Abstract
Recently, convolutional neural networks (CNNs) and self-attention mechanisms have been widely applied in plant disease identification tasks, yielding significant successes. Currently, the majority of research models for tomato leaf disease recognition rely solely on traditional convolutional models or Transformer architectures and fail to capture both local and global features simultaneously. This limitation may result in biases in the model’s focus, consequently impacting the accuracy of disease recognition. Consequently, models capable of extracting local features while attending to global information have emerged as a novel research direction. To address these challenges, we propose an Eff-Swin model that integrates the enhanced features of the EfficientNetV2 and Swin Transformer networks, aiming to harness the local feature extraction capability of CNNs and the global modeling ability of Transformers. Comparative experiments demonstrate that the enhanced model has achieved a further increase in training accuracy, reaching an accuracy rate of 99.70% on the tomato leaf disease dataset, which is 0.49~3.68% higher than that of individual network models and 0.8~1.15% higher than that of existing state-of-the-art combined approaches. The results show that integrating attention mechanisms into convolutional models can significantly enhance the accuracy of tomato leaf disease recognition while also offering the great potential of the Eff-Swin backbone with self-attention in plant disease identification.
1. Introduction
Tomatoes, as one of the three major vegetables traded globally, play a pivotal role in the global vegetable trade [1]. With the increasing demand for tomatoes, the global production and planting area of tomatoes continue to expand. Concurrently, the growing consumer demand for food quality, including ecofriendly and high-quality produce, is increasing. However, tomato production and quality are frequently compromised by various diseases, resulting not only in diminished yields and quality but also in substantial economic losses for farmers [2]. Therefore, ensuring tomato yield and quality is highly important for the global population and even global economic trade. Traditionally, the identification of tomato leaf diseases has heavily relied on visual observation by workers, a method fraught with subjectivity, low accuracy, and inefficiency. Therefore, it is particularly important to achieve efficient automatic identification of tomato leaf diseases and pests using modern technology.
With the rise of computer network technology, research on pest identification using machine learning has made significant advancements. Machine learning categorizes and labels external image features of crops, revealing associative information among data through techniques such as data analysis and data mining. Consequently, it automatically detects the extent of damage, greatly improving the efficiency and accuracy of recognition. However, due to the reliance of machine learning on manually extracted features and the varied performance of different disease characteristics in different scenarios, along with significant differences in symptoms of the same disease at different stages of onset, in complex environmental backgrounds, it is unable to autonomously capture advanced semantic features, resulting in low learning efficiency and weak generalization, making it difficult to achieve satisfactory recognition results. Compared to traditional machine learning methods, deep-learning-based disease recognition methods do not require complex image preprocessing or feature extraction operations. Instead, they automatically extract image features by introducing operations such as convolutional layers, pooling layers, and fully connected layers. Their deep network structures can accommodate rich semantic information, enabling them to quickly and accurately complete target localization and image classification tasks even in complex application scenarios, demonstrating excellent recognition performance in pest and disease image analysis. For example, Sladojevic et al. [3] proposed a recognition model for leaf diseases utilizing deep convolutional neural network technology and successfully identifying 13 types of plant diseases. Liu et al. [4] developed a plant disease detection model based on AlexNet for recognizing apple leaf diseases. Latif et al. [5] improved the ResNet model for pest and disease detection and increased the detection accuracy. Yao et al. [6] proposed an enhanced model based on VGG16 for apple leaf disease identification, where they substituted the fully connected layer with a global average pooling layer to reduce the number of parameters and integrated batch normalization layers to improve the convergence speed. Zeng et al. [7] presented a group multiscale attention network (GMA-Net) for rubber tree leaf disease image recognition by devising group multiscale dilated convolution (GMDC) modules for multiscale feature extraction and cross-scale attention feature fusion (CAFF) modules for integrating multiscale attention features. Huang et al. [8] proposed an automatic identification and detection method for crop leaf disease detection based on a fully convolutional–switchable normalized dual-path network (FC-SNDPN); this was used to reduce the influence of complex backgrounds on the image identification of crop diseases and pests.
In the field of natural language processing (NLP), the Transformer [9] has achieved significant success in many tasks due to its self-attention mechanism. With the deepening of research on Transformers, this architecture has shown enormous application prospects in computer vision tasks. For example, Dosovitskiy et al. [10]., inspired by Transformers in the NLP field, directly applied the standard Transformer to image tasks, leading to the proposal of the Vision Transformer (ViT). The Swin Transformer [11] architecture adopts a hierarchical approach commonly used in CNNs for model construction, facilitating the capture of relevant dataset features across different layers.
Although the aforementioned studies have robustly demonstrated the effectiveness of CNNs and Transformer structures in the field of plant pest and disease recognition, there are still some limitations, such as the potential for further improvement in model recognition accuracy and the inability to simultaneously focus on both local and global features. Therefore, we advocate for the fusion of CNNs and Swin Transformers for tomato disease recognition. Our objective is to harness CNNs’ strengths in image feature extraction alongside the rich feature representations acquired by Swin Transformers for processing long sequential data and establishing inter-feature dependencies. This approach is intended to enhance model recognition accuracy and bolster the model’s generalization capability.
2. Methods
2.1. Overview
CNNs excel at feature extraction due to their inherent localized perception. Similarly, the self-attention mechanism in Transformer structures also demonstrates excellent capabilities for extracting global features. Our aim is to leverage the strengths of both CNNs and Transformers by establishing a hybrid network. Furthermore, owing to its faster training speed and fewer parameters compared to other CNN architectures, EfficientNetV2 is well suited for the efficient recognition and classification of agricultural diseases.
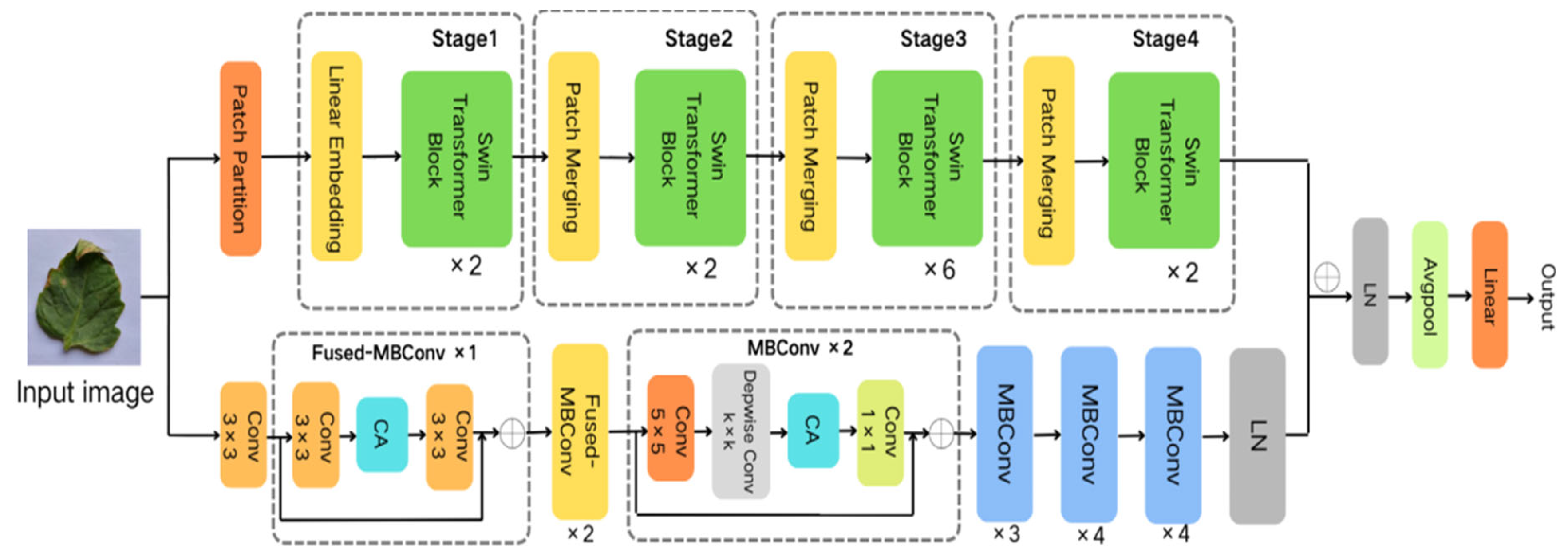
Thus, this study employs EfficientNetV2 and the Swin Transformer to construct distinct branches for capturing local and global features, respectively. Building upon this foundation, we propose the Eff-Swin model, which consists of three main components: a CNN branch primarily based on the improved EfficientNetV2 module, a Transformer branch primarily based on the Swin Transformer module, and a feature fusion classification module. The network architecture of the model is depicted in Figure 1.
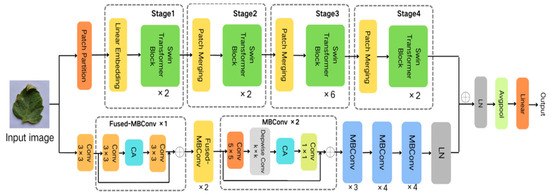
Figure 1.
Network architecture of the Eff-Swin model.
2.2. Convolutional Neural Network (CNN) Backbone
To minimize overfitting and improve performance, the model’s CNN backbone should employ a concise structure with fewer parameters, such as the VGG model [12] or EfficientNetV2. The CNN backbone proposed in this paper is inspired by EfficientNetV2. Subsequently, the CNN branch of the model was designed based on EfficientNetV2, yielding promising results.
EfficientNetV2, acting as the CNN branch of Eff-Swin, primarily focuses on extracting shallow features from input images and iteratively refining them to capture local characteristics. To further enhance the accuracy of tomato disease image recognition, this study introduces the channel attention (CA) mechanism to further enhance EfficientNetV2. This enhancement strengthens the learning of crucial positional information within tomato leaf disease images.
EfficientNetV2 provides three initial models: S, M, and L. To minimize overfitting and achieve optimal performance, we chose the lightest initial model, EfficientNetV2-S, as the basis for our improvements. Although EfficientNetV2-S is the lightest among the initial models provided by the original authors, it still encounters challenges during training, including high computational demands, lengthy training times, and significant model space occupation. To address these issues, we made enhancements to the original network structure layers.
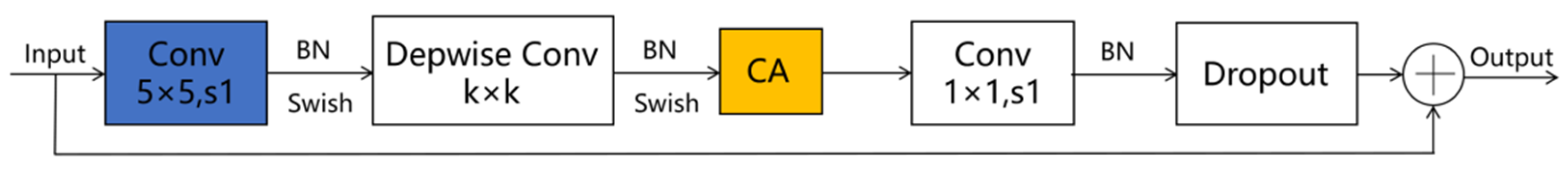
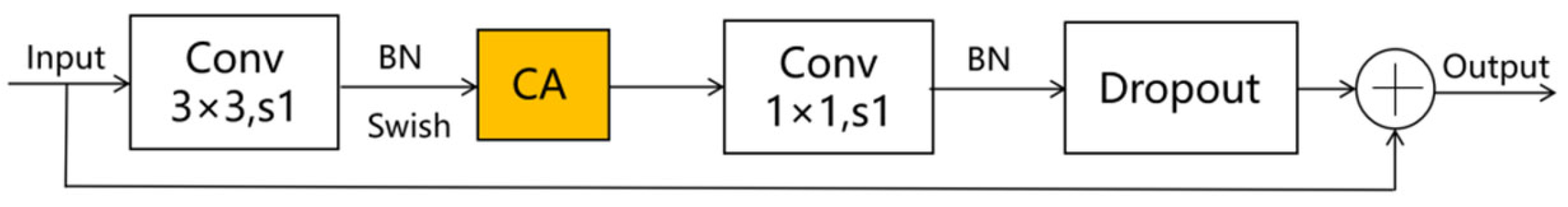
Given the smaller expansion ratio of MBConv versus Fused-MBConv, which results in less memory access overhead, we employ Fused-MBConv in only the initial two stages. In contrast, MBConv modules are utilized in the latter four stages to enhance the model’s lightweight characteristics. Furthermore, we replace the SE module in both the MBConv and Fused-MBConv modules with a CA module, as illustrated in Figure 2 and Figure 3, respectively. This modification allows the CA module to grasp long-term dependencies across network channels, thereby enabling the network to concentrate on target-relevant regions without sacrificing precise positional details. Subsequently, we adopt 5 × 5 convolutional kernels over the original 3 × 3 kernels within MBConv to improve the accuracy and reduce the number of convolutional layers. By modifying the quantity of MBConv modules and substituting certain convolutional layers with larger 5 × 5 kernels, we expand this branch’s receptive field. Consequently, this leads to a reduced need for convolutional layers to sustain model accuracy, decreasing the network layers in stages 1 through 6 to 1, 2, 2, 3, 4, and 4 layers, respectively. By employing fewer convolutional layers than EfficientNetV2, this branch architecture notably enhances the feature extraction capability. In the final stage, we normalize the output from the sixth stage, apply global average pooling, flatten it, and subsequently integrate it with the Swin Transformer branch.
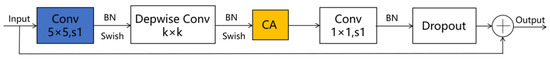
Figure 2.
The improved MBConv schematic diagram. Note: ⊕ stands for channel-level addition.
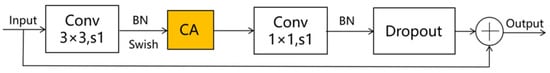
Figure 3.
The improved Fused-MBConv schematic diagram. Note: ⊕ stands for channel-level addition.
2.3. Swin Transformer Backbone
While the EfficientNetV2 branch leverages the enhanced EfficientNetV2 network for extracting local feature information, it struggles to adequately capture global feature information, particularly concerning large areas of disease in tomato images. In contrast to convolutional operations, which are renowned for their efficacy in extracting local features, the Swin Transformer excels in global modeling, thereby adeptly capturing global dependencies. Consequently, to preserve spatial information more effectively in tomato disease images, we introduce the Swin Transformer branch to complement the feature extraction process.
The Swin Transformer is a visual model based on the Transformer architecture that enhances the applicability and generalizability of Transformers through techniques such as blockwise self-attention, hierarchical structure, and multiscale fusion. Its primary feature lies in its adoption of a hierarchical Transformer structure, where feature maps at various scales are constructed by merging image blocks. Moreover, it employs a shifted window mechanism to limit the computational scope of self-attention, facilitating cross-window connections. This hierarchical structure enables the Swin Transformer to adapt to various scale visual tasks, while the shifted window mechanism maintains linear computational complexity proportional to the image size.
In the initial stage of this process, a 224 × 224 × 3 image is input into the patch partition and divided into multiple nonoverlapping patches of size 4 × 4, where each patch has the dimensions of 4 × 4 × 3. In Stage 1, the partitioned patches are input into a linear embedding layer to transform the feature dimension to C and then fed into a Swin Transformer block. The Swin Transformer block performs self-attention calculations to extract image features, and the resulting output is passed to the next stage. For Stages 2 to 4, the process involves patch merging, where the feature maps input from the previous stage are merged into 2 × 2 sizes to combine adjacent windows. These merged feature maps are then input into the Swin Transformer block to generate hierarchical feature representations. During this process, the patch merging layer handles downsampling and dimensionality increase, whereas the Swin Transformer block extracts features from the images.
The Swin Transformer block module consists of a window-based multi-head self-attention (W-MSA) module, a shifted window-based multi-head self-attention (SW-MSA) module, a multilayer perceptron (MLP) module with a nonlinear GELU activation function, a residual connectivity layer, a normalization function, and the Swin Transformer block itself. The SW-MSA module is the core of the Swin Transformer block, which calculates self-attention by alternating between standard windows and shifted windows at different stages. This design enables the model to effectively capture global information while ensuring computational efficiency. In the process of self-attention computation, the model employs relative positional encoding to compute similarity and confines the attention calculation within each window, thereby mitigating computational overheads. The formula for self-attention calculation with relative positional encoding is as follows:
where B is the relative position code B∈. M2 indicates the number of patches in the window. Q, K, V∈, where d is the dimension of the matrix. is the scale factor used to avoid the influence of dot product variance, and k is the number of queries.
The formula for the overall block is as follows:
where represents the characteristics of (S) the output of the W-MSA module, and Zl represents the characteristics of the output of the MLP module. L indicates the number of blocks.
3. Experimental Results and Analysis
3.1. Datasets
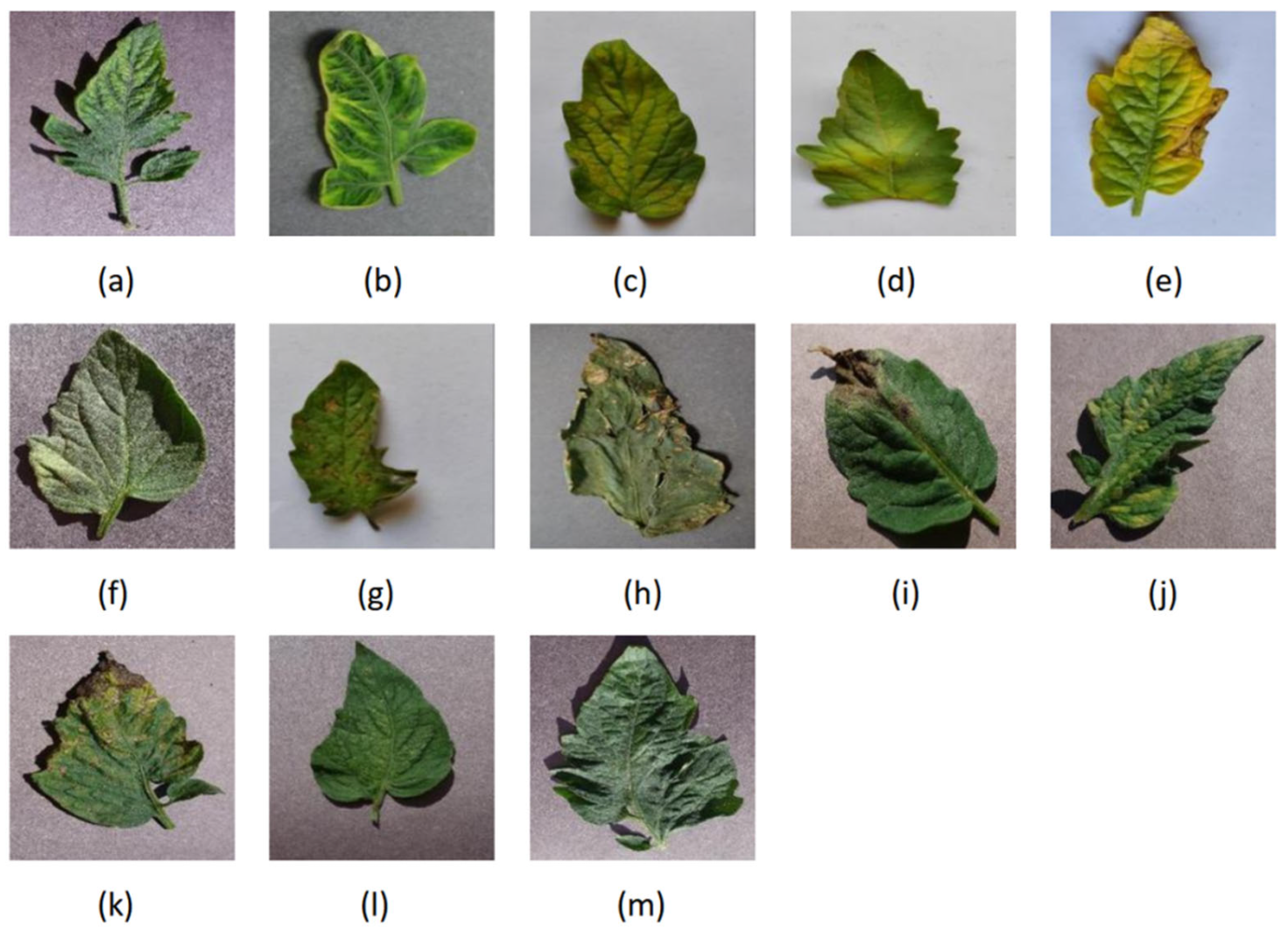
The dataset used in this study is sourced from the PlantVillage dataset [13] and the Tomato-Village dataset [14]. The PlantVillage dataset contains a large number of images depicting plant diseases and pests. There are ten different classes in this dataset of 18,161 tomato leaves from PlantVillage. Nine of those ten classes are for diseases, while the final class is for health. The ten classes are healthy, bacterial spot, early blight, late blight, leaf mold, Septoria leaf spot, spider mites, target spot, tomato mosaic virus, and tomato yellow leaf curl virus. In tomato villages, there are eight different classes in this dataset of 4064 tomato leaves. Seven of eight classes are for diseases, while the final class is for health. The eight classes are early blight, late blight, leaf minor, magnesium deficiency, nitrogen deficiency, potassium deficiency, and healthy. To enhance the reliability of the model and the generalization of training, this study utilizes data from both the PlantVillage and Tomato-Village datasets, mixing images of the same categories of diseases and pests. We selected the 13 most common types of tomato leaf disease images and healthy tomato leaf images and randomly selected images from each category at a ratio of 4:1. These categories included early blight, bacterial spot, late blight, leaf mold, Septoria leaf spot, spider mites, target spot, tomato mosaic virus, tomato yellow leaf curl virus, magnesium deficiency, nitrogen deficiency, potassium deficiency, and healthy, totaling 17,043 images, as illustrated in Figure 4.
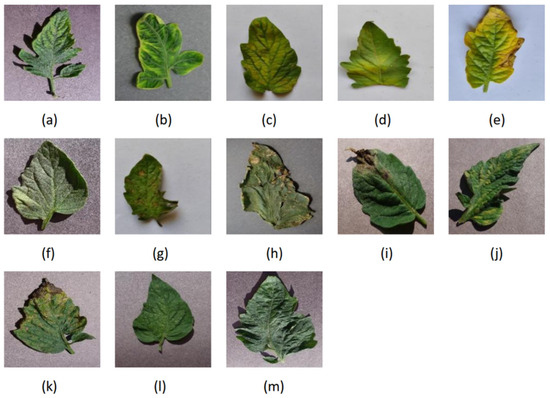
Figure 4.
Sample plant–disease pairs of tomato leaf disease: (a) early blight, (b) bacterial spot, (c) late blight, (d) leaf mold, (e) Septoria leaf spot, (f) spider mites, (g) target spot, (h) tomato mosaic virus, (i) tomato yellow leaf curl virus, (j) magnesium deficiency, (k) nitrogen deficiency, (l) potassium deficiency, (m) healthy.
3.2. Image Preprocessing
To achieve better detection performance, preprocessing is applied to the tomato leaf images. Due to variations in image sizes and the presence of redundant information, the images of tomato leaf diseases are first resized to 224 × 224 × 3 pixels. Subsequently, the processed images are normalized by subtracting the mean value, serving as the final experimental dataset for the tomato disease samples. Consequently, a total of 17,043 images are utilized for classification, with the dataset divided into training, validation, and testing phases at a ratio of 70:15:15. The data used in the experiment are all the same dataset, that is, the final dataset obtained after image processing.
3.3. Evaluation Metrics
Evaluating the proposed model is crucial, and employing various evaluation metrics is essential for this purpose. The metrics used for evaluation are vital for ensuring the performance of the model. The predictive or classification performance of the model can be measured using metrics such as accuracy, precision, recall, and F1-score.
3.3.1. Accuracy
The accuracy is the proportion of accurately classified predictions made. The calculation method is as follows:
The abbreviations “TP”, “TN”, “FP”, and “FN” stand for “true positives”, “true negatives”, “false positives”, and “false negatives”.
3.3.2. Precision
Precision represents the proportion of true positive results. The calculation method is as follows:
3.3.3. Recall
Recall represents the proportion of true positives successfully detected. The calculation method is as follows:
3.3.4. F1-Score
The F1-score is computed as the harmonic mean of precision and recall, defined as follows:
3.4. Ablation Experiment
Regarding the effectiveness of the attention mechanism selected for the CNN branch, this study conducted experiments on various attention mechanisms on tomato leaf disease and pest image datasets to compare their impacts on the network, including the initial SE, CBAM, ECA, and CA methods used in this paper, as shown in Table 1.

Table 1.
Comparison of the results of different attention mechanism methods.
The experimental results indicate that the CA exhibits different improvements in recognition accuracy compared to the CBAM, SE, and ECA attention modules. As the CA attention mechanism not only captures inter-channel information but also considers directionally relevant positional information; it demonstrates greater efficiency and better performance in handling global contextual dependencies and channel spatial relationships than SE, CBAM, and ECA. Therefore, the CA attention mechanism shows better effectiveness in classifying tomato leaf diseases and pests with the Eff-Swin dual-stream network, achieving a network model accuracy of 99.70%, which was 1.16% higher than that of the original SE method.
Furthermore, to further validate the effectiveness of the CNN branch and Swin Transformer branch proposed in this paper, detailed ablation experiments were conducted on tomato leaf disease and pest image datasets. Table 2 presents the results of the ablation experiments for the proposed methods, where the CNN branch is based on the improved EfficientNetV2 framework, and the backbone network of the Transformer branch is the Swin Transformer. The experimental results demonstrate that the accuracy of the Eff-Swin model, which combines the improved EfficientNetV2 and Swin Transformer, is 0.49%~3.68% higher than the accuracy of the individual branches. This indicates that the Eff-Swin algorithm can effectively extract and integrate both local and global information from tomato disease and pest images, making the feature representation more discriminative and further improving the accuracy of tomato disease and pest classification.

Table 2.
Ablation experiment of the Eff-Swin algorithm.
3.5. Result Analysis
To validate the effectiveness and advancement of the algorithm proposed in this paper, comparisons were made between the proposed algorithm and existing mainstream image classification methods on the tomato disease and pest dataset, as shown in Table 3. All models were trained 300 epochs to provide a fair comparison. In addition, for fair contrast, the batch size was kept the same between comparators. It is important to note that all algorithms were evaluated under the same dataset and experimental conditions.

Table 3.
Comparative experiment results of different models.
From the comparative experimental results, it can be observed that the performance of the proposed dual-branch network model in tomato leaf disease and pest identification outperforms that of the classical convolutional neural networks ResNet18 and EfficientNetv2. Compared to the lightweight convolutional neural network model MobileNet-V2, the classification accuracy of the proposed model is improved by 7.45%. Compared to Transformer-based models such as Vision Transformer and Swin Transformer, the proposed model achieves classification accuracy improvements of 4.51% and 0.58%, respectively. This indicates that the combined dual-stream network model performs better in tomato leaf disease and pest classification than single-stream network models.
Moreover, compared to mainstream feature fusion networks, including Conformer [15] and CMT [16], which also integrate features extracted from CNN and Transformer branches, the experimental results show that the proposed model achieves classification accuracy improvements of 1.15% and 0.8%, respectively. This suggests that the CA attention mechanism enhances the generalization ability of the network model, and the combination of a CNN and a Swin Transformer achieves superior classification accuracy compared to current mainstream methods.
Although the training computational time of Eff-Swin model is a little longer than some lightweight models, the accuracy is much higher than them such as MobileNet-V2 and VGG-19 and so on. We believe that the high performance of Eff-Swin model makes it worth training for longer than others to a certain extent. In addition, for the same type of binding model, with the concise CNN backbone we designed, our model not only trained faster than other CNN branches but also had higher accuracy.
4. Discussion
Table 4 summarizes recent relevant studies on tomato leaf disease classification, presenting the employed architectures, publication years, number of considered classes and samples, and the best-performing model based on achieved accuracy. Compared with other single-stream models, the proposed model in this paper exhibits a significant advantage of 1.7–9%. In addition to CNN-based and Transformer-based methods, Fuzzy SVM [17], DVT [18], and R–CNN [17] have all been used to carry out the classification task. (In addition to methods based on CNNs or Transformers, Fuzzy support vector machines and R-CNNs have also been utilized for classification tasks.) To the best of our knowledge, few studies have classified nine diseases and health, and none have classified the thirteen diseases and health. Additionally, our study presents a relatively compact and efficient model that not only eliminates restrictions posed by other factors but also outperforms other single-stream models in terms of accuracy, making it a valuable tool for tomato leaf disease classification.

Table 4.
Result comparisons with related studies.
However, our study also has certain limitations. First, it exclusively focuses on the task of tomato leaf disease recognition and does not consider other types of recognition tasks. Second, the study is based only on two datasets and does not consider the differences and impacts among multiple datasets.
In summary, the results of this study indicate that the proposed combined algorithm theoretically exhibits significant effectiveness and superiority in the task of tomato leaf disease recognition. These findings provide useful information for future research and practical solutions for application. Future studies could further explore how to apply the proposed improved algorithm to other types of disease recognition tasks and conduct experimental validation, as well as enhance the real-time performance of the model and better handle the differences and impacts among different datasets.
5. Conclusions
This study primarily focused on classifying tomato leaf diseases by proposing a custom fusion model and comparing its performance with that of existing mainstream image classification models. To extract features from tomato disease images more effectively, we first apply a self-attention-based Transformer structure to tomato leaf disease image classification and detection and propose the Eff-Swin model, which integrates the EfficientNetv2 and Swin Transformer structures. To improve the effectiveness of CNN feature extraction on tomato disease image features and avoid overfitting caused by model fusion, we replaced the SE module in EfficientNetv2 with a CA module in the Eff-Swin model. Additionally, we redesigned the network architecture of EfficientNetv2 as the CNN backbone of Eff-Swin while employing the Swin-Transformer-based structure to construct the Transformer backbone.
The experimental results demonstrate that the Eff-Swin model achieves a remarkable accuracy of 99.70% on the tomato leaf disease dataset. We also conducted comprehensive experiments using other state-of-the-art methods. The results indicate that for the tomato disease dataset, the Eff-Swin model outperforms the best-performing CNN model in the experiments by a significant margin of 1.16%, the best-performing Transformer model (2021) by 3.68%, and the best-performing fusion model by 0.8%. These results underscore the exceptional classification performance of the Eff-Swin model in tomato leaf disease image classification and recognition tasks.
Furthermore, the model evaluation reveals that the incorporation of appropriate attention mechanisms significantly enhances the robustness of the Eff-Swin model. Among the models employed in this study, the improved EfficientNetv2 model achieves a high accuracy of 99.21%, notably outperforming other studies that have attempted the same task.
In conclusion, the Eff-Swin model represents a pioneering attempt to combine EfficientNetV2 with the Swin Transformer and apply it to the challenging task of tomato leaf disease recognition. The model’s outstanding feature extraction capabilities and extensive potential for application in such tasks have been clearly demonstrated.
The outcomes of this research can provide advanced methodological guidance for researchers dedicated to real-time classification tasks in the agricultural domain. Moreover, the practical application of this work is not limited solely to tomato leaf disease recognition but can also be extended to the classification of leaf diseases in other plants.
Furthermore, by integrating the Eff-Swin model with other techniques, such as image segmentation and feature extraction, the fusion of these methods holds promise for achieving optimal results in plant disease recognition tasks. This line of research has the potential to revolutionize the field of plant disease diagnosis and contribute to the development of more efficient and effective agricultural practices.
Author Contributions
Conceptualization, L.N.; methodology, L.N.; software, Y.S.; validation, Y.S.; formal analysis, Y.S.; data curation, Y.S.; writing—original draft preparation, Y.S.; writing—review and editing, Y.S., J.Y., L.N. and B.Z.; visualization, Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a project of Shandong Province Higher Educational Program for Introduction and Cultivation of Young Innovative Talents in 2021 and National College Students’ innovation and entrepreneurship training program (No. 202310434201).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Li, T.; Cui, J.; Guo, W.; She, Y.; Li, P. The Influence of Organic and Inorganic Fertilizer Applications on Nitrogen Transformation and Yield in Greenhouse Tomato Cultivation with Surface and Drip Irrigation Techniques. Water 2023, 15, 3546. [Google Scholar] [CrossRef]
- Yu, Y.H. Research Progress of Crop Disease Image Recognition Based on Wireless Network Communication and Deep Learning. Wirel. Commun. Mob. Comput. 2021, 2021, 7577349. [Google Scholar] [CrossRef]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, Y.; He, D.J.; Li, Y.X. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Latif, G.; Alghazo, J.; Maheswar, R.; Vijayakumar, V.; Butt, M. Deep learning based intelligence cognitive vision drone for automatic plant diseases identification and spraying. J. Intell. Fuzzy Syst. 2020, 39, 8103–8114. [Google Scholar] [CrossRef]
- Yao, J.B.; Liu, J.H.; Zhang, Y.N.; Wang, H.S. Identification of winter wheat pests and diseases based on improved convolutional neural network. Open Life Sci. 2023, 18, 20220632. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.W.; Li, C.M.; Zhang, B.; Wang, R.R.; Fu, W.; Wang, J.; Zhang, X.R. Rubber Leaf Disease Recognition Based on Improved Deep Convolutional Neural Networks With a Cross-Scale Attention Mechanism. Front. Plant Sci. 2022, 13, 829479. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.B.; Chen, A.B.; Zhou, G.X.; Zhang, X.; Wang, J.W.; Peng, N.; Yan, N.; Jiang, C.H. Tomato Leaf Disease Detection System Based on FC-SNDPN. Multimed. Tools Appl. 2023, 82, 2121–2144. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.Q.; Liu, M.J.X.; Liu, T.Y.; Lin, H.; Huang, C.B.; Ning, L. Attention is all you need: Utilizing attention in AI-enabled drug discovery. Brief. Bioinform. 2024, 25, bbad467. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. arXiv 2021, arXiv:2103.14030. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef]
- Gehlot, M.; Saxena, R.K.; Gandhi, G.C. “Tomato-Village”: A dataset for end-to-end tomato disease detection in a real-world environment. Multimed. Syst. 2023, 29, 3305–3328. [Google Scholar] [CrossRef]
- Peng, Z.L.; Guo, Z.H.; Huang, W.; Wang, Y.W.; Xie, L.X.; Jiao, J.B.; Tian, Q.; Ye, Q.X. Conformer: Local Features Coupling Global Representations for Recognition and Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 9454–9468. [Google Scholar] [CrossRef]
- Guo, J.; Han, K.; Wu, H.; Tang, Y.; Chen, X.; Wang, Y.; Xu, C. CMT: Convolutional Neural Networks Meet Vision Transformers. arXiv 2021, arXiv:2107.06263. [Google Scholar] [CrossRef]
- Nagamani, H.S.; Sarojadevi, H. Tomato leaf disease detection using deep learning techniques. Int. J. Adv. Comput. Sci. Appl. 2022, 13. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, G.; Lv, T.; Zhang, X. Using a Hybrid Convolutional Neural Network with a Transformer Model for Tomato Leaf Disease Detection. Agronomy 2024, 14, 673. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, Q.; Liu, A.; Meng, X.; Loui, A. Can Deep Learning Identify Tomato Leaf Disease? Adv. Multimed. 2018, 2018, 6710865. [Google Scholar] [CrossRef]
- Agarwal, M.; Singh, A.; Arjaria, S.; Sinha, A.; Gupta, S. ToLeD: Tomato leaf disease detection using convolution neural network. Procedia Comput. Sci. 2020, 167, 293–301. [Google Scholar] [CrossRef]
- Elhassouny, A.; Smarandache, F. Smart mobile application to recognize tomato leaf diseases using Convolutional Neural Networks. In Proceedings of the 2019 International Conference of Computer Science and Renewable Energies (ICCSRE), Agadir, Morocco, 22–24 July 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Ahmad, I.; Hamid, M.; Yousaf, S.; Shah, S.T.; Ahmad, M.O. Optimizing Pretrained Convolutional Neural Networks for Tomato Leaf Disease Detection. Complexity 2020, 2020, 8812019. [Google Scholar] [CrossRef]
- Nithish Kannan, E.; Kaushik, M.; Prakash, P.; Ajay, R.; Veni, S. Tomato leaf disease detection using convolutional neural network with data augmentation. In Proceedings of the 2020 5th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 10–12 June 2020; pp. 1125–1132. [Google Scholar] [CrossRef]
- Basavaiah, J.; Arlene Anthony, A. Tomato leaf disease classification using multiple feature extraction techniques. Wirel. Pers. Commun. 2020, 115, 633–651. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, G.; Chen, A.; Yi, J.; Zhang, W.; Hu, Y. Identification of tomato leaf diseases based on combination of ABCK-BWTR and B-ARNet. Comput. Electron. Agric. 2020, 178, 105730. [Google Scholar] [CrossRef]
- Thangaraj, R.; Anandamurugan, S.; Kaliappan, V.K. Automated tomato leaf disease classification using transfer learning-based deep convolution neural network. J. Plant Dis. Prot. 2021, 128, 73–86. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Peng, Y.; Liu, J.Z.; Wu, S. Tomato Leaf Disease Diagnosis Based on Improved Convolution Neural Network by Attention Module. Agriculture 2021, 11, 651. [Google Scholar] [CrossRef]
- Chen, H.C.; Widodo, A.M.; Wisnujati, A.; Rahaman, M.; Lin, J.C.W.; Chen, L.K.; Weng, C.E. AlexNet Convolutional Neural Network for Disease Detection and Classification of Tomato Leaf. Electronics 2022, 11, 951. [Google Scholar] [CrossRef]
- Peng, D.; Li, W.J.; Zhao, H.M.; Zhou, G.X.; Cai, C. Recognition of Tomato Leaf Diseases Based on DIMPCNET. Agronomy 2023, 13, 1812. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).