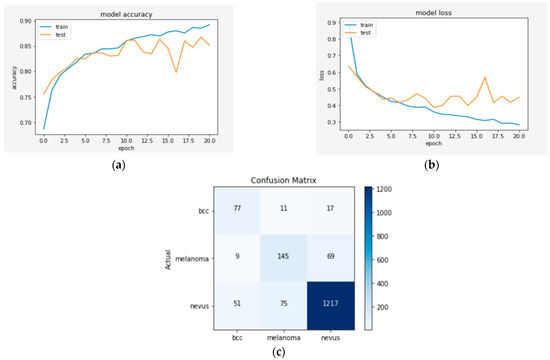
Figure 4.
(a) The validation accuracy curve of Model 1; (b) the validation loss curve of Model 1; and (c) the confusion matrix of Model 1.
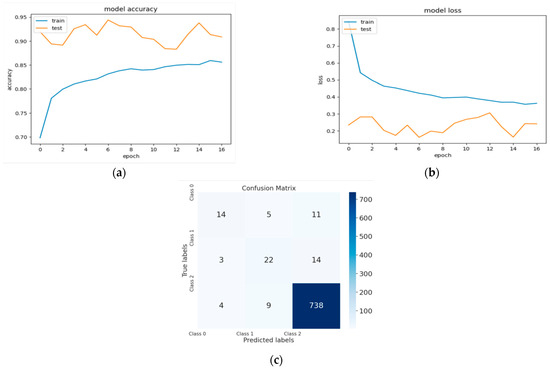
Figure 5.
(a) The validation accuracy curve of Model 2; (b) the validation loss curve of Model 2; and (c) the confusion matrix of Model 2.
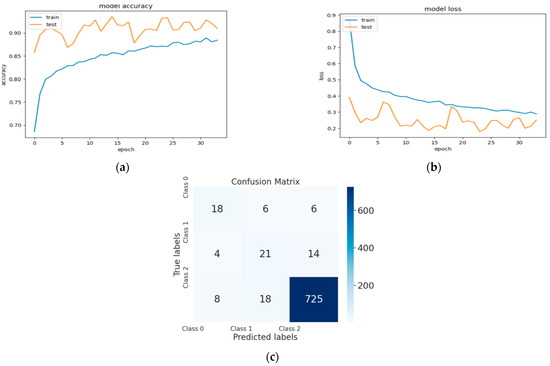
Figure 6.
(a) The validation accuracy curve of Model 3; (b) the validation loss curve of Model 3; and (c) the confusion matrix of Model 3.
5.2. Quality Insights
Based on the obtained results of the comparison phase of the three models, and considering the lingering ambiguity surrounding deep learning, it can be hypothesized that in the field of medicine, the quality of the images used for training deep learning models may be more crucial than the quantity.
A number of ideas and techniques, including preprocessing and the technique used for obtaining the images, are intended to increase quality. In this context, certain preprocessing techniques that can improve quality were mentioned by Madinakhon et al. [43]. Based on these data, it can be concluded that while the concept of quality has been discussed generally, it has not received the required attention. In fact, some researchers in the field overlook quality and bias image selection due to the continued emphasis on quantity in deep learning models.
5.3. Results of Testing Phase for Model 2
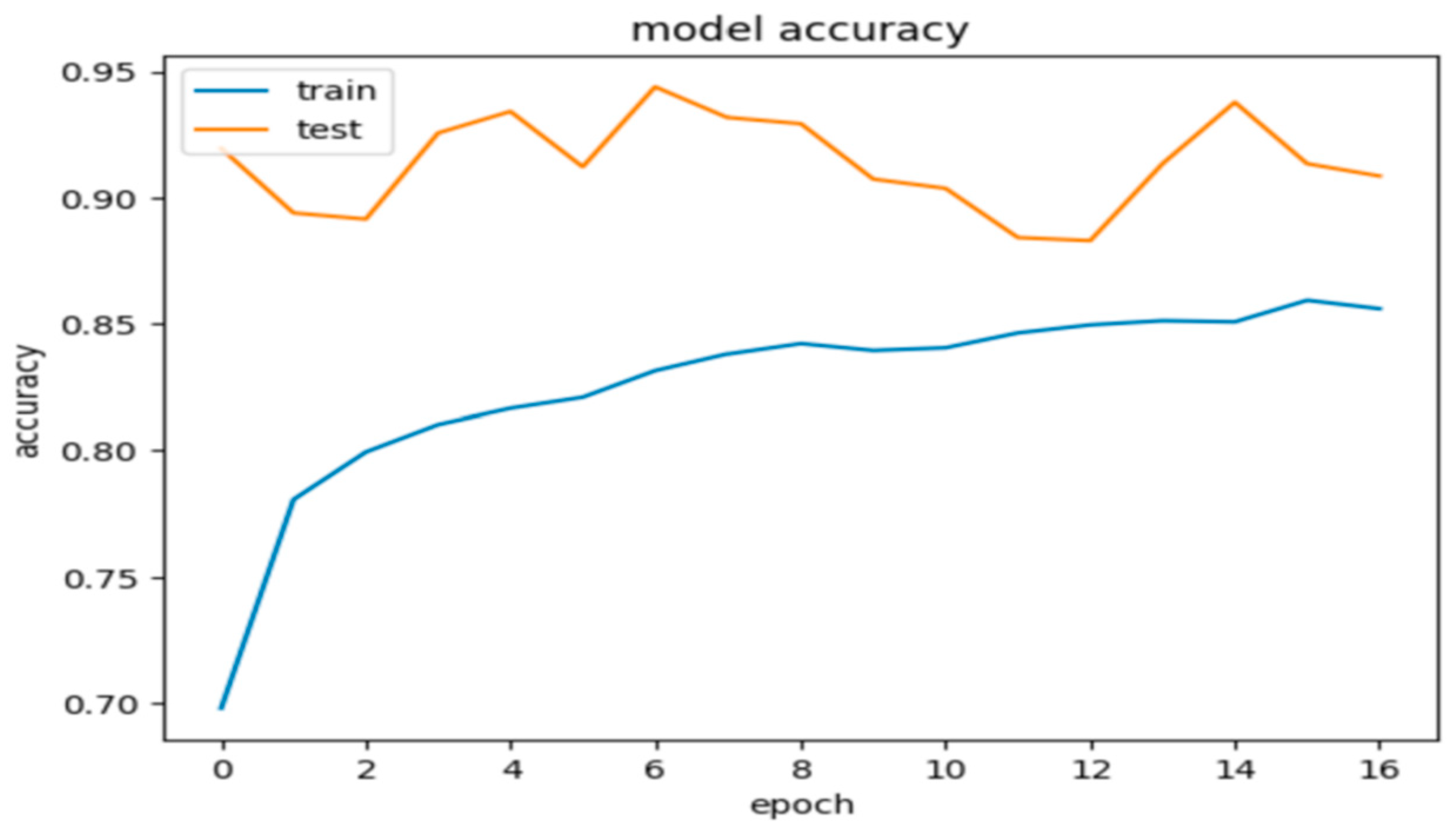
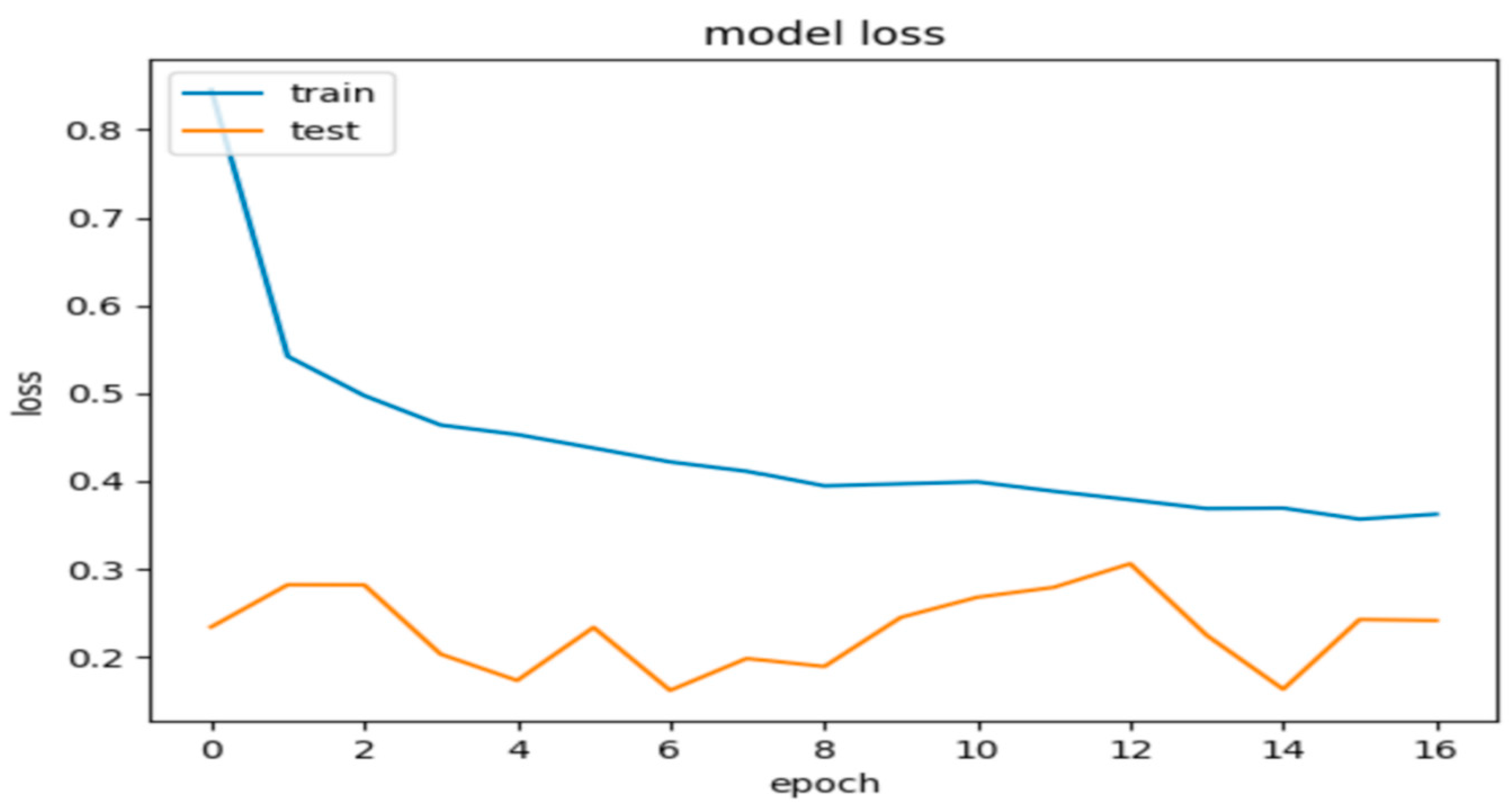
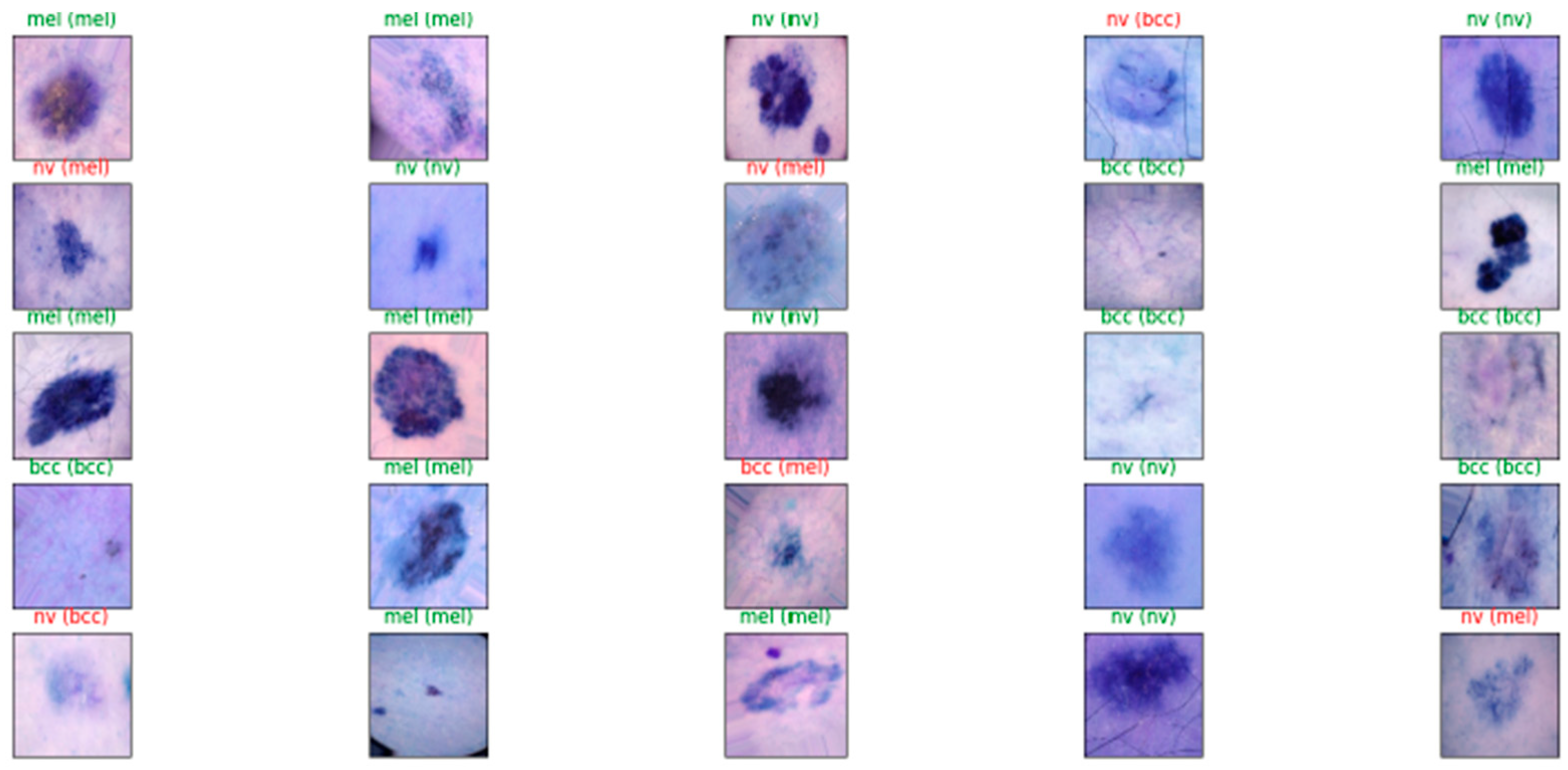
Model 2, when evaluated on the test dataset, yielded promising results with an accuracy of 84.5%. These metrics suggest that the model’s predictions are generally close to the true labels. Additionally, the precision, recall, and F1 score were calculated as 0.86%, 0.85%, and 0.845, respectively, further indicating the model’s promising performance. Figure 7 and Figure 8 show these results using learning curves, and Figure 9 illustrates some predictions of Model 2.
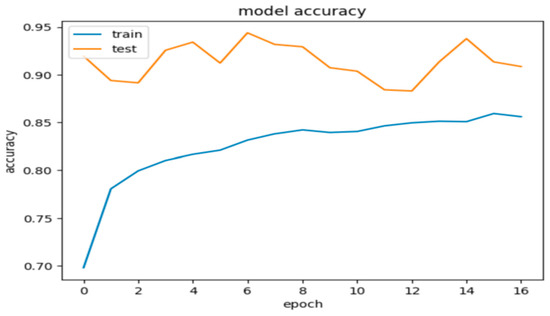
Figure 7.
Model 2’s accuracy during the training and validation phases.
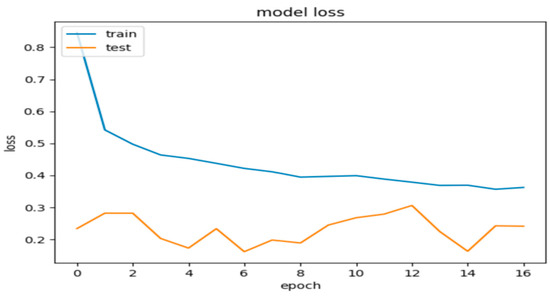
Figure 8.
Model 2’s loss during the training and validation phases.
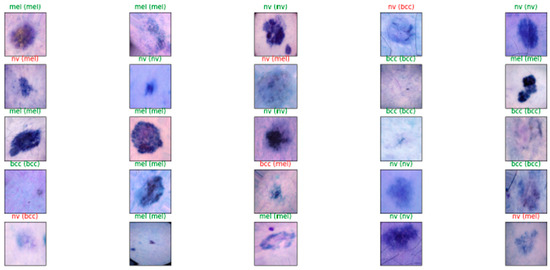
Figure 9.
Results of Model 2’s diagnosis process for some images.
5.4. Comparison with Related Work
In our study, we compared our VGG16 model to other related methods, and we mainly focused on types of cancers, datasets, and results as shown in Table 3. This comparative analysis was essential to evaluate the efficiency and contribution of our approach in terms of existing methodologies in skin cancer diagnosis. Our VGG16 model showed promising performance in the results compared to previous work.

Table 3.
Comparative analysis of different methods used for skin cancer diagnosis.
While this study presents promising results in utilizing the VGG-16 model for skin cancer diagnosis, several limitations must be acknowledged. One significant concern is the potential bias in dataset selection, as the images were sourced primarily from Kaggle [47], which may not fully represent the diverse spectrum of skin types and lesions encountered in clinical practice. This limitation may affect the generalizability of the model, particularly when applied to less common skin lesions or those from diverse demographic groups. Additionally, variations in imaging techniques, lighting conditions, and image quality may introduce inconsistencies that impact the model’s performance. Future research should aim to include more varied datasets and address these biases to enhance the robustness and applicability of the model across a broader range of skin conditions.
6. Conclusions
This work was conducted based on the problem revolving around the doubtful diagnosis of skin anomalies, namely NV and two types of skin cancer, BCC and MEL. Using the selected methodology, VGG16, three models were developed, Model 1, Model 2, and Model 3, with diagnostic validation accuracies of 86%, 94%, and 93%, respectively. These values are satisfactory. We found that the second model is the best of the three by comparing these values. We used validation accuracy for comparison purposes. It is not advisable to use it to determine the performance of a model. To learn more about the diagnostic behavior and performance of Model 2, it was tested on an additional unpublished dataset of 4500 photos. After the learning phase, the following results were obtained: 84%, 84.5%, 84.6%, and 86% for the accuracy, f1 score, recall, and accuracy, respectively. We found that the model works well when comparing these results with similar work, particularly by comparing the precision values.
In this study, we also varied the quantity of the data for each model in order to understand how these changes impact the model’s performance. During the comparison phase between the three models, we kept the same code base. This means that the variables used during the training and validation phases, which could affect the model’s performance, were consistent for all three models. This approach ensured a fair comparison, with the results being influenced only by a single variable, which is the amount of data used to form each model. Model 1 was formed with 14,454 images, the second with 13,232 images, and the third with 10,232 images. The images in Models 2 and 3 come from a single dataset, while Model 1 was trained with another dataset to observe the results. The comparison step demonstrated the critical importance of utilizing high-quality images to develop a successful model. By highlighting this in training profound learning models for the classification of medical images, our work contributes to the field of deep learning models. Indeed, these models require a substantial amount of data, which has prompted extensive research efforts to collect additional data in pursuit of improved performance. However, a significant challenge remains due to the considerable lack of data in most disciplines, particularly in the medical field. In summary, to achieve the desired results, researchers must place greater emphasis on the preprocessing steps of the images.
This research shows the effectiveness of the VGG-16 model in skin cancer diagnosis, and several avenues for future research could enhance its performance and applicability. Exploring alternative architectures, such as ResNet and VGG-19, may improve accuracy and robustness due to their advanced feature extraction capabilities. Incorporating ensemble methods that combine predictions from multiple models could further enhance diagnostic accuracy. However, it is crucial to note that these models must be trained on pre-processed data to ensure optimal performance.
Future research should focus on expanding the dataset to include a wider variety of skin types, lesions, and imaging conditions to improve generalizability. Implementing techniques such as data augmentation and synthetic image generation could simulate diverse scenarios and strengthen model adaptability. Additionally, integrating explainable AI methods would provide insights into the model’s decision-making processes, increasing trust among healthcare professionals. Lastly, conducting longitudinal studies and real-world clinical trials to evaluate the model’s performance over time and in diverse healthcare settings would be invaluable for assessing its practical utility and efficacy in improving patient outcomes.
In the future, it may be possible to achieve better results using other deep learning networks like ResNET, VGG-19, and RCNN. Our work was conducted using the VGG-16 model, which is regarded as an effective architecture. Using a faster, more robust model is not enough to diagnose skin cancer reliably from complex images. However, this does not preclude the possibility that other networks may yield superior results. The model with the highest validation score was selected for further testing using a separate test dataset that it had not previously encountered in order to accurately evaluate the model’s performance. In context related to training medical models, we hope there will be wider interest and a greater focus on the importance of data quality, whether in the preprocessing phases or the acquisition phase of images, i.e., the acquisition method and the tools used.
Our research aims to advance general deep learning models, specifically for medical applications. Additionally, we want to improve the model’s ability to identify more abnormalities and malignancies and determine whether they are connected to other illnesses or the skin.
Author Contributions
Conceptualization, K.D.; Methodology, K.D.; Software, R.B.B. and H.M.; Validation, K.D.; Formal analysis, K.D.; Investigation, K.D.; Resources, K.D.; Data curation, K.D.; Writing—review & editing, K.D., R.B.B. and H.M.; Supervision, P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are openly available on Kaggle at https://www.kaggle.com/datasets/kmader/skin-cancer-mnist-ham10000, accessed in the period of 8 June 2024.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rogers, H.W.; Weinstock, M.A.; Feldman, S.R.; Coldiron, B.M. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA Dermatol. 2015, 151, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts and Figures 2024. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2024-cancer-facts-figures.html (accessed on 17 January 2024).
- Mansouri, B.; Housewright, C. The treatment of actinic keratoses—The rule rather than the exception. J. Am. Acad. Dermatol. 2017, 153, 1200. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat. Rev. Clin. Oncol. 2021, 18, 170–186. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Thammasitboon, S.; Singhal, G.; Epner, P. Diagnostic Error. In Patient Safety; Agrawal, A., Bhatt, J., Eds.; Springer: Cham, Switzerland, 2023; pp. 225–239. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Luna, S.A.; Siddique, Z. Machine-Learning-Based Disease Diagnosis: A Comprehensive Review. Healthcare 2022, 10, 541. [Google Scholar] [CrossRef]
- Galić, I.; Habijan, M.; Leventić, H.; Romić, K. Machine Learning Empowering Personalized Medicine: A Comprehensive Review of Medical Image Analysis Methods. Electronics 2023, 12, 4411. [Google Scholar] [CrossRef]
- Groh, M.; Badri, O.; Daneshjou, R.; Koochek, A.; Harris, C.; Soenksen, L.R.; Doraiswamy, P.M.; Picard, R. Deep learning-aided decision support for diagnosis of skin disease across skin tones. Nat. Med. 2024, 30, 573–583. [Google Scholar] [CrossRef]
- Singh, B.; Malhotra, H.; Kumar, D.; Mujtaba, S.F.; Upadhyay, A.K. Understanding Cellular and Molecular Events of Skin Aging and Cancer: An Integrative Perspective. In Skin Aging & Cancer; Dwivedi, A., Agarwal, N., Ray, L., Tripathi, A., Eds.; Springer: Singapore, 2019; pp. 27–46. [Google Scholar] [CrossRef]
- Swathi, B.; Kannan, K.; Chakravarthi, S.S.; Ruthvik, G.; Avanija, J.; Reddy, C.C.M. Skin Cancer Detection using VGG16, InceptionV3 and ResUNet. In Proceedings of the 2023 4th International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 6–8 July 2023; pp. 812–818. [Google Scholar] [CrossRef]
- Aljohani, K.; Turki, T. Automatic Classification of Melanoma Skin Cancer with Deep Convolutional Neural Networks. AI 2022, 3, 512–525. [Google Scholar] [CrossRef]
- Lomas, A.; Leonardi-Bee, J.; Bath-Hextall, F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br. J. Dermatol. 2012, 166, 1069–1080. [Google Scholar] [CrossRef] [PubMed]
- Christenson, L.J.; Borrowman, T.A.; Vachon, C.M.; Tollefson, M.M.; Otley, C.C.; Weaver, A.L.; Roenigk, R.K. Incidence of basal cell and squamous cell carcinomas in a population younger than 40 years. JAMA 2005, 294, 681–690. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma recognition in Dermoscopy images via aggregated deep convolutional features. IEEE Trans. Biomed. Eng. 2019, 66, 1006–1016. [Google Scholar] [CrossRef]
- Lodde, G.; Zimmer, L.; Livingstone, E.; Schadendorf, D.; Ugurel, S. Malignant melanoma. Hautarzt 2020, 71, 63–77. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Abeni, D.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur. J. Cancer 2005, 41, 28–44. [Google Scholar] [CrossRef]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Greenspan, H.; van Ginneken, B.; Summers, R.M. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans. Med. Imaging 2016, 35, 1153–1159. [Google Scholar] [CrossRef]
- Ghanem, N.M.; Attallah, O.; Anwar, F.; Ismail, M.A. Artificial Intelligence in Cancer Diagnosis and Prognosis, Volume 2: Breast and Bladder Cancer; IOP Publishing: Bristol, UK, 2022. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; 2012; pp. 1097–1105. Available online: https://dl.acm.org/doi/10.1145/3065386 (accessed on 1 August 2024).
- Dong, Y.; Hu, Z.; Uchimura, K.; Murayama, N. Driver inattention monitoring system for intelligent vehicles: A review. IEEE Trans. Intell. Transp. Syst. 2011, 12, 596–614. [Google Scholar] [CrossRef]
- Vincent, P.; Larochelle, H.; Lajoie, I.; Bengio, Y.; Manzagol, P.-A. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 2010, 11, 3371–3408. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Bengio, Y. Generative adversarial nets. In Advances in Neural Information Processing Systems; 2014; pp. 2672–2680. Available online: https://dl.acm.org/doi/10.1145/3422622 (accessed on 1 August 2024).
- Kingma, D.P.; Welling, M. Auto-encoding variational bayes. arXiv 2013, arXiv:1312.6114. [Google Scholar]
- Dinh, L.; Sohl-Dickstein, J.; Bengio, S. Density estimation using real nvp. arXiv 2016, arXiv:1605.08803. [Google Scholar]
- Lipton, Z.C.; Berkowitz, J.; Elkan, C. A critical review of recurrent neural networks for sequence learning. arXiv 2015, arXiv:1506.00019. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; 2017; pp. 5998–6008. Available online: https://arxiv.org/abs/1706.03762 (accessed on 1 August 2024).
- Chamberlain, A.J.; Fritschi, L.; Kelly, J.W. Nodular melanoma: Patients’ perceptions of presenting features and implications for earlier detection. J. Am. Acad. Dermatol. 2003, 48, 694–701. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; Cai, W.; Pujol, S.; Kikinis, R.; Feng, D. Early diagnosis of Alzheimer’s disease with deep learning. In Proceedings of the International Symposium on Biomedical Imaging, Beijing, China, 29 April–2 May 2014; pp. 1015–1018. [Google Scholar]
- Brosch, T.; Tam, R. Manifold learning of brain MRIs by deep learning. Med. Image Comput. Comput. Assist. Interv. 2013, 16, 633–640. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Williams, R.J.; Zipser, D. A learning algorithm for continually running fully recurrent neural networks. Neural Comput. 1989, 1, 270–280. [Google Scholar] [CrossRef]
- Smolensky, P. Information Processing in Dynamical Systems: Foundations of Harmony Theory; Colorado University at Boulder, Department of Computer Science: Boulder, CO, USA, 1986. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A Survey of the Recent Architectures of Deep Convolutional Neural Networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Nazari, S.; Garcia, R. Automatic Skin Cancer Detection Using Clinical Images: A Comprehensive Review. Life 2023, 13, 2123. [Google Scholar] [CrossRef]
- Dildar, M.; Akram, S.; Irfan, M.; Khan, H.U.; Ramzan, M.; Mahmood, A.R.; Alsaiari, S.A.; Saeed, A.H.M.; Alraddadi, M.O.; Mahnashi, M.H. Skin Cancer Detection: A Review Using Deep Learning Techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, M.; Gilani, S.Q.; Syed, T.; Marques, O.; Kim, H.-C. Skin Cancer Detection Using Deep Learning—A Review. Diagnostics 2023, 13, 1911. [Google Scholar] [CrossRef]
- Abadi, M. TensorFlow: Learning functions at scale. In Proceedings of the 21st ACM SIGPLAN International Conference on Functional Programming, Nara, Japan, 18–24 September 2016; Volume 51, p. 1. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Madinakhon, R.; Mukhtorov, D.; Cho, Y.-I. Integrating Principal Component Analysis and Multi-Input Convolutional Neural Networks for Advanced Skin Lesion Cancer Classification. Appl. Sci. 2024, 14, 5233. [Google Scholar] [CrossRef]
- Saini, A.; Guleria, K.; Sharma, S. Skin Cancer Classification Using Transfer Learning-Based Pre-Trained VGG 16 Model. In Proceedings of the 2023 IEEE International Conference on Computer Communication and Information Systems (ICCCIS), Greater Noida, India, 3–4 November 2023; pp. 305–310. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Jin, Z. A Visually Interpretable Deep Learning Framework for Histopathological Image-Based Skin Cancer Diagnosis. IEEE J. Biomed. Health Inform. 2021, 25, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Khamsa, D.; Pascal, L.; Zakaria, B.; Lokman, M.; Zakaria, M.Y. Skin Cancer Diagnosis and Detection Using Deep Learning. In Proceedings of the 2023 International Conference on Electrical Engineering and Advanced Technology (ICEEAT), Batna, Algeria, 5–7 November 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset: A large collection of multi-source dermatoscopic images of common pigmented skin lesions. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).