Computational and Mathematical Methods for Neuroscience
1. Introduction
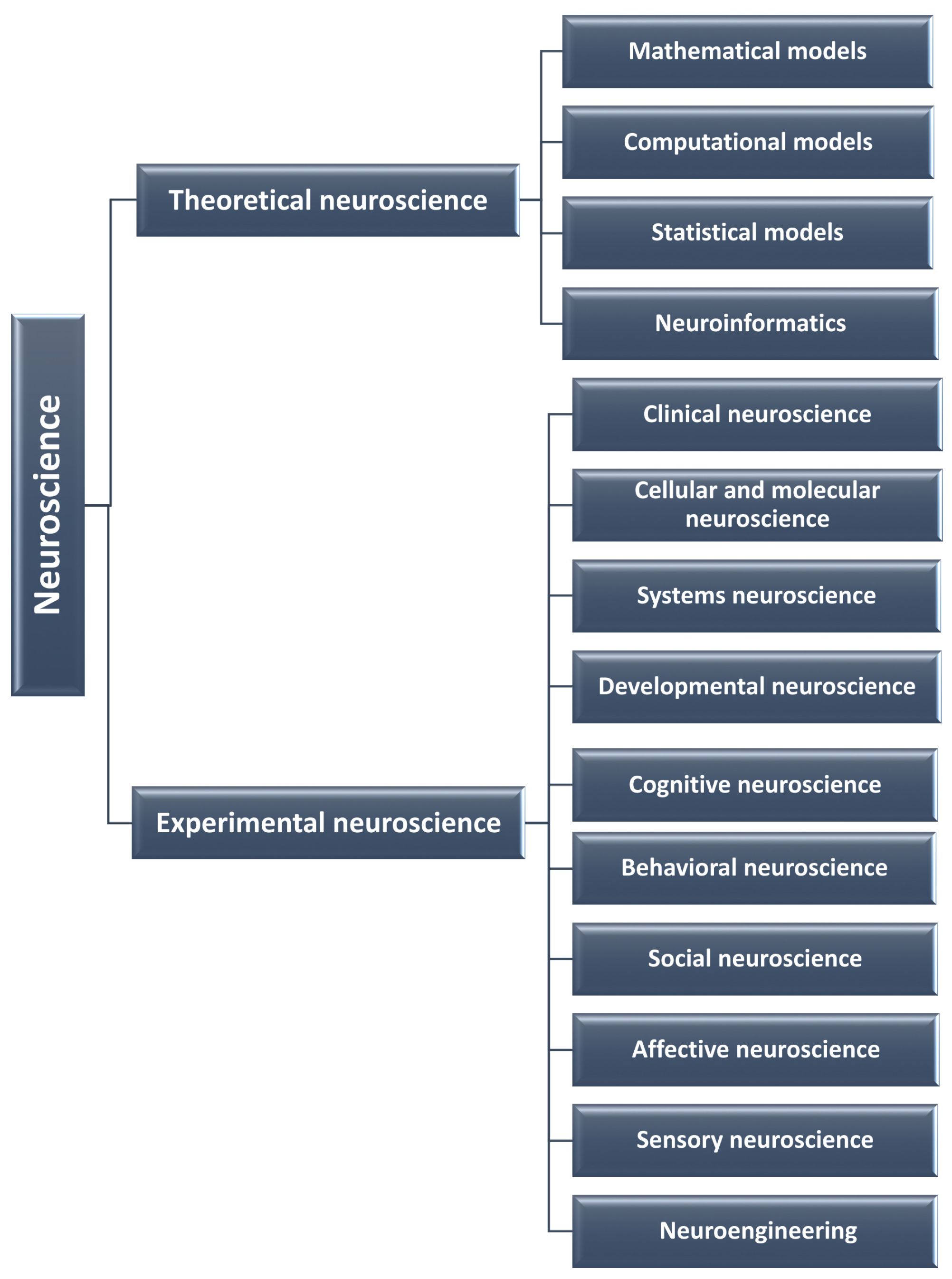
2. Fields of Neuroscience
2.1. Theoretical Neuroscience
2.2. Experimental Neuroscience
3. Highlights and Key Contributions of Published Articles
3.1. Medical Applications
3.2. Cognitive Neuroscience
3.3. Machine Learning
3.4. Statistical Methods
4. Conclusions
Conflicts of Interest
List of Contributions
- Tomás, D.; Pais-Vieira, M.; Pais-Vieira, C. Sensorial Feedback Contribution to the Sense of Embodiment in Brain–Machine Interfaces: A Systematic Review. Appl. Sci. 2023, 13, 13011. https://doi.org/10.3390/app132413011.
- Altwijri, O.; Alanazi, R.; Aleid, A.; Alhussaini, K.; Aloqalaa, Z.; Almijalli, M.; Saad, A. Novel Deep-Learning Approach for Automatic Diagnosis of Alzheimer’s Disease from MRI. Appl. Sci. 2023, 13, 13051. https://doi.org/10.3390/app132413051.
- Kołodziej, M.; Majkowski, A.; Rak, R.; Wiszniewski, P. Convolutional Neural Network-Based Classification of Steady-State Visually Evoked Potentials with Limited Training Data. Appl. Sci. 2023, 13, 13350. https://doi.org/10.3390/app132413350.
- Petzold, A. Partial Parallelism Plots. Appl. Sci. 2024, 14, 602. https://doi.org/10.3390/app14020602.
- Roy, S.; Ehrlich, S.; Lampe, R. Somatosensory Mismatch Response in Patients with Cerebral Palsy. Appl. Sci. 2024, 14, 1030. https://doi.org/10.3390/app14031030.
- Rossi, D.; Aricò, P.; Di Flumeri, G.; Ronca, V.; Giorgi, A.; Vozzi, A.; Capotorto, R.; Inguscio, B.; Cartocci, G.; Babiloni, F.; et al. Analysis of Head Micromovements and Body Posture for Vigilance Decrement Assessment. Appl. Sci. 2024, 14, 1810. https://doi.org/10.3390/app14051810.
- Peña Serrano, N.; Jaimes-Reátegui, R.; Pisarchik, A.N. Hypergraph of Functional Connectivity Based on Event-Related Coherence: Magnetoencephalography Data Analysis. Appl. Sci. 2024, 14, 2343. https://doi.org/10.3390/app14062343.
- Yao, L.; Lu, Y.; Wang, M.; Qian, Y.; Li, H. Exploring EEG Emotion Recognition through Complex Networks: Insights from the Visibility Graph of Ordinal Patterns. Appl. Sci. 2024, 14, 2636. https://doi.org/10.3390/app14062636.
- Koźmiński, P.; Gniazdowska, E. Design, Synthesis and Molecular Modeling Study of Radiotracers Based on Tacrine and Its Derivatives for Study on Alzheimer’s Disease and Its Early Diagnosis. Appl. Sci. 2024, 14, 2827. https://doi.org/10.3390/app14072827.
- Chen, Y.; Zhang, L.; Xue, X.; Lu, X.; Li, H.; Wang, Q. PadGAN: An End-to-End dMRI Data Augmentation Method for Macaque Brain. Appl. Sci. 2024, 14, 3229. https://doi.org/10.3390/app14083229.
- Sait, A. A LeViT–EfficientNet-Based Feature Fusion Technique for Alzheimer’s Disease Diagnosis. Appl. Sci. 2024, 14, 3879. https://doi.org/10.3390/app14093879.
- Gómez, C.; Rodríguez-Martínez, E.; Altahona-Medina, M. Unavoidability and Functionality of Nervous System and Behavioral Randomness. Appl. Sci. 2024, 14, 4056. https://doi.org/10.3390/app14104056.
- Billat, V.; Berthomier, C.; Clémençon, M.; Brandewinder, M.; Essid, S.; Damon, C.; Rigaud, F.; Bénichoux, A.; Maby, E.; Fornoni, L.; et al. Electroencephalography Response during an Incremental Test According to the O2max Plateau Incidence. Appl. Sci. 2024, 14, 5411. https://doi.org/10.3390/app14135411.
- Mattie, D.; Peña-Castillo, L.; Takahashi, E.; Levman, J. MRI Diffusion Connectomics-Based Characterization of Progression in Alzheimer’s Disease. Appl. Sci. 2024, 14, 7001. https://doi.org/10.3390/app14167001.
- Cedron, F.; Alvarez-Gonzalez, S.; Ribas-Rodriguez, A.; Rodriguez-Yañez, S.; Porto-Pazos, A. Efficient Implementation of Multilayer Perceptrons: Reducing Execution Time and Memory Consumption. Appl. Sci. 2024, 14, 8020. https://doi.org/10.3390/app14178020.
- Ferri, L.; Mason, F.; Di Vito, L.; Pasini, E.; Michelucci, R.; Cardinale, F.; Mai, R.; Alvisi, L.; Zanuttini, L.; Martinoni, M.; et al. Cortical Connectivity Response to Hyperventilation in Focal Epilepsy: A Stereo-EEG Study. Appl. Sci. 2024, 14, 8494. https://doi.org/10.3390/app14188494.
References
- Hodgkin, A.L.; Huxley, A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952, 117, 500–544. [Google Scholar] [CrossRef] [PubMed]
- FitzHugh, R. Impulses and physiological states in theoretical models of nerve membrane. Biophys. J. 1961, 1, 445–466. [Google Scholar] [CrossRef] [PubMed]
- Nagumo, J.; Arimoto, S.; Yoshizawa, S. An active pulse transmission line simulating nerve axon. Proc. IRE 1962, 50, 2061–2070. [Google Scholar] [CrossRef]
- Hindmarsh, J.L.; Rose, R.M. A model of neuronal bursting using three coupled first order differential equations. Proc. R. Soc. Lond. 1984, 221, 87–102. [Google Scholar]
- Wilson, H.R.; Cowan, J.D. Excitatory and inhibitory interactions in localized populations of model neurons. Biophys. J. 1972, 12, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Izhikevich, E.M. Simple model of spiking neurons. IEEE Trans. Neural Netws. 2003, 14, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Abbott, L.F. Lapicque’s introduction of the integrate-and-fire model neuron (1907). Brain Res. Bull. 2003, 50, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Rulkov, N.F. Modeling of spiking-bursting neural behavior using two-dimensional map. Phys. Rev. E 2002, 65, 041922. [Google Scholar] [CrossRef] [PubMed]
- Lytton, W.W. Computer modelling of epilepsy. Nat. Rev. Neurosci. 2008, 9, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Frolov, N.; Grubov, V.V.; Maksimenko, V.A.; Lüttjohann, A.; Makarov, V.V.; Pavlov, A.N.; Sitnikova, E.; Pisarchik, A.N.; Kurths, J.; Hramov, A.E. Statistical properties and predictability of extreme epileptic events. Sci. Rep. 2019, 9, 7243. [Google Scholar] [CrossRef] [PubMed]
- Nicolas-Alonso, L.F.; Gomez-Gil, J. Brain computer interfaces, a review. Sensors 2012, 12, 1211–1279. [Google Scholar] [CrossRef] [PubMed]
- Hramov, A.E.; Maksimenko, V.A.; Pisarchik, A.N. Physical principles of brain-computer interfaces and their applications for rehabilitation, robotics and control of human brain states. Phys. Rep. 2021, 918, 1–133. [Google Scholar] [CrossRef]
- World Health Organization. Ageing and Health, 1 October 2024. Available online: https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (accessed on 2 December 2024).
- American Parkinson Disease Association. Parkinson’s Disease, 2024. Available online: https://www.apdaparkinson.org/what-is-parkinsons (accessed on 2 December 2024).
- Bretto, A. Hypergraph Theory: An Introduction; Springer: Cham, Switzerland, 2013. [Google Scholar]
- Ogura, A.; Koyama, D.; Hayashi, N.; Hatano, I.; Osakabe, K.; Yamaguchi, N. Optimal b values for generation of computed high-b-value DW images. AJR Am. J. Roentgenol. 2016, 206, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Destexhe, A.; Rudolph-Lilith, M. Neuronal Noise; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Pisarchik, A.N.; Hramov, A.E. Coherence resonance in neural networks: Theory and experiments. Phys. Rep. 2023, 1000, 1–57. [Google Scholar] [CrossRef]
- Jaimes-Reátegui, R.; Huerta-Cuellar, G.; García-López, J.H.; Pisarchik, A.N. Multistability and noise-induced transitions in the model of bidirectionally coupled neurons with electrical synaptic plasticity. Eur. Phys. J. Spec. Top. 2022, 231, 255–265. [Google Scholar] [CrossRef]
- Pisarchik, A.N.; Hramov, A.E. Multistability in Physical and Living Systems: Characterization and Applications; Springer: Cham, Switzerland, 2022. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pisarchik, A.N. Computational and Mathematical Methods for Neuroscience. Appl. Sci. 2024, 14, 11296. https://doi.org/10.3390/app142311296
Pisarchik AN. Computational and Mathematical Methods for Neuroscience. Applied Sciences. 2024; 14(23):11296. https://doi.org/10.3390/app142311296
Chicago/Turabian StylePisarchik, Alexander N. 2024. "Computational and Mathematical Methods for Neuroscience" Applied Sciences 14, no. 23: 11296. https://doi.org/10.3390/app142311296
APA StylePisarchik, A. N. (2024). Computational and Mathematical Methods for Neuroscience. Applied Sciences, 14(23), 11296. https://doi.org/10.3390/app142311296




