Abstract
Malaria is a leading cause of morbidity and mortality in tropical and sub-tropical regions. This research proposed a malaria diagnosis system based on the you only look once algorithm for malaria parasite detection and the convolutional neural network algorithm for malaria parasite life stage classification. Two public datasets are utilized: MBB and MP-IDB. The MBB dataset includes human blood smears infected with Plasmodium vivax (P. vivax). While the MP-IDB dataset comprises 4 species of malaria parasites: P. vivax, P. ovale, P. malariae, and P. falciparum. Four distinct stages of life exist in every species, including ring, trophozoite, schizont, and gametocyte. For the MBB dataset, detection and classification accuracies of 0.92 and 0.93, respectively, were achieved. For the MP-IDB dataset, the proposed algorithms yielded the accuracies for detection and classification as follows: 0.84 and 0.94 for P. vivax; 0.82 and 0.93 for P. ovale; 0.79 and 0.93 for P. malariae; and 0.92 and 0.96 for P. falciparum. The detection results showed the models trained by P. vivax alone provide good detection capabilities also for other species of malaria parasites. The classification performance showed the proposed algorithms yielded good malaria parasite life stage classification performance. The future directions include collecting more data and exploring more sophisticated algorithms.
1. Introduction
Red blood cells (RBCs) play an important role in oxygen transmission of organs; thus, they must traverse narrow blood vessels without undergoing destruction. During the intra-erythrocytic stage of malaria parasite infection, red blood cells undergo remarkable changes [1]. Such parasitic invasion supports survival of the parasite within the host red blood cell and often leads to an increased incidence rate of the disease that includes cerebral malaria and anaemia [2]. Malaria cannot be treated until it is diagnosed. Typical symptoms of malaria infection include fever, muscle fatigue, and headaches [3]. If a delayed diagnosis or treatment occurs, it will lead to anaemia and block the capillaries that carry blood to the brain [4].
Malaria is a disease transmitted by the female Anopheles mosquito and ranks among the foremost global public health concerns. This severe parasitic infection stands as a primary contributor to illness and death in tropical and sub-tropical regions worldwide. According to the World Health Organization (WHO), in the past 2 decades, the number of malaria infections has seen a remarkable decline, plummeting by 76% from 22.8 million cases in 2000 to 5.4 million cases in 2021. Additionally, the incidence rate has substantially dropped by 82%, falling from 17.9 to 3.2 cases per 1000 population at risk in the Southeast Asia region [5]. Five different species of plasmodium parasite can infect humans, including P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi. Each of these undergoes four distinct stages in their life cycle, comprising four distinct phases in their development: ring, trophozoite, schizont, and gametocyte [6]. Among them, only four are considered real parasites that can infect humans, while P. knowlesi is still considered a zoonotic malaria. P. vivax stands as the prevailing parasite in the WHO Region of the Americas, contributing to 64% of malaria cases. Its prevalence exceeds 30% in the WHO Southeast Asia region and reaches 40% in the Eastern Mediterranean regions [7].
Malaria is a treatable illness, with available medications for its management, including preventive drugs for travelers who visit malaria-endemic areas. However, despite active research and field studies, there is currently no effective malaria vaccine. Once infected by malaria, it advances swiftly, underscoring the critical significance of timely diagnosis [8]. Due to the varying morphologies exhibited by different species of plasmodium parasites throughout their distinct life stages, to classify a certain class of a malaria-infected red blood cell in laboratory diagnosis, an experienced technician is required.
Currently, the most common method for malaria diagnosis involves the use of light microscopy [9]. While the microscopic examination method is conventional, it does come with the drawback of being time consuming and requiring the expertise of a well-trained technician. It is common to have well-trained microscopists who can diagnose malaria accurately and effectively in well-funded medical facilities in large cities where malaria is rarely encountered. While on the other hand, the situation takes a different turn in developing countries, especially in some remote villages where malaria is more prevalent. The diagnostic method in these regions is often inaccurate and inefficient. The potential problems of this diagnosis arise from (a) the high cost of training microscopists and (b) inaccurate diagnosis results due to duplication work, limited experience, or mental and physical exhaustion [10]. Moreover, malaria cannot be treated until it is diagnosed. The delayed diagnosis or treatment will lead to anaemia and block the capillaries that carry blood to the brain. As a result, the infected and destroyed red blood cells will kill malaria-infected patients [3,4,11]. Thus, fast diagnosis and acute treatment of this illness are very helpful to reduce mortality.
Meanwhile, to address the limitations of manual microscopic examination, a cost-saving, accurate, and fast computer diagnostic system is required. There are many reviews for malaria diagnosis and its automated approaches in microscopic images [12,13,14,15]. A deep learning image processing approach has the potential to provide rapid and consistent estimates of examination outcomes. Deep learning has been applied in many medical applications, for example, magnetic resonance imaging (MRI) images [16], computed tomography (CT) images [17], positron emission tomography (PET)/CT images [18], microscopic images [19], fundus images [20], text-based medical data [21], etc. In an early study of automatic malaria examination, morphological methods were considered for red blood cell and malaria parasite detection [22]. In contrast, image processing has shown greater advancements when compared to the manual observation of blood smears by technicians. The introduction of deep learning algorithms has ushered in significant progress in the realm of computer vision, particularly in the detection of abnormalities within medical images. Notably, algorithms such as the YOLO series (V1 [23], V2 [24], V3 [25], V4 [26], V5), SSD [27], and some others, which also refer to one-stage object detection algorithms, approach the object detection process as a unified regression problem rather than proposing regions of interest separately [28].
To aid malaria diagnosis, besides the image processing-based or artificial intelligence-based technologies like what is proposed here, there are some other technologies recently proposed to solve this problem as well. A graphene metasurface biosensor was developed to detect early-stage malaria-infected red blood cells [29]. The bio-impedance signature of infected red blood cells was utilized to detect malaria infection using a capacitive micro-sensor [30]. The differences between electrical impedance spectroscopy of the healthy and the malaria-infected red blood cells were used to distinguish them [31]. The near infrared spectroscopy was applied to diagnose malaria due to P. falciparum and P. vivax through the patients’ skin [32]. The malaria-infected red blood cells were detected using one-dimensional photonic crystals [33]. A combination of a polystyrene-based microfluidic device with an immunoassay and surface enhanced Raman spectroscopy was also applied to the malaria detection problem [34]. Even though these are state-of-the-art methods to detect malaria red blood cells, they are still in the early stages of development.
In this research, the automatic methods are proposed to detect and classify infected red blood cells in human blood smear images. Two public benchmark datasets, i.e., the Malaria Bounding Boxes (MBB) dataset and the Malaria Parasite Image Database for Image Processing and Analysis (MP-IDB) dataset, are used for testing detection and classification performances. For the malaria parasite detection problem, the task is to scan each multi-cell image pixel by pixel and find infected cells in that image. This problem is not straightforward because there are also other objects, i.e., non-infected red blood cells, white blood cells, background, debris, etc., in the image. The outputs are the bounded boxes surrounding the infected cells. In this detection stage, only the MBB dataset (P. vivax only) is trained by the proposed methods based on the YOLO algorithm. Meanwhile, the MP-IDB dataset is only utilized as a role of testing to verify how the models trained by P. vivax can only work on datasets of other malaria parasite species. In order to improve the detection performance, five different preprocessing methods are applied. Also, to lessen the impact of Giemsa staining in the training set on the unstained dataset of blood smears, all color images are converted into grayscale before training. For the malaria parasite life stage classification problem, each single-cell image is classified into 1 of 5 classes, i.e., red blood cell, ring, trophozoite, schizont, and gametocyte, corresponding to the non-infected cells and four life stages, respectively. In the classification stage, single-cell images are cropped using three cropping methods. The problem of dataset imbalance caused by too many red blood cells can be solved by using image augmentation. Then, the models based on CNN LeNet-5 are used to evaluate the classification performance of the data.
This paper is organized as follows. The following section describes the datasets used in this research. The proposed malaria parasite detection and life stage classification are elaborated in Section 3. The evaluation measures are also given in the same section. Section 4 provides the experimental results. The results and findings are discussed in Section 5, and the limitations and future directions are given in Section 6. Finally, the conclusion is drawn in Section 7.
2. Data Descriptions
There are two datasets utilized in this research. The first dataset, namely the Malaria Bounding Boxes (MBB) dataset [35] (P. vivax only), used in this research is available at https://www.kaggle.com/kmader/malaria-bounding-boxes (accessed on 21 June 2022). There are six classes of cell images, i.e., red blood cell, ring, trophozoite, schizont, gametocyte, and leukocyte. The “Difficult” label in the original dataset is not really the ground truth, but rather the expert’s opinion that expresses the difficultly of labeling those images. Hence, there is no ground truth for them, and so they are discarded here. Meanwhile, the leukocytes (white blood cells) are discarded here as well. This dataset has a total of 1328 Giemsa-stained blood smear images, including 1208 training images and 120 test images. There are four distinct life stages of P. vivax, i.e., ring, trophozoite, schizont, and gametocyte. This dataset is called “MBB (P. vivax)”.
The second dataset is the Malaria Parasite Image Database for Image Processing and Analysis (MP-IDB) dataset [36], available at https://link.springer.com/chapter/10.1007/978-3-030-13835-6_7 (accessed on 10 March 2023). There are four different species of malaria parasites in this dataset, i.e., P. vivax (40 images), P. ovale (29 images), P. malariae (37 images), and P. falciparum (104 images). For each species, there are four distinct stages of life, i.e., ring, trophozoite, schizont, and gametocyte. The four sub-datasets are named as “MP-IDB (P. vivax)”, “MP-IDB (P. ovale)”, “MP-IDB (P. malariae)”, and “MP-IDB (P. falciparum)”.
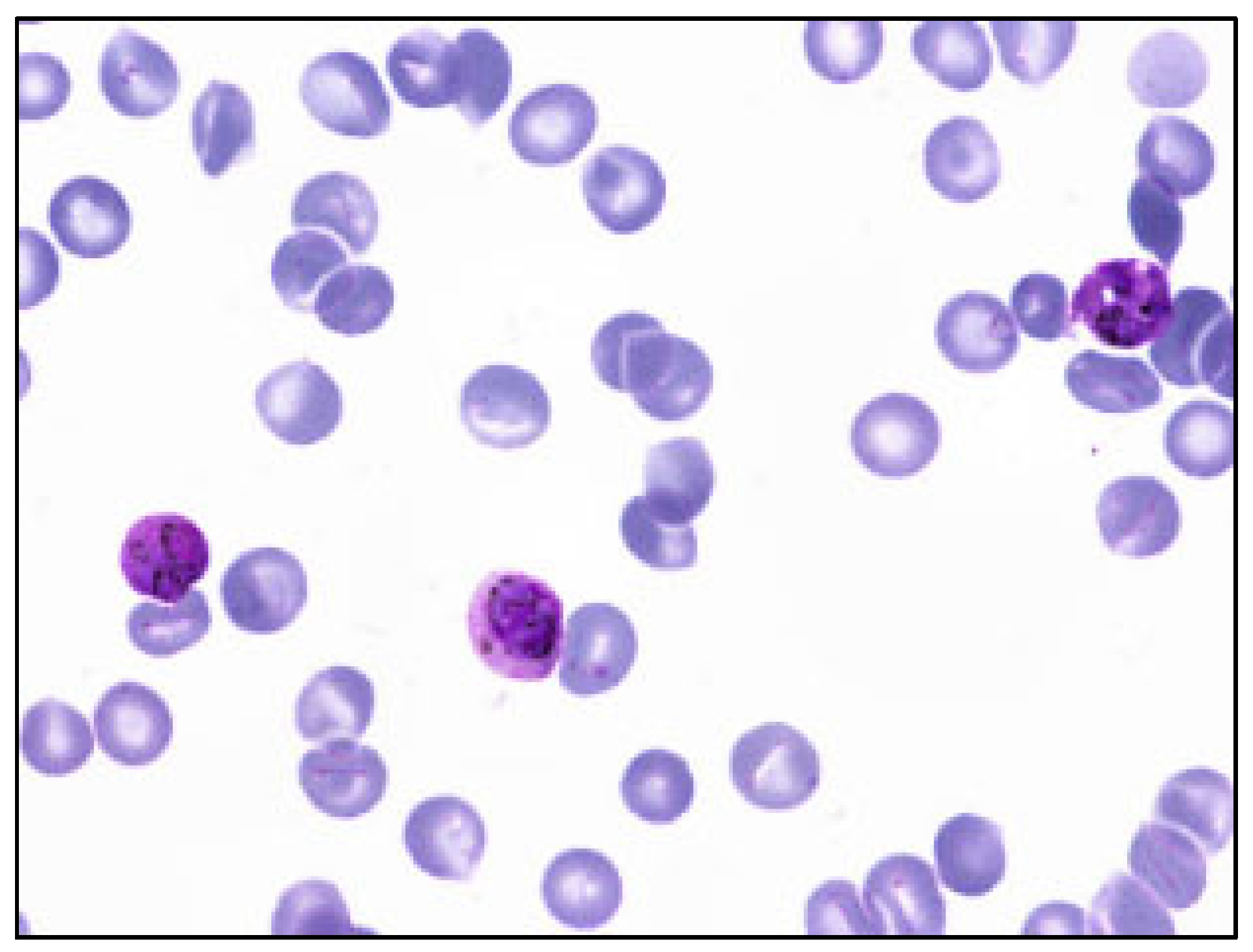
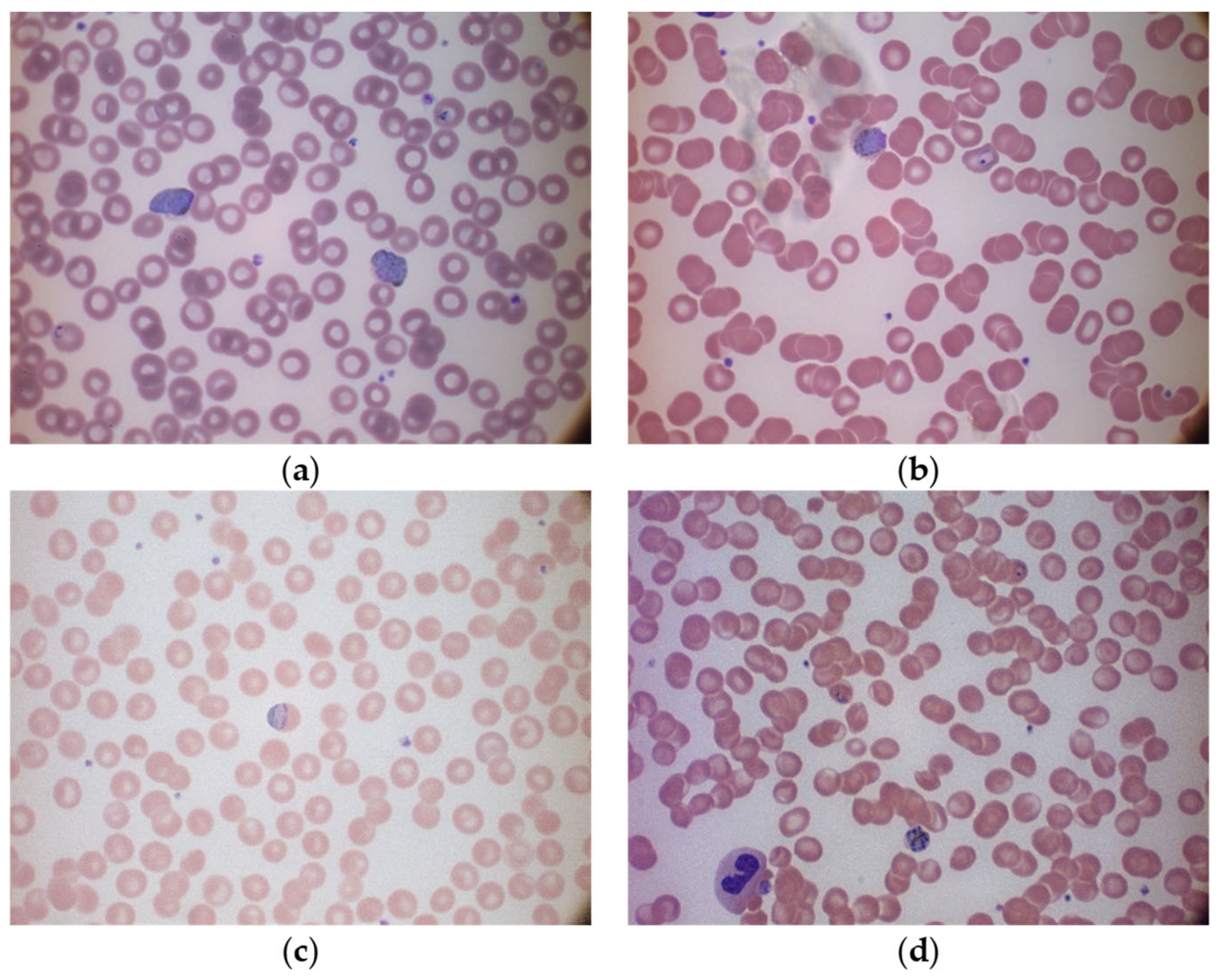
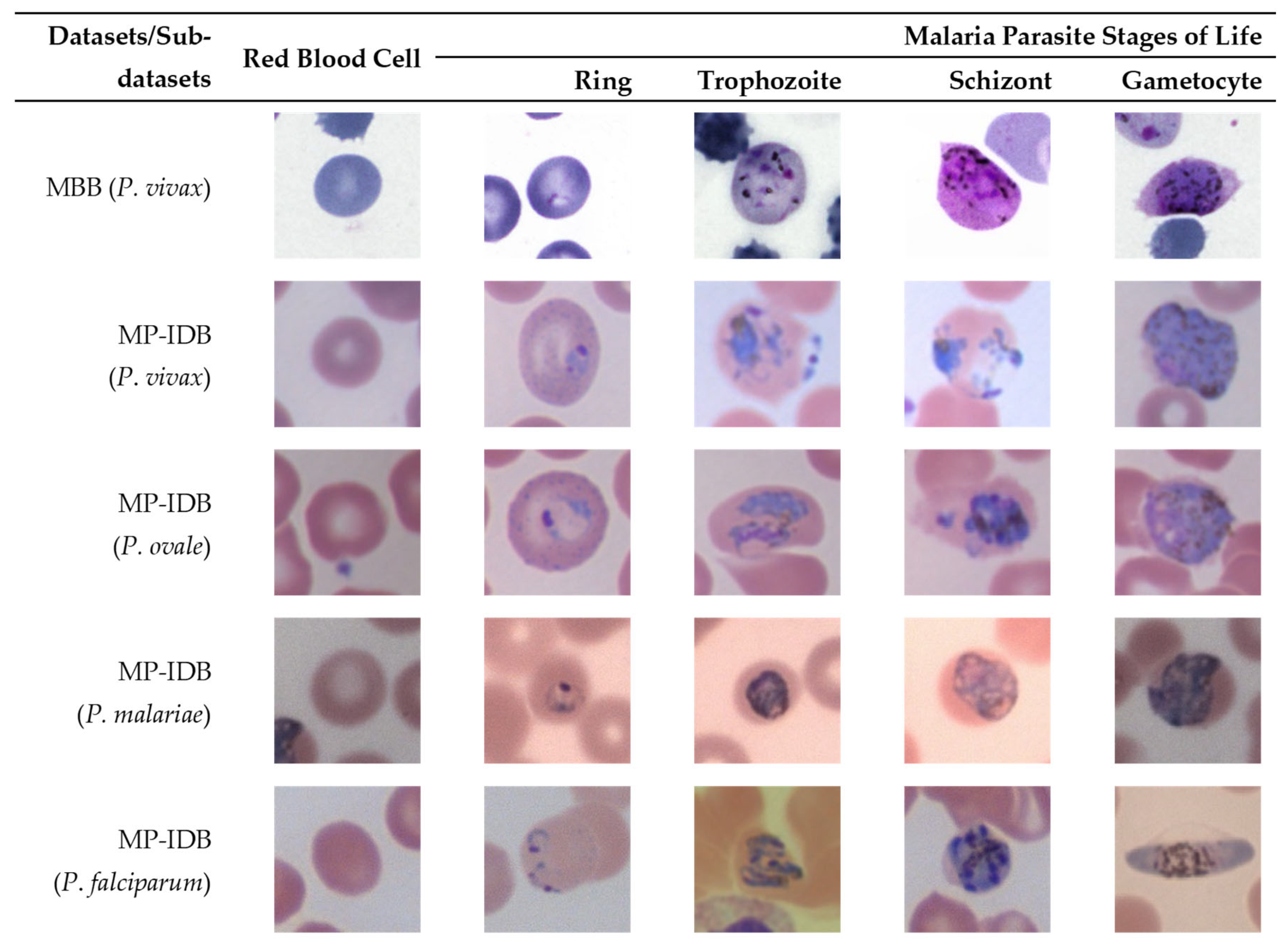
To show the variation in these two datasets, the multi-cell images of the MBB dataset and the MP-IDB dataset are illustrated in Figure 1 and Figure 2, respectively. Furthermore, to show how difficult it is to distinguish different malaria parasite life stages in each species, sample images from both datasets are shown accordingly in Figure 3. The figure shows images from different malaria parasites’ stages of life, i.e., ring, trophozoite, schizont, and gametocyte, in different species of plasmodium parasites, i.e., P. vivax, P. ovale, P. malariae, and P. falciparum, from the MBB and MP-IDB datasets. Some sample red blood cells are also illustrated in the figure. More details of the characteristics of each life stage of each species are elaborated as follows [6,37].
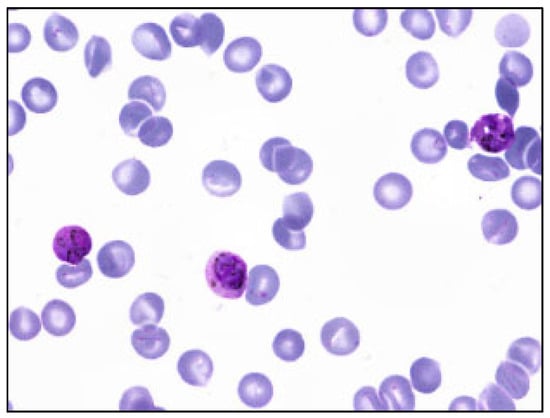
Figure 1.
Sample multi-cell image of MBB dataset. Only 1 species (P. vivax) exists in this dataset.
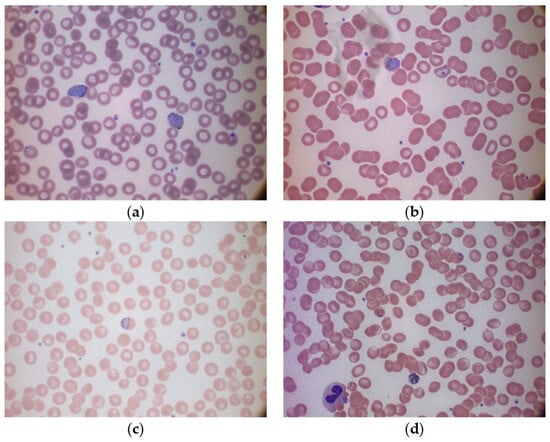
Figure 2.
Sample multi-cell images in different species of the MP-IDB dataset: (a) P. vivax, (b) P. ovale, (c) P. malariae, and (d) P. falciparum.
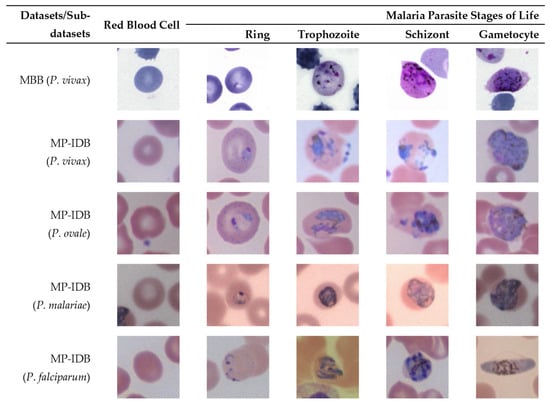
Figure 3.
Sample images from different malaria parasites’ stages of life (ring, trophozoite, schizont, gametocyte) in different species of plasmodium parasites (P. vivax, P. ovale, P. malariae, P. falciparum) from the MBB and MP-IDB datasets. The cell of interest is at the center of each image.
For P. vivax, the infected red blood cell is slightly enlarged and has an irregular shape. The appearance of P. vivax parasites in different stages is as follows:- ring: large cytoplasm with occasional pseudopods and large chromatin dot; trophozoite: amoeboid in shape with yellowish-brown pigment inside; schizont: enlarged and may almost fill RBC, one matured schizont has 12–24 merozoites with yellowish-brown and coalesced pigment inside; gametocyte: round to oval shape, may almost fill RBC. It has compacted chromatin, with scattered brown pigment inside.
For P. ovale, the infected red blood cell is slightly enlarged and has an oval shape with tufted ends. The appearance of P. ovale parasites in different stages is as follows: ring: sturdy cytoplasm and large chromatin; trophozoite: compacted with large chromatin with dark-brown pigment inside; schizont: matured schizont cell has 6–14 merozoites with large nuclei that cluster around mass of dark-brown pigment; gametocyte: round to oval shape, may almost fill RBC, compacted chromatin, with brown malarial pigment inside.
For P. malariae, the infected red blood cell is not enlarged. The appearance of P. ovale parasites in different stages is as follows: ring: sturdy cytoplasm, large chromatin; trophozoite: occasional band form with large chromatin, dark-brown pigment inside; schizont: occasional rosettes, mature schizont have 6–12 merozoites with large nuclei that cluster around a mass of coarse, dark-brown pigment inside; gametocyte: round to oval shape, may almost fill RBC, compacted chromatin, with scattered brown pigment inside.
For P. falciparum, the infected blood cell is not enlarged. The appearance of P. falciparum parasites in different stages is as follows: ring: multiple infections are more common than in other species, visible cytoplasm, and 1 or 2 small chromatin dots; trophozoite: more dense cytoplasm, round shape with brown malarial pigment inside; schizont: more than 2 and up to 32 nuclei (merozoites) with dark brown pigment clumped in the middle; gametocyte: crescent or sausage shape, visible chromatin as a single mass or diffuse.
3. Proposed Methods
The proposed methods are mainly divided into two parts, i.e., malaria parasite detection and malaria parasite life stage classification. To make the proposed detection method robust to variation in staining methods, appropriate image preprocessing is required. In this research, a set of image preprocessing techniques is performed to determine a choice with good detection performance. Different cropping methods are also implemented to generate input images for classification models. For both the MBB dataset and MP-IDB dataset (i.e., its four sub-datasets), their malaria parasite detection performance and malaria parasite life stage classification performance are evaluated separately.
3.1. Malaria Parasite Detection
For malaria parasite detection, in order to improve detection performance, preprocessing the original data is essential and efficacious. Upon visual inspection, it becomes apparent that the cellular components within the images exhibit a darker contrast relative to the background. Thus, when the contrast limited adaptive histogram equalization (CLAHE) [38,39,40] or contrast stretching [41] are applied, they create obvious effects by tuning the brightness levels of various image regions. Consequently, the background region becomes brighter while the cell regions become darker. This preprocessing step significantly facilitates the differentiation between cells and their background surroundings. In addition, due to the different staining methods between the MBB dataset and the MP-IDB dataset, the MBB dataset is stained by Giemsa reagent while the MP-IDB dataset is not. To subside the effect of image color on detection and to simplify the detection process for the algorithm, the grayscale image converting process is indispensable here.
The you only look once (YOLO) algorithm is applied to train the proposed malaria parasite detection models. The YOLO algorithm used in this research is the 5th version of it. Because the descriptions of the YOLO algorithm are widely available in the literature [42], only brief descriptions are provided here. Among several versions of YOLO algorithms, the key differences between different versions are that each version has been built on top of the previous version with enhanced features such as improved accuracy, faster processing (by reducing the number of parameters and the amount of calculations), and better handling of small objects. However, compared with some new algorithms released recently, like the YOLOv8 (2023) and YOLOv9 (2024) algorithms, the YOLOv5 algorithm has stood the test of time and is still a relatively stable and reliable algorithm. The YOLOv5x algorithm is a popular object detection algorithm in the computer vision field and is known for its high-speed performance and high accuracy in object detection tasks. It has simpler implementation and training procedures due to its integration with PyTorch version 1.12.0 [43].
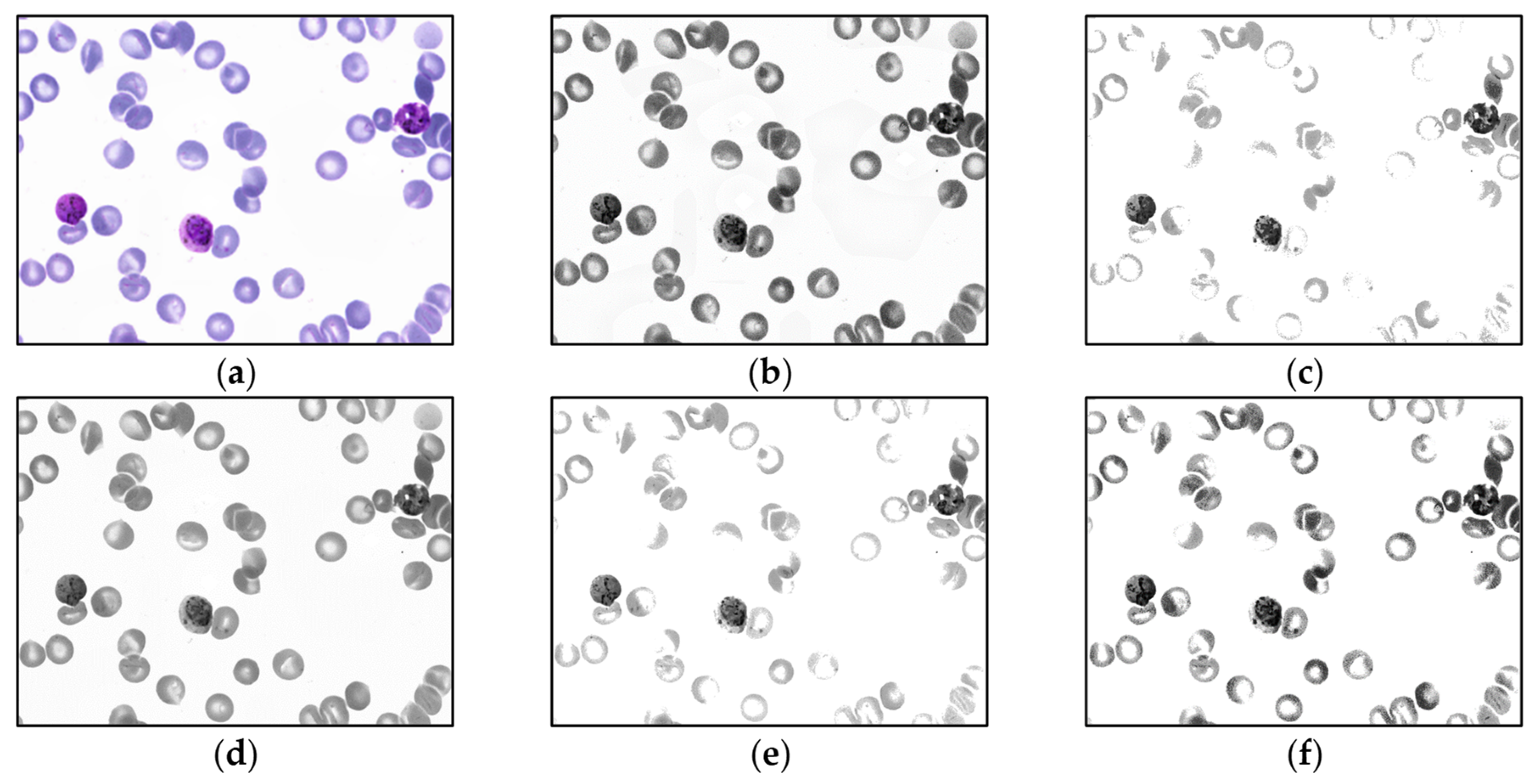
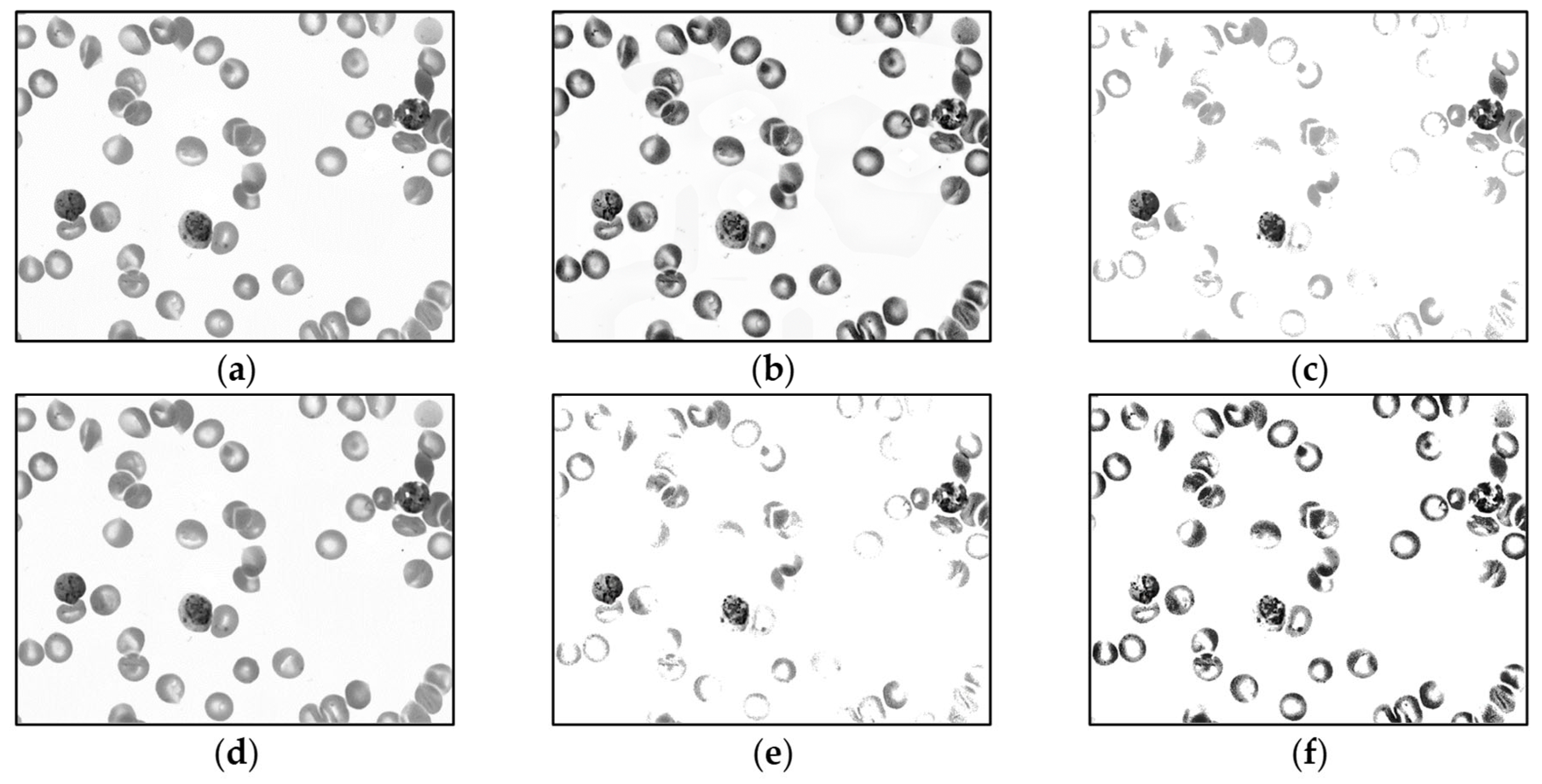
For malaria parasite detection experiments, only the MBB dataset is trained by the YOLO algorithm. The MP-IDB dataset is only utilized as a test group. Before the training process, the input images are preprocessed by each of 5 different operations, i.e., (1) contrast limited adaptive histogram equalization (CLAHE), (2) contrast stretching, (3) median blur [43], and 2 other methods that combined CLAHE with contrast stretching together, (4) contrast stretching then CLAHE, and (5) CLAHE then contrast stretching. To lessen the impact of Giemsa staining in the training set on the unstained dataset of blood smears, all images are converted into grayscale version [44] before training. Thus, for the first stage, there are totally 11 different input image groups after preprocessing. Sample output images of these 5 preprocessing methods on the MBB dataset are shown in Figure 4 and Figure 5. In Figure 4, each input color image is preprocessed, and then each output is converted into the grayscale image. Meanwhile, in Figure 5, each input color image is converted into the grayscale space, and then the grayscale image is preprocessed accordingly. Moreover, it is straightforward that the model with the highest accuracy will be selected as the optimal model on the MBB dataset. Then, the optimal model is used to test the malaria detection performance of the MP-IDB dataset, respectively.
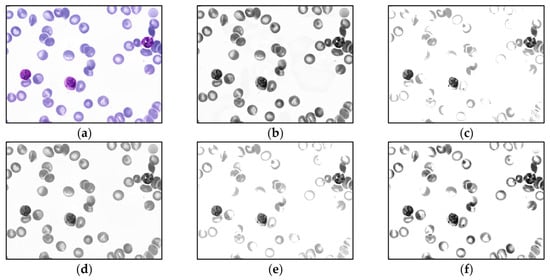
Figure 4.
Sample output images of 5 preprocessing methods of MBB dataset. Each input color image is preprocessed by 1 of the 5 methods in the color space, and then each output is converted into the grayscale image: (a) original color image, (b) contrast limited adaptive histogram equalization (CLAHE), (c) contrast stretching (CS), (d) median blur, (e) CS then CLAHE, and (f) CLAHE then CS.
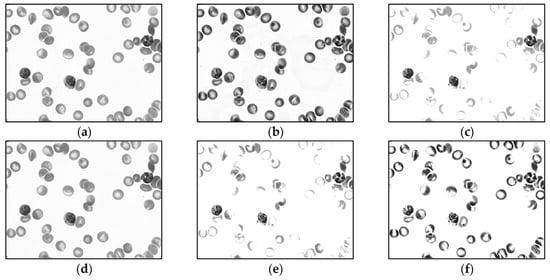
Figure 5.
Sample output images of different preprocessing methods of MBB dataset. Each input color image is converted into the grayscale space, and then the corresponding grayscale image is preprocessed by 1 of the 5 methods: (a) grayscale image, (b) contrast limited adaptive histogram equalization (CLAHE), (c) contrast stretching (CS), (d) median blur, (e) CS then CLAHE, and (f) CLAHE then CS.
To fine-tune the detection outputs, the non-maximum suppression is applied, as it is commonly used in object detection to filter out redundant or overlapping bounding boxes [45]. Thus, to reduce a large number of duplicate bounding boxes detected by the YOLO algorithm here, the non-maximum suppression is applied. Consider the value of intersection over union (IOU) as follows:
only the detection regions with IOU above a thresholding value are kept. In this research, the thresholding value is set to 0.8. This approach can filter the original detected regions and then increase the malaria parasite detection performance. It is worthwhile noting that, for the MBB dataset, 1208 training images are used to train the training models while 120 test images are used in testing. Meanwhile, all multi-cell images in the MP-IDB dataset are used for testing purposes only.
3.2. Malaria Parasite Life Stage Classification
For malaria parasite life stage classification, the convolutional neural network (CNN) algorithm is utilized. Like that of the YOLO, the descriptions of CNN are widely available in the literature [46]. Hence, only brief descriptions are given here. The CNN algorithm used in this research is LeNet-5. LeNet-5 is a pioneering CNN architecture. It is considered a fundamental model in the field of deep learning and computer vision. It has also been adapted for various other image classification tasks, including cell classification in biomedical image processing [46]. To prepare input images for CNN LeNet-5, the malaria parasite life stage classification encompasses 3 distinct cell cropping methods, including (1) direct cropping of a single cell, (2) zero padding method [47], and (3) cell cropping with background. All of these methods are applied on both datasets. Sample cell images cropped by the aforementioned 3 methods are shown in Figure 6.
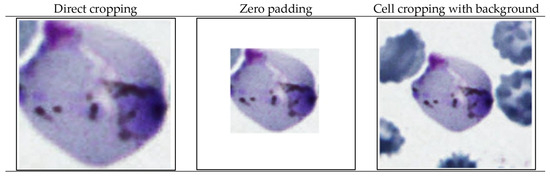
Figure 6.
Sample cropped single cell images to prepare inputs to CNN-based malaria parasite classification.
According to the composition of different datasets shown in Table 1, both of the 2 datasets have the same heavily imbalanced class towards red blood cell class. This is not surprising because the normal red blood cells are much more abundant in nature than the infected cells. This issue of imbalanced classes poses a significant challenge in this research, particularly when applying machine learning algorithms to detect these infrequent occurrences within sizable datasets. As a result of the unequal distribution of classes, the algorithms tend to favor categorization into the more prevalent class, the majority class. This inclination can misleadingly convey the impression of a highly accurate weight, ultimately resulting in poor classification performance [48].

Table 1.
Number of images in each class before and after image augmentation.
Thus, instead of reducing the number of cells in red blood cell groups, the numbers of other infected cell classes are increased to the same level as that of red blood cells. This can be done via image augmentation, i.e., top-down, left-right translation, vertical, horizontal reflection, and rotation [49]. The comparison of the numbers of images before and after data augmentation is shown in Table 1. Each single-cell image is resized to 250 × 250 pixels. The images are randomly divided into training and test sets with a ratio of 4:1.
3.3. Evaluation Measures for Detection and Classification
To evaluate detection and classification performances in this research, precision, recall, F1-score, and accuracy are utilized. These evaluation criteria are listed as follows:
where TP, TN, FP, and FN are the numbers of true positives, true negatives, false positives, and false negatives, respectively. The performances of the proposed methods, either for detection or classification, are evaluated by comparing their outputs to the ground truths, i.e., infected cell positions or stages of life, provided by skilled experts in both public benchmark datasets.
4. Experimental Results
The performance of malaria parasite detection and life stage classification are evaluated separately and shown in this section. As mentioned in Section 3.2, to solve the unbalanced data problem, the number of each infected cell class was increased to the same level as that of red blood cells via image augmentation. The numbers of images in each class before and after image augmentation are shown in Table 1. Table 2 and Table 3 illustrate the malaria parasite detection performance, including the precision, recall, F1-score, and accuracy for 2 approaches: (1) preprocessing each input color image in the color space and then converting each output into a grayscale image, and (2) converting each input color image into the grayscale space and then preprocessing the corresponding grayscale image, respectively. The sample output images of 5 different image preprocessing methods using the 2 approaches are illustrated in Figure 4 and Figure 5, respectively. Table 4 shows malaria parasite detection performance on the MP-IDB dataset by using the best training model from the MBB dataset. For malaria parasite life stage classification, the sample cropped single cell images using 3 cropping methods, i.e., direct cropping, zero padding, and cell cropping with background, are shown in Figure 6. The malaria parasite life stage classification results on the MBB test sets using 3 different cell cropping methods are shown in Table 5. The corresponding life stage classification results on the MP-IDB test sets for 4 different species of malaria parasites, i.e., P. vivax, P. ovale, P. malariae, and P. falciparum, are shown in Table 6, Table 7, Table 8 and Table 9, respectively. It is worthwhile noting that, for each species, a 5-class classification problem is considered for red blood cells and 4 distinct stages of life of malaria parasites, i.e., ring, trophozoite, schizont, and gametocyte.

Table 2.
Malaria parasite detection results for MBB dataset using different preprocessing methods (mean ± S.D.) when each input color image is preprocessed by 1 of the 5 methods in the color space, and then each output is converted into a grayscale image.

Table 3.
Malaria parasite detection results for MBB dataset using different preprocessing methods (mean ± S.D.) when each input color image is converted into the grayscale space, and then the corresponding grayscale image is preprocessed by 1 of the 5 methods.

Table 4.
Malaria parasite detection performance on MP-IDB dataset by using the best training model from MBB dataset (mean ± S.D.).

Table 5.
Malaria parasite life stage classification results on MBB dataset.

Table 6.
Malaria parasite life stage classification results on MP-IDB (P. vivax) dataset.

Table 7.
Malaria parasite life stage classification results on MP-IDB (P. ovale) dataset.

Table 8.
Malaria parasite life stage classification results on MP-IDB (P. malariae) dataset.

Table 9.
Malaria parasite life stage classification results on MP-IDB (P. falciparum) dataset.
The first malaria parasite detection results are for the MBB dataset using different preprocessing methods when each input color image was preprocessed by 1 of the 5 methods in the color space, and then each output was converted into a grayscale image. As shown in Table 2, the proposed detection method achieves precision, recall, F1-score, and accuracy scores of 0.97, 0.94, 0.95, and 0.92, respectively, on the test set of the MBB dataset when the CLAHE was applied as the preprocessing method. Next, the second malaria parasite detection results are for the MBB dataset using different preprocessing methods when each input color image was converted into the grayscale space, and then the corresponding grayscale image was preprocessed by 1 of the 5 methods. Table 3 shows a general decline in the performance of all methods compared to that shown in Table 2, notably the precision and accuracy. Among them, the performances using the CLAHE for preprocessing had the most significant performance decrease. The use of contrast stretching for preprocessing maintained relatively stable performance across both sets of results, with only minor variations. Anyhow, the CLAHE yielded the best overall performance in terms of the precision, F1-score, and accuracy. Its performance on the recall was also among the highest. For the median blur and the CLAHE then contrast stretching preprocessing methods, they also performed well, particularly in the recall and F1-score, showing their potential to be good alternatives to the CLAHE. All in all, the results suggested that using the first approach, i.e., preprocessing each input color image in the color space and converting each output into a grayscale image, was a better choice. Hence, this approach was applied to the proposed detection method with the CLAHE as the preprocessing method throughout the rest of the experiments.
It is very interesting that when the best trained model was applied to another dataset (MP-IDB) with different staining and image acquisition methods, the test results were still very promising. These results are not surprising because the results in Table 2 and Table 3 are from the test images with the same staining and image acquisition methods as the training dataset (MBB). Meanwhile, the results in Table 4 are from another dataset (MP-IDB) with different staining and image acquisition methods from the training data. The proposed detection methods yielded an accuracy of 0.84 for P. vivax, 0.82 for P. ovale, 0.79 for P. malariae, and 0.92 for P. falciparum on the MP-IDB dataset. Meanwhile, the F1-scores for these 4 species were 0.90, 0.89, 0.85, and 0.96, respectively. The accuracy for P. falciparum reached 0.92 with the largest number of 104 multi-cell images. While considering P. ovale, the detection performances were reasonably high. The accuracy reached 0.82, ranked 3rd among the 4 species. However, with the number of 29 multi-cell images, which is the lowest among the species, the small sample size may affect the reliability of the results. For P. malariae, which contains 37 multi-cell images, it yielded the lowest detection performance among them. It suggested that increasing the dataset size might enhance the detection performance for this species. Of course, another possible reason may be that the images of this species itself contain a large number of overlapping cells, resulting in a much more difficult malaria parasite detection problem. Therefore, collecting related and high-quality data might be a key to improving the detection performance.
Another point worth noting is that since the training malaria parasite detection model was trained only on the MBB dataset (P. vivax species only), it was a bit surprising that the P. vivax test images did not become the ones with the highest accuracy. Due to the model being trained solely on P. vivax images, it could be speculated that the feature representations learned from the model were more appropriate for P. vivax. However, the precision, recall, F1-score, and accuracy achieved by the trained model on P. falciparum were exceptional. This might suggest that P. falciparum shared more distinguishable features with P. vivax or that the dataset used for evaluation had a more representative sample of P. falciparum. Conversely, the model struggled with P. malariae, suggesting that P. malariae might have more subtle differences from P. vivax or less distinct features that the model could not extend from the P. vivax data alone. The ability that this detection model was generalized from the P. vivax data to other species like P. falciparum, P. ovale, or P. malariae suggested some level of transferable feature learning or generalization ability. As mentioned earlier, however, the poor detection performance on P. malariae highlights the need for more diverse training data to improve its detection performance. Thus, it shows the potential to diversify training data is worth trying again in the future.
For malaria parasite stage of life classification, all classification models trained and tested on single-cell images with the 3 different cropping methods yielded quite good classification performance. They achieved accuracies of around 0.90. By comparing the results from Table 5, Table 6, Table 7, Table 8 and Table 9, it can be found that the direct cropping method is generally the best-performing method, especially for P. vivax and P. malariae species. It demonstrated the highest overall accuracies and F1-scores in most of the 5 scenarios, i.e., MBB (P. vivax), MP-IDB (P. vivax), MP-IDB (P. ovale), MP-IDB (P. malariae), and MP-IDB (P. falciparum). This cropping method performed well on the P. vivax data, especially for red blood cells and schizont stage among all 5 scenarios. The classification of red blood cells yielded very high precision, recall, and F1-score (close to 1.00 in most cases). The performances might not be that good in ring and trophozoite classes in the MP-IDB (P. falciparum) data. However, the F1-scores of 0.78 and 0.85 were still quite good. The zero padding method still performed reasonably well but yielded lower accuracies and F1-scores for certain classes, particularly for trophozoites and schizonts classes. It also provided high performance for red blood cell classification and achieved perfect or near-perfect precision and recall. It yielded slightly lower overall accuracy compared to the direct cropping and cropping with background methods. Meanwhile, compared to the direct cropping method, the cropping with background method often performed particularly well on the P. falciparum data with the highest accuracy of 0.96. It also showed a good balance between different malaria parasite species and stages. Compared with direct cropping and zero padding methods on the other classification datasets, the precision and recall for other datasets were slightly lower in the red blood cell classification. Enhancing its classification performance can be achieved by focusing on the challenges imposed by trophozoite and ring classes across different methods and datasets.
5. Discussion
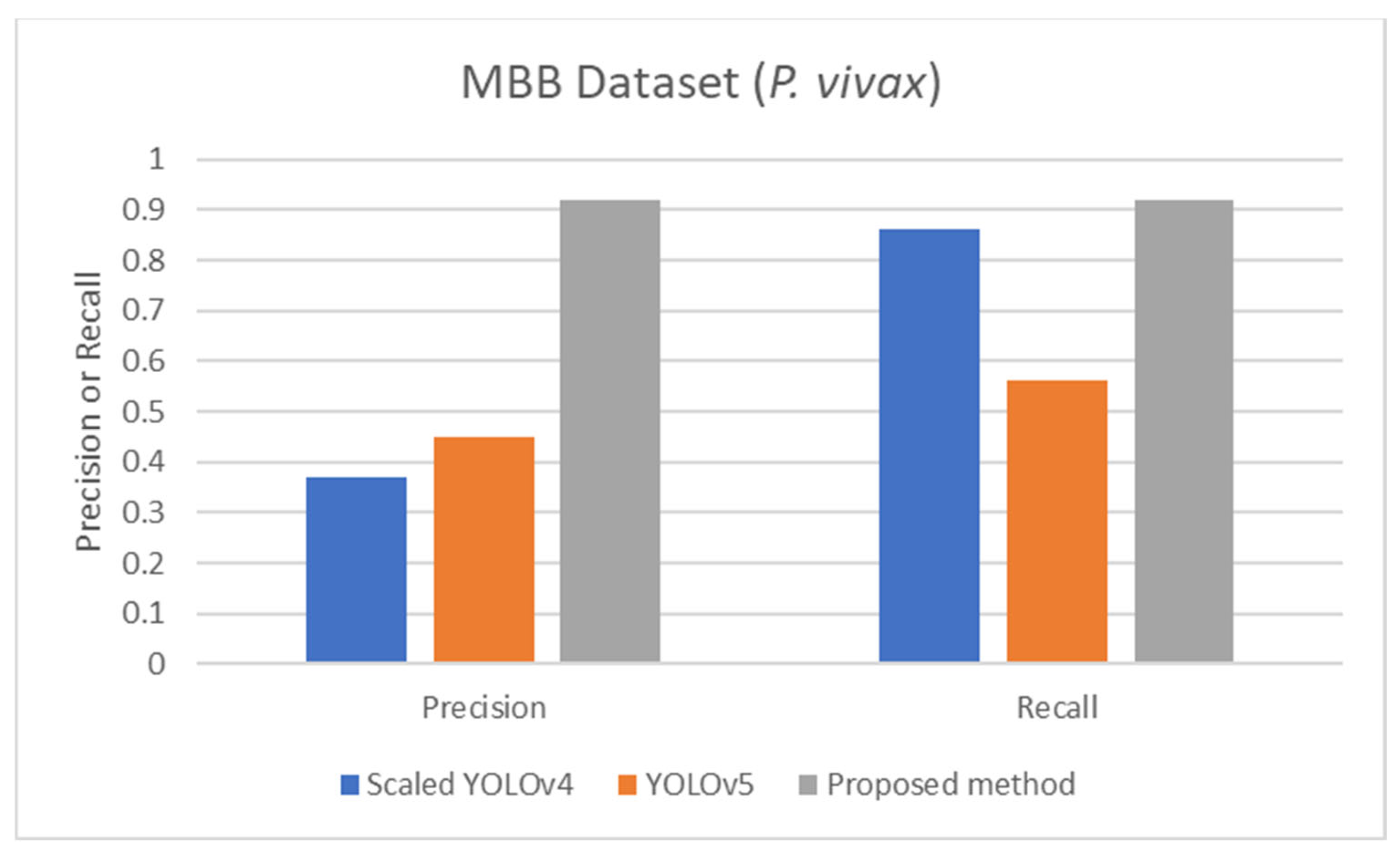
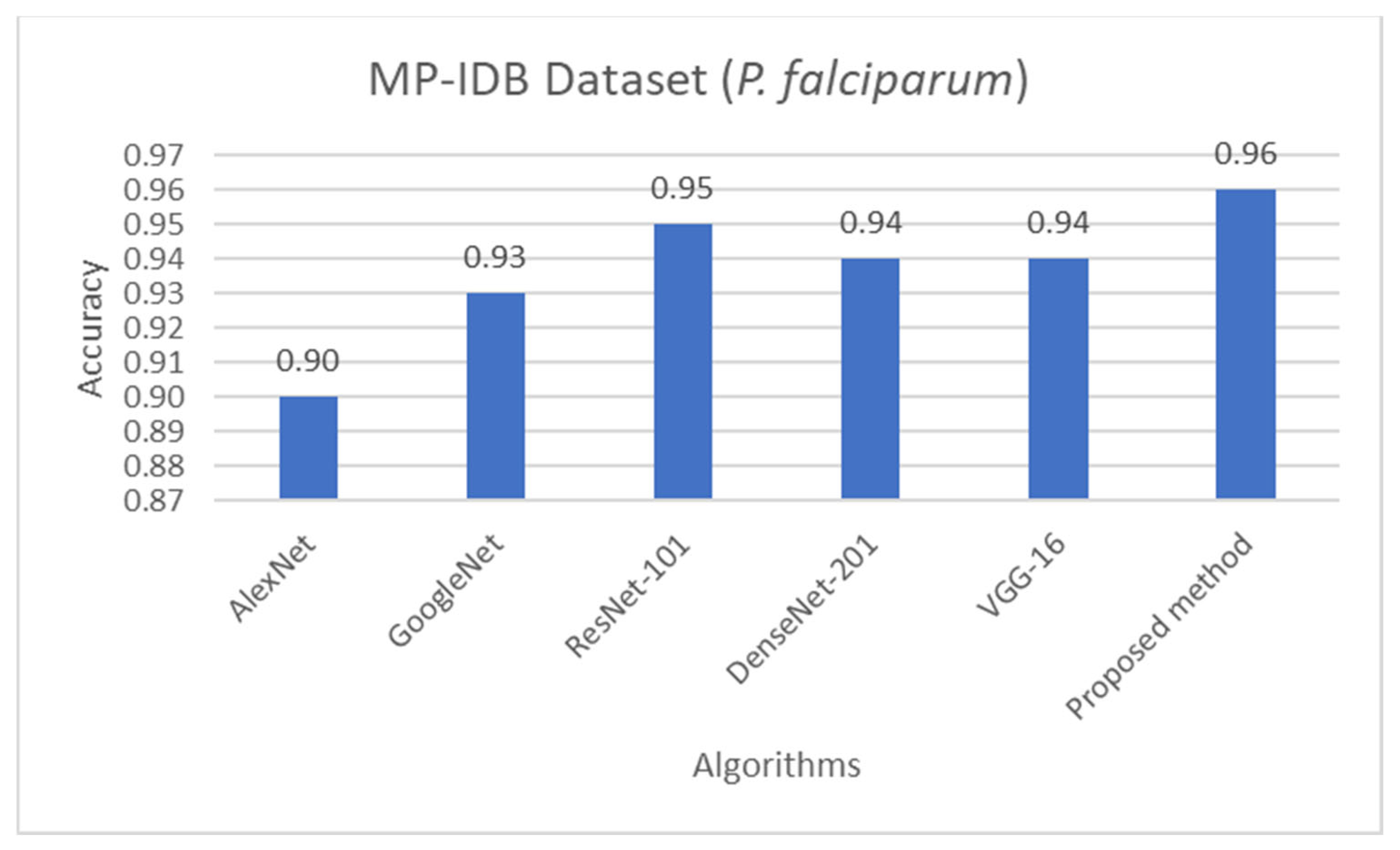
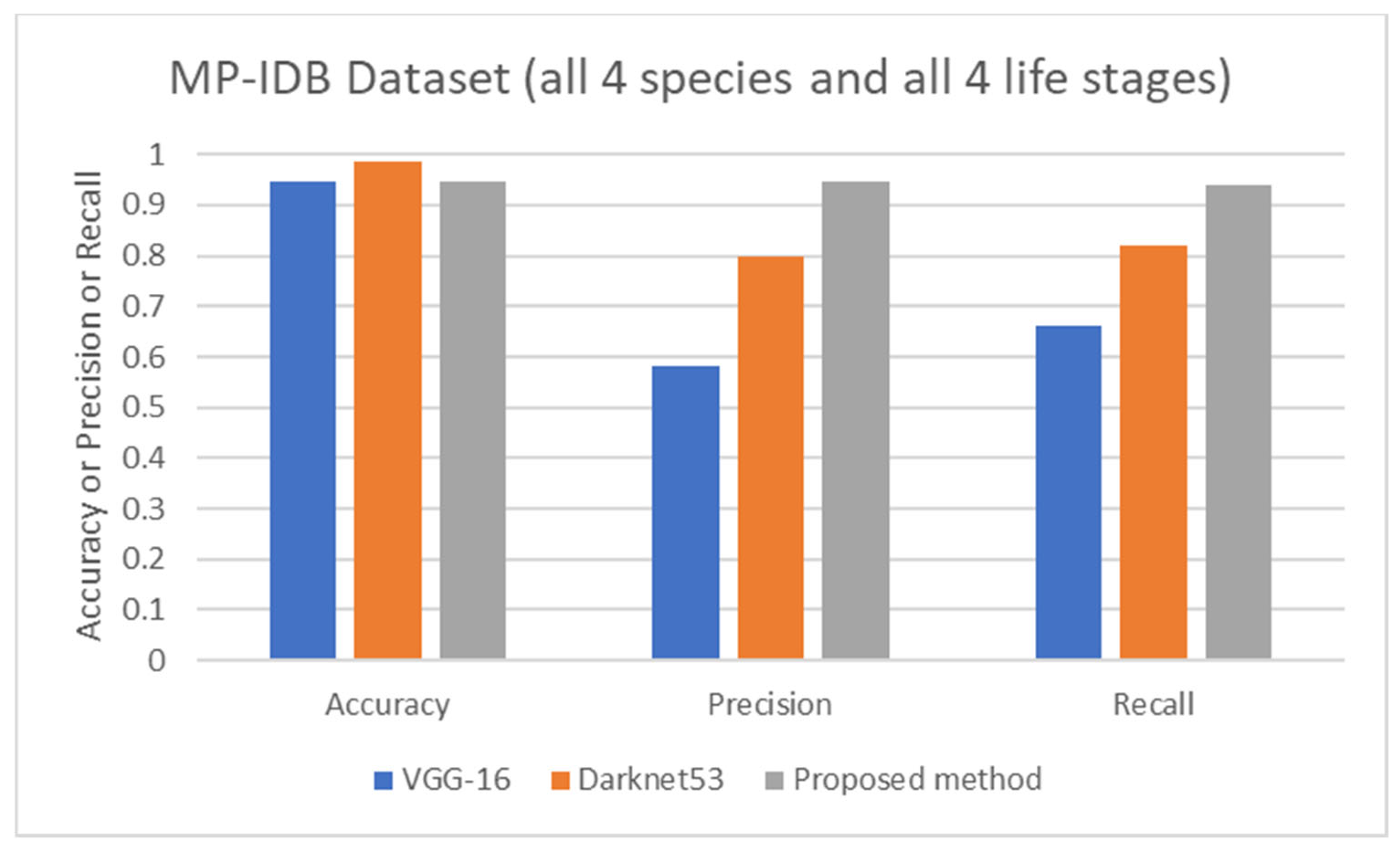
The proposed detection method was compared to other existing works, it was found that the proposed method achieved comparable performance to them. To be more specific, while the proposed detection method achieved 0.92 in accuracy on the MBB dataset, the detection performances of previous works ranged from 0.53 average precision (AP) using the mask R-CNN with ResNet101 as the backbone [50], 0.939 accuracy using the integrated CNN [51], to 0.997 accuracy using a semantic segmentation CNN [52]. It is worthwhile noting that it is not really possible to compare the results directly because different approaches used different setups. The results were on different subsets of test data, and their generalizations are always questionable. Many methods might perform extremely well only on a particular dataset or subset. As shown here, the proposed detection method was able to be further applied efficiently to another dataset (MP-IDB) even with other plasmodium species. Also, for the malaria parasite life stage classification, the performances of the proposed methods are compared to results achieved by the previous works. Unfortunately, there are only a few works that worked on malaria parasite life stage classification. Most of the previous works were on species classification, like that in [53,54,55], which is different from the current problem of interest. There is a study on malaria parasite life stage classification on a different dataset that achieved 0.588 accuracy using a random forest classifier [56]. For the works on malaria parasite life stage classification on the MP-IDB dataset, they did not report the detailed results for each species. Some of them considered only subsets of some species of the MP-IDB dataset. The classification performances achieved by different methods [57,58,59,60,61,62] on the MP-IDB dataset for different plasmodium species, including P. vivax, P. ovale, P. malariae, P. falciparum, or some subsets of it, are shown in Table 10. Even though it is difficult to compare the results from different works because they used different ways of reporting the results, to provide clearer comparison through visualization, the plots of related results that could be compared are shown in Figure 7, Figure 8 and Figure 9. In Figure 7, the precision and recall of life stage classification on the MBB dataset, which contains only P. vivax, are shown. It can be clearly seen that the proposed method performed better than its counterparts. The life stage classification accuracies on the MP-IDB dataset when considering only P. falciparum are shown in Figure 8. Once again, it can be seen that the proposed method performed better than previous works that used the AlexNet, GoogleNet, ResNet-101, DenseNet-201, and VGG-16. When all four species and all four life stages are considered, the average accuracy, average precision, and average recall are exploited for comparison. It should be noted that the averages are calculated across four life stages for the results from [62] and across four species for the results from the proposed method. It is believed that the average evaluation measures would be good representatives to evaluate the classification performance when the entire dataset is considered. The average accuracy, average precision, and average recall are shown in Figure 9. The figure shows that the three methods achieve similar average accuracy, in which the Darknet53 might yield a little bit better performance. However, it is clear that the proposed method yielded better average precision and average recall than the VGG-16 and Dartket53. It can be seen from Table 10 and Figure 7, Figure 8 and Figure 9 that the proposed malaria parasite life stage classification method achieved similar or better results compared to its counterparts. As mentioned in the detection performance comparison, the results cannot be compared directly because of different setups in different works. Some extremely good performance achieved by some methods might not be applied in general. The bottom line is that the proposed malaria parasite detection and stage of life classification methods are very promising and achieved comparable results to the existing methods. However, the image preprocessing methods that convert each input color image into a grayscale version make the proposed methods robust to variation in staining methods.

Table 10.
Comparison of malaria parasite life stage classification results achieved by different methods on MP-IDB dataset for different plasmodium species.
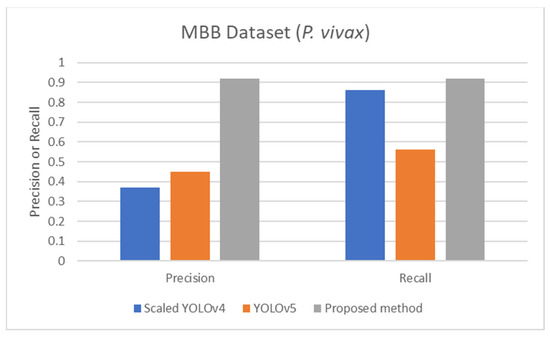
Figure 7.
Comparison of precision and recall of life stage classification achieved by different works including the scaled YOLOv4 [57], YOLOv5 [57], and the proposed method, on the MBB dataset.
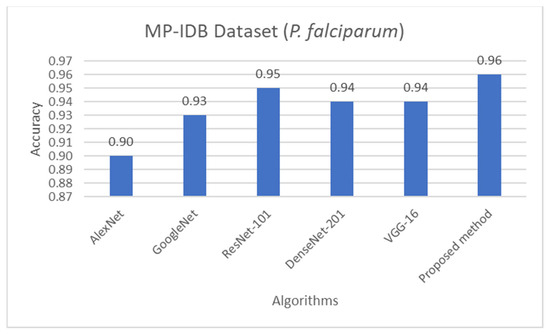
Figure 8.
Comparison of accuracy of life stage classification achieved by different works including AlexNet [58], GoogleNet [58], ResNet-101 [58], DenseNet-201 [58], VGG-16 [58], and the proposed method, on the MP-IDB dataset when only P. falciparum is considered.
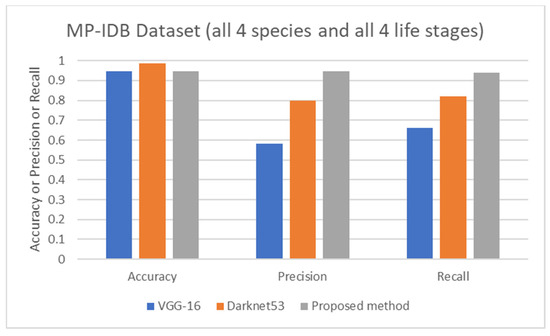
Figure 9.
Comparison of average accuracy, average precision, and average recall of life stage classification achieved by different works including VGG-16 [62], Darknet53 [62], and the proposed method, on the MP-IDB dataset when all 4 species and 4 life stages are considered.
The computational complexity of the YOLO algorithms and CNN algorithms has been improved all the time. If different methods utilize the same algorithm, then their complexities are not much different. In real-world applications, a factor that certainly imposes the difference in computation time is the input image size. Even with the same dataset of images, the use of higher resolution images renders a larger number of pixels, which would force a longer computation time. Due to different experimental settings in different methods or different experimental settings even within one method, it is hard to compare them based on the actual computation time. Moreover, the systems used in the experiments are the crucial factor. The high-end cloud computing systems would definitely yield shorter computation times than personal computers or lower end cloud systems. In this research, the YOLO and CNN algorithms are utilized because they provide fast computation among the deep learning-based algorithms. In the experiments, a cloud computing service from Tencent Cloud was utilized. The information about the service is as follows: CPU: Intel Xeon Platinum 8255C CPU, with a clock rate of 2.5 GHz; GPU: NVIDIA Tesla T4, 8.1 TFLOPS; GPU video memory: 16 GB HBM2; vCPU: 20 cores; Memory: 80 GB DDR4 RAM. Here are the actual computation times for training. For the detection problem, the YOLOv5 algorithm took approximately 2.5 min per epoch to train the MBB dataset. A total of 50 epochs was needed for training the detection models. Meanwhile, for the classification problem, the CNN LeNet-5 algorithm used approximately 15 s per epoch in each (sub)dataset for training. A total of 50 epochs was needed in the training of the classification models. On the testing, the computation times are as follows. The detection of infected cells required about 4 s for each multi-cell image. For classification, only about 10 milliseconds are needed for each single-cell image. The speeds in the testing stages indicate that the proposed methods can be applied in real-world situations.
6. Limitations and Future Directions
For future work, combining the strengths of direct cropping and cropping with background methods could be explored to create a more robust method. Furthermore, there are still a lot of parameters in many steps in the experiments that can be optimized. In addition, besides the YOLOv5 and CNN LeNet-5 algorithms used in this research, some other more new algorithms can be used as well, for example, YOLOv8 in malaria parasite detection and VGG-16 in life stage classification [63]. Except for the YOLO series, the other complicated algorithms can also be used to improve the detection results.
It can be seen from the classification results that the classification performances of some malaria parasite species can still be improved, especially schizont and trophozoite. Even though data augmentation had been performed, these two classes still yielded the lowest classification performances among other species. Although this work has proved that the models based on the YOLO-based models trained by P. vivax solely worked for different species of malaria parasite detection, due to the relatively small number of images included in both datasets, continued data collection is imperative. Especially for machine learning methods, a lack of sufficient data support can lead to unreliable results [64]. Furthermore, for future research, the collection and expansion of datasets cannot only be to expand schizont and trophozoite stages but also to collect more data of P. falciparum, P. malariae, P. ovale, P. vivax, and P. knowlesi species with some other staining method [65]. Then, in this way, the algorithm may not only detect whether a cell is infected or not but can also distinguish the plasmodium species.
For an implementation issue, during the training process for malaria parasite life stage classification by CNN LeNet-5 models on the MBB dataset, there was a difficulty arising from the lack of GPU memory. It was unfortunate that the MBB dataset after image augmentation, containing about 400,000 images, was too large to be trained. It was almost impossible to train the dataset directly. The CNNs could only be trained after size reduction via image downsampling. Even though the downsampled images still looked visually acceptable, some image information will be lost, such as blurring or generating artifacts [66]. Thus, it is believed that if the image size was not reduced, the training performance of the MBB dataset should be better.
As mentioned earlier, the proposed methods were executed online using a commercial cloud computing service and achieved fast analysis, i.e., approximately 4 s to analyze each multi-cell image for the detection problem and approximately 10 milliseconds to analyze each single-cell image for the classification problem. This approach imposes a limitation that internet connections are required. In the scenario where no internet connection is available, one or more high-performance computers are required on-site to achieve the real-time analysis. Fortunately, malaria screening does not require such a real-time capability, i.e., completing the execution in microseconds or milliseconds. So, this limitation is not very crucial in this problem.
Besides plasmodium parasite detection and the parasite stage of life classification, which are the main themes of this work, there were also some other efforts proposed to apply YOLO algorithms or their variations to automated malaria diagnosis problems. It is interesting that there are several different aspects to consider problems in the same common problem of malaria diagnosis. To a low-resource setting utilization, the YOLOv3 algorithm was applied to detect malaria in images taken from a mobile phone camera [67]. Considering the variation in parasite size, the YOLOv8 algorithm was utilized, focusing on detecting several parasite sizes [68]. The YOLOv4 algorithm was modified to reduce the computational complexity via layer pruning [69]. The YOLOv8 algorithm was applied to detect parasites and leukocytes, and parasite density estimation was ultimately achieved [70]. This short list of works demonstrates that the YOLO algorithm can be applied to many problems in automated malaria diagnosis.
Finally, it is worthwhile discussing that technology as a whole could eventually be applied to give an alternative solution beyond the conventional method. However, in the real-world applications, there are some other factors to take into account, for example, the availability of microscopic image acquisition systems and computers, the personnel’s computer and software use skills, and so on. In some scenarios, the conventional method might be the best solution. So, the deployment of a new technology to real settings requires such thorough considerations.
7. Conclusions
In this research, an automatic malaria parasite detection method and malaria parasite life stage classification method are proposed, leveraging the YOLO and CNN LeNet-5 algorithms. From the experimental results, it was evident that in the detection stage, the optimal solution to the MBB dataset was achieved by converting each color image into a grayscale image, then using the CLAHE for preprocessing. Its detection accuracy reached 0.91 and 0.92 on the MBB dataset and the MP-IDB (P. falciparum) dataset, respectively. It is interesting that the detection models were trained and tested using images with totally different staining and image acquisition methods. However, the results still demonstrated high detection performance.
In malaria parasite life stage classification, all classification datasets achieved impressive classification accuracies around 0.90. The cropping with background method yielded 0.96 accuracy in the MP-IDB (P. falciparum) dataset, accomplished by using the CNN LeNet-5 algorithm. Looking ahead, there is ample opportunity to further enhance performance by fine-tuning numerous parameters. Collecting and expanding datasets are also the top priorities. There are five species of plasmodium parasites that can infect human beings. In addition to the four species that have been involved in this research, there is one more still, Plasmodium knowlesi. In actual applications, in addition to correctly detecting and classifying the malaria parasite and its life stage, it is also an important step to correctly distinguish its species. Additionally, exploring more sophisticated algorithms holds the potential to achieve even greater accuracy when applying the same data manipulation techniques as demonstrated in this study.
Author Contributions
Conceptualization, T.X., S.A. and N.T.-U.; methodology, T.X. and N.T.-U.; software, T.X.; validation, T.X., S.A. and N.T.-U.; formal analysis, T.X., S.A. and N.T.-U.; investigation, S.A. and N.T.-U.; resources, S.A. and N.T.-U.; data curation, T.X. and N.T.-U.; writing—original draft preparation, T.X., S.A. and N.T.-U.; writing—review and editing, T.X., S.A. and N.T.-U.; visualization, T.X. and N.T.-U.; supervision, N.T.-U.; project administration, N.T.-U.; funding acquisition, S.A. and N.T.-U. All authors have read and agreed to the published version of the manuscript.
Funding
This research has received funding support from Chiang Mai University (RG31/2566).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available on the Malaria Bounding Boxes Database at https://www.kaggle.com/kmader/malaria-bounding-boxes (accessed on 21 June 2022), and the Malaria Parasite Image Database for Image Processing and Analysis at https://link.springer.com/chapter/10.1007/978-3-030-13835-6_7 (accessed on 10 March 2023).
Acknowledgments
Tong Xu would like to thank Biomedical Engineering Institute, Chiang Mai University, for a graduate student scholarship.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cooke, B.M.; Mohandas, N.; Coppel, R.L. The malaria-infected red blood cell: Structural and functional changes. Adv. Parasitol. 2001, 50, 1–86. [Google Scholar] [PubMed]
- Mohandas, N.; An, X. Malaria and human red blood cells. Med. Microbiol. Immunol. 2012, 201, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; MacDonald, A.; Liang, S.Y.; Brady, W.J.; Koyfman, A.; Gottlieb, M.; Do, S.C. Malaria: A focused review for the emergency medicine clinician. Am. J. Emerg. Med. 2024, 77, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Eijk, A.M.v.; Mannan, A.S.; Sullivan, S.A.; Carlton, J.M. Defining symptoms of malaria in India in an era of asymptomatic infections. Malar. J. 2020, 19, 237. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Malaria Report 2022; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Poostchi, M.; Silamut, K.; Maude, R.J.; Jaeger, S.; Thoma, G. Image analysis and machine learning for detecting malaria. Transl. Res. 2018, 194, 36–55. [Google Scholar] [CrossRef] [PubMed]
- Savkare, S.S.; Narote, S.P. Automated system for malaria parasite identification. In Proceedings of the 2015 International Conference on Communication, Information & Computing Technology, Mumbai, India, 15–17 January 2015. [Google Scholar]
- Tek, F.B.; Dempster, A.G.; Kale, İ. Parasite detection and identification for automated thin blood film malaria diagnosis. Comput. Vis. Image Underst. 2010, 114, 21–32. [Google Scholar] [CrossRef]
- World Health Organization. Basic Malaria Microscopy—Part I: Learner’s Guide, 2nd ed.; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Sunarko, B.; Djuniadi; Bottema, M.; Iksan, N.; Hudaya, K.A.N.; Hanif, M.S. Red blood cell classification on thin blood smear images for malaria diagnosis. J.Phys. Conf. Ser. 2022, 1444, 012036. [Google Scholar] [CrossRef]
- Aikawa, M.; Iseki, M.; Barnwell, J.W.; Taylor, D.; Oo, M.M.; Howard, R.J. The pathology of human cerebral malaria. The American J. Trop. Med. Hyg. 1990, 43, 30–37. [Google Scholar] [CrossRef]
- Fitri, L.E.; Widaningrum, T.; Endharti, A.T.; Prabowo, M.H.; Winaris, N.; Nugraha, R.Y.B. Malaria diagnostic update: From conventional to advanced method. J. Clin. Lab. Anal. 2022, 36, e24314. [Google Scholar] [CrossRef]
- Kavanaugh, M.J.; Azzam, S.E.; Rockabrand, D.M. Malaria rapid diagnostic tests: Literary review and recommendation for a quality assurance, quality control algorithm. Diagnostics 2021, 11, 768. [Google Scholar] [CrossRef]
- Das, D.K.; Mukherjee, R.; Chakraborty, C. Computational microscopic imaging for malaria parasite detection: A systematic review. J Microsc. 2015, 260, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rubio, C.M.; de Oliveira, A.D.; Nadal, S.; Bilalli, B.; Zarzuela, F.S.; Espasa, M.S.; Sulleiro, E.; Bosh, M.; Veiga, A.L.l.; Abelló, A.; et al. Advances and challenges in automated malaria diagnosis using digital microscopy imaging with artificial intelligence tools: A review. Front. Microbiol. 2022, 13, 1006659. [Google Scholar]
- Gassenmaier, S.; Küstner, T.; Nickel, D.; Herrmann, J.; Hoffmann, R.; Almansour, H.; Afat, S.; Nikolaou, K.; Othman, A.E. Deep learning applications in magnetic resonance imaging: Has the future become present? Diagnostics 2021, 11, 2181. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, H.; Lee, D. Deep learning-based computed tomographic image super-resolution via wavelet embedding. Radiat. Phys. Chem. 2023, 205, 110718. [Google Scholar] [CrossRef] [PubMed]
- Fallahpoor, M.; Chakraborty, S.; Pradhan, B.; Faust, O.; Datta Barua, P.; Chegeni, H.; Acharya, R. Deep learning techniques in PET/CT imaging: A comprehensive review from sinogram to image space. Comput. Methods Programs Biomed. 2024, 243, 107880. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Dai, W.; Wu, T.; Wang, M.; Wan, S.; Liu, J. AIMIC: Deep learning for microscopic image classification. Computer Methods and Programs in Biomedicine 2022, 226, 107162. [Google Scholar] [CrossRef]
- Theera-Umpon, N.; Poonkasem, I.; Auephanwiriyakul, S.; Patikulsila, D. Hard exudate detection in retinal fundus images using supervised learning. Neural Comput. Appl. 2020, 32, 13079–13096. [Google Scholar]
- Amarbayasgalan, T.; Pham, V.H.; Theera-Umpon, N.; Piao, Y.; Ryu, K.H. An efficient prediction method for coronary heart disease risk based on two deep neural networks trained on well-ordered training datasets. IEEE Access 2021, 9, 135210–135223. [Google Scholar] [CrossRef]
- Ruberto, C.D.; Dempster, A.; Khan, S.; Jarra, B. Automatic thresholding of infected blood images using granulometry and regional extrema. In Proceedings of the International Conference on Pattern Recognition, Barcelona, Spain, 3–8 September 2000. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA, 27–30 June 2016. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Rosado, L.; Correia da Costa, J.M.; Elias, D.; Cardoso, J.S. Automated detection of malaria parasites on thick blood smears via mobile devices. Procedia Comput. Sci. 2016, 90, 138–144. [Google Scholar] [CrossRef]
- Abdurahman, F.; Fante, K.A.; Aliy, M. Malaria parasite detection in thick blood smear microscopic images using modified YOLOV3 and YOLOV4 models. BMC Bioinform. 2021, 22, 112. [Google Scholar] [CrossRef] [PubMed]
- Qiuyang, W.; Yani, Z.; Zhe, G.; Zhongtian, Y.; Jia, X.; JiaQin, G.; Yiming, Y.; Pinna, W.; Yongkang, W. Surface plasmon resonance microstructure optical fiber biosensor for malaria cell detections in the terahertz band. Diam. Relat. Mater. 2024, 139, 110401. [Google Scholar]
- Guin, S.; Chowdhury, D.; Chattopadhyay, M. A novel methodology for detection of malaria. Microsyst. Technol. 2024, 1–14. [Google Scholar] [CrossRef]
- Panklang, N.; Techaumnat, B.; Tanthanuch, N.; Chotivanich, K.; Horprathum, M.; Nakano, M. On-chip impedance spectroscopy of malaria-infected red blood cells. Sensors 2024, 24, 3186. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.; Kariyawasam, T.; Lord, A.; Costa, C.; Chaves, L.; Lima-Junior, J.; Maciel-de-Freitas, R.; Sikulu-Lord, M. First report of rapid, non-invasive, and reagent-free detection of malaria through the skin of patients with a beam of infrared light. Res. Sq. 2022, 1, 1–19. [Google Scholar]
- Saini, S.K.; Awasthi, S.K. Sensing and detection capabilities of one-dimensional defective photonic crystal suitable for malaria infection diagnosis from preliminary to advanced stage: Theoretical study. Crystals 2023, 13, 128. [Google Scholar] [CrossRef]
- Oliveira, M.J.; Caetano, S.; Dalot, A.; Sabino, F.; Calmeiro, T.R.; Fortunato, E.; Martins, R.; Pereira, E.; Prudêncio, M.; Byrne, H.J.; et al. A simple polystyrene microfluidic device for sensitive and accurate SERS-based detection of infection by malaria parasites. Analyst 2023, 148, 4053–4063. [Google Scholar] [CrossRef]
- Ljosa, V.; Sokolnicki, K.L.; Carpenter, A.E. Annotated high-throughput microscopy image sets for validation. Nat. Methods 2012, 9, 637. [Google Scholar] [CrossRef]
- Loddo, A.; Di Ruberto, C.; Kocher, M.; Prod’Hom, G. Processing and Analysis of Biomedical Information; MP-IDB: The Malaria Parasite Image Database for Image Processing and Analysis; Springer: Berlin/Heidelberg, Germany, 2019; pp. 57–65. [Google Scholar]
- Malaria Comparison Chart v4. Available online: https://www.cdc.gov/dpdx/resources/pdf/benchaids/malaria/malaria_comparison_p1-2.pdf (accessed on 31 August 2024).
- Zuiderveld, K. Contrast Limited Adaptive Histogram Equalization, Graphics Gems IV; Heckbert, P.S., Ed.; Academic Press Professional: San Diego, CA, USA, 1994; pp. 474–485. [Google Scholar]
- Reza, A.M. Realization of the contrast limited adaptive histogram equalization (CLAHE) for real-time image enhancement. J. VLSI Signal Process. Syst. Signal Image Video Technol. 2004, 38, 35–44. [Google Scholar] [CrossRef]
- Yadav, G.; Maheshwari, S.; Agarwal, A. Contrast limited adaptive histogram equalization based enhancement for real time video system. In Proceedings of the 2014 International Conference on Advances in Computing, Communications and Informatics, Delhi, India, 24–27 September 2014. [Google Scholar]
- Yang, C.-C. Image enhancement by modified contrast-stretching manipulation. Opt. Laser Technol. 2006, 38, 196–201. [Google Scholar] [CrossRef]
- Terven, J.; Cordova-Esparza, D.-M.; Romero-González, J.-A. A comprehensive review of YOLO architectures in computer vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- George, G.; Oommen, R.M.; Shelly, S.; Philipose, S.S.; Varghese, A.M. A survey on various median filtering techniques for removal of impulse noise from digital image. In Proceedings of the Conference on Emerging Devices and Smart Systems, Tamilnadu, India, 2–3 March 2018. [Google Scholar]
- Saravanan, C. Color image to grayscale image conversion. In Proceedings of the International Conference on Computer Engineering and Applications, Bali, Indonesia, 19–21 March 2010. [Google Scholar]
- Neubeck, A.; Gool, L.V. Efficient non-maximum suppression. In Proceedings of the International Conference on Pattern Recognition, Hong Kong, China, 20–24 August 2006. [Google Scholar]
- Zhou, L.; Yu, W. Improved convolutional neural image recognition algorithm based on LeNet-5. J. Comput. Netw. Commun. 2022, 2022, 1636203. [Google Scholar] [CrossRef]
- Han, S.; Niu, P.; Luo, S.; Li, Y.; Zhen, D.; Feng, G.; Sun, S. A novel deep convolutional neural network combining global feature extraction and detailed feature extraction for bearing compound fault diagnosis. Sensors 2023, 23, 8060. [Google Scholar] [CrossRef]
- Prati, R.; Batista, G.; Monard, M.-C. Data mining with imbalanced class distributions: Concepts and methods. In Proceedings of the Indian International Conference on Artificial Intelligence, Karnataka, India, 16–18 December 2009. [Google Scholar]
- Shaheed, K.; Szczuko, P.; Abbas, Q.; Hussain, A.; Albathan, M. Computer-aided diagnosis of COVID-19 from chest X-ray images using hybrid-features and random forest classifier. Healthcare 2023, 11, 837. [Google Scholar] [CrossRef]
- Lin, C.-C.; Liao, J.-X.; Chiu, M.-S.; Yeh, M.-T.; Chung, Y.-N.; Hsu, C.-H. Applying deep learning algorithm to cell identification. J. Netw. Intell. 2021, 6, 401–410. [Google Scholar]
- Turuk, M.; Sreemathy, R.; Kadiyala, S.; Kotecha, S.; Kulkarni, V. CNN based deep learning approach for automatic malaria parasite detection. IAENG Int. J. Comput. Sci. 2022, 49, 745–753. [Google Scholar]
- Silka, W.; Wieczorek, M.; Silka, J.; Wozniak, M. Malaria detection using advanced deep learning architecture. Sensors 2023, 23, 1501. [Google Scholar] [CrossRef]
- Yang, Z.; Benhabiles, H.; Hammoudi, K.; Windal, F.; He, R.; Collard, D. A generalized deep learning-based framework for assistance to the human malaria diagnosis from microscopic images. Neural Comput. Appl. 2022, 34, 14223–14238. [Google Scholar] [CrossRef]
- Ohdar, K.; Nigam, A. A robust approach for malaria parasite identification with CNN based feature extraction and classification using SVM. In Proceedings of the International Conference on Computing Communication and Networking Technologies, Delhi, India, 6–8 July 2023. [Google Scholar]
- Rahman, A.; Zunair, H.; Reme, T.R.; Rahman, M.S.; Mahdy, M.R.C. A comparative analysis of deep learning architectures on high variation malaria parasite classification dataset. Tissue Cell 2021, 69, 101473. [Google Scholar] [CrossRef]
- Abbas, S.S.; Dijkstra, T.M.H. Detection and stage classification of Plasmodium falciparum from images of Giemsa stained thin blood films using random forest classifiers. Diagn. Pathol. 2020, 15, 130. [Google Scholar] [CrossRef] [PubMed]
- Krishnadas, P.; Chadaga, K.; Sampathila, N.; Rao, S.; Swathi, K.S.; Prabhu, S. Classification of malaria using object detection models. Informatics 2022, 9, 76. [Google Scholar] [CrossRef]
- Loddo, A.; Fadda, C.; Di Ruberto, C. An empirical evaluation of convolutional networks for malaria diagnosis. J. Imaging 2022, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Maity, M.; Jaiswal, A.; Gantait, K.; Chatterjee, J.; Mukherjee, A. Quantification of malaria parasitaemia using trainable semantic segmentation and capsnet. Pattern Recognit. Lett. 2020, 138, 88–94. [Google Scholar] [CrossRef]
- Sifat, M.M.H.; Islam, M.M. A fully automated system to detect malaria parasites and their stages from the blood smear. In Proceedings of the IEEE Region 10 Symp., Dhaka, Bangladesh, 5–7 June 2020. [Google Scholar]
- Chen, S.; Zhao, S.; Huang, C. An automatic malaria disease diagnosis framework integrating blockchain-enabled cloud-edge computing and deep learning. IEEE Internet Things J. 2023, 10, 21544–21553. [Google Scholar] [CrossRef]
- Acula, D.D.; Carlos, J.A.P.; Lumacad, M.M.; Minano, J.C.L.O.; Reodica, J.K.R. Detection and classification of plasmodium parasites in human blood smear images using Darknet with YOLO. In Proceedings of the International Conference on Green Energy, Computing and Intelligent Technology, Iskandar Puteri, Malaysia, 10–12 July 2023. [Google Scholar]
- Madhu, G.; Mohamed, A.W.; Kautish, S.; Shah, M.A.; Ali, I. Intelligent diagnostic model for malaria parasite detection and classification using imperative inception-based capsule neural networks. Sci. Rep. 2023, 13, 13377. [Google Scholar] [CrossRef]
- Dou, B.; Zhu, Z.; Merkurjev, E.; Ke, L.; Chen, L.; Jiang, J.; Zhu, Y.; Liu, J.; Zhang, B.; Wei, G.-W. Machine learning methods for small data challenges in molecular science. Chem. Rev. 2023, 123, 8736–8780. [Google Scholar] [CrossRef]
- Jajosky, R.P.; Wu, S.C.; Jajosky, P.G.; Stowell, S.R. Plasmodium knowlesi (Pk) malaria: A review & proposal of therapeutically rational exchange (T-REX) of Pk-resistant red blood cells. Trop. Med. Infect. Dis. 2023, 8, 478. [Google Scholar] [CrossRef]
- Chen, L.; Fu, Y.; Wei, K.; Zheng, D.; Heide, F. Instance segmentation in the dark. Int. J. Comput. Vis. 2023, 131, 2198–2218. [Google Scholar] [CrossRef]
- Chibuta, S.; Acar, A.C. Real-time malaria parasite screening in thick blood smears for low-resource setting. J. Digit. Imaging 2020, 33, 763–775. [Google Scholar] [CrossRef]
- Zedda, L.; Loddo, A.; Di Ruberto, C. YOLO-PAM: Parasite-attention-based model for efficient malaria detection. J. Imaging 2023, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Sukumarran, D.; Hasikin, K.; Khairuddin, A.S.M.; Ngui, R.; Sulaiman, W.Y.W.; Vythilingam, I.; Divis, P.C.S. An optimised YOLOv4 deep learning model for efficient malarial cell detection in thin blood smear images. Parasit Vectors 2024, 17, 188. [Google Scholar] [CrossRef] [PubMed]
- Hoyos, K.; Hoyos, W. Supporting malaria diagnosis using deep learning and data augmentation. Diagnostics 2024, 14, 690. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).