Abstract
Increasing awareness of the problems caused by synthetic agrochemicals, such as chemical fertilizers, pesticides, and herbicides, makes it crucial to discover substitute approaches that can guarantee competitive plant production and protect the environment while maintaining the natural balance in agroecosystems. One of the leading alternatives is utilizing rhizobacterial strains named plant growth-promoting rhizobacteria (PGPR). The utilization of PGPR-based biofertilizers for advancement in the sustainability of farming productions has received considerable critical attention all over the world because of their contribution to not only improving plant growth but also inducing biotic and abiotic stress tolerance. This review updates the aforementioned eco-friendly strategy in sustainable agroecosystems and provides new insights into the phytostimulation and bioprotection ability of lactic acid bacteria (LAB), an emerging taxon of PGPR. In this regard, the ability of LAB to synthesize metabolites, including organic acids, phenolic acids and their flavonoid derivatives, phytohormones, and antimicrobial substrates, is presented. The use of LAB provides a bridge between PGPR and environmentally friendly crop productivity, which can lead to sustainable production systems by reducing the use of agrochemicals, improving soil quality, and minimizing environmental pollution. All the beneficial aspects of LAB need to be addressed by future research to plan systematic methodologies for their use and/or to combine the use of PGPR along with other organic or inorganic inputs in sustainable production systems.
1. Introduction
Supplying adequate agrifood products and byproducts, the demand for which has increased as the global population rises, requires diverse strategies, including (i) incrementing the cultivation area and (ii) improving the production per unit area. However, although the first approach (e.g., land use change) increased production, the conversion of natural landforms into farming land led to an environmental challenge through land degradation [1,2,3]. This problem has become increasingly important in the Mediterranean basin, which demonstrates obvious movements of degradation, especially in areas where climate changes and meteorological conditions contribute extremely to it [4]. As an alternative strategy, the application of agrochemicals (e.g., artificial fertilizers, herbicides, etc.) and intensive farming management practices to increase crop production has brought the major disadvantage of the increasing contamination of agricultural products and the environment [5,6]. Moreover, the strong demand for agrochemicals from domestic and global markets has driven up their prices and caused economic challenges in the agricultural sector [7].
One of the most significant current discussions is a reconsideration of technologies to boost plant production, focusing on alternative strategies, mainly the application of beneficial biological approaches and bio-based products. The application of beneficial microbial consortia, mainly plant growth-promoting rhizobacteria (PGPR), has become one of the most widely used biological alternatives in sustainable agriculture. Such beneficial rhizobacteria can be considered plant biostimulants, which, according to Regulation (EU) No. 2019/1009 of the European Parliament [8], can be used as fertilizing products to promote the plant nutritional value, increase the nutrient locked-up availability in the rhizosphere/soil, and improve plant tolerance against abiotic and biotic stresses [9]. Moreover, the presented sustainable alternatives of agrochemicals receive considerable critical attention in fulfilling part of the United Nations Sustainable Development Goal 15, including how microbial-based biofertilization can promote the sustainable use of agroecosystems and preserve farmlands from degradation.
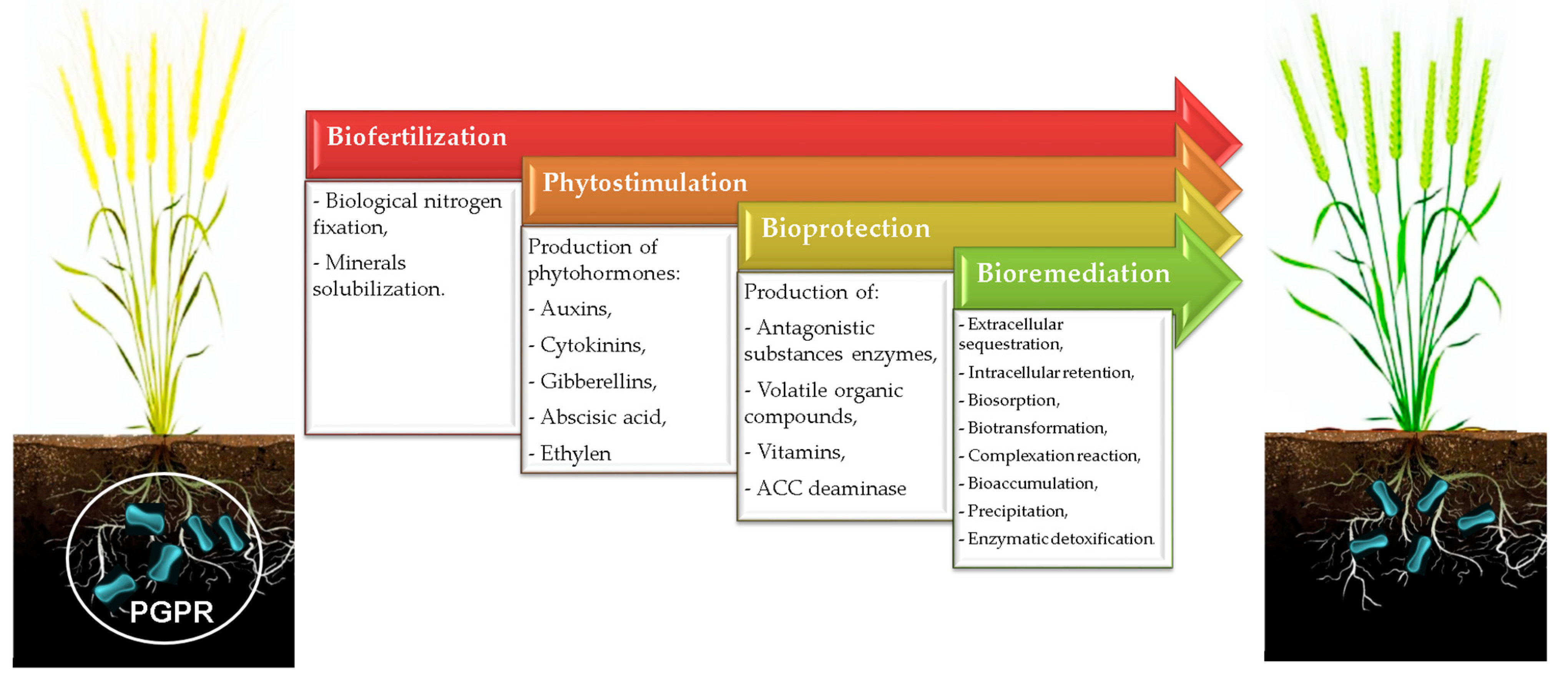
Recently, investigators have attempted to evaluate the potential of identified PGPR and their mechanisms of action as agents of biofertilization, phytostimulation, bioremediation, and bioprotection (Figure 1). Table 1 presents an overview of some identified PGPR and their functional attributes, as further discussed in Section 2 and Section 3.
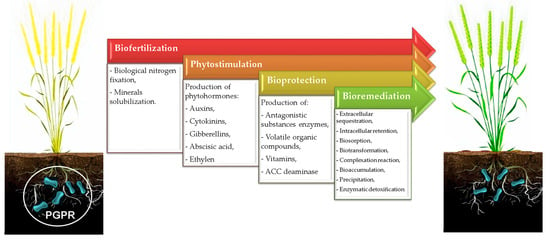
Figure 1.
Potential roles of PGPR in agroecosystems.

Table 1.
Some identified PGPR and their mechanisms as the agents of biofertilization, phytostimulation, bioremediation, and bioprotection.
Among the most potentially beneficial rhizobacteria, lactic acid bacteria (LAB) are suggested as a new promising group of PGPR. However, their functions in agroecosystems, including their role as biofertilizers, biocontrol agents against pathogens, and biostimulants in plant production, have received far too little attention, apart from intensive studies on their traditional role in food processing sectors. Understanding the functional attributes of LAB and their mechanisms is important to use them as a potential way to improve soil health and sustainable plant production. Therefore, this review aims to update the recognized potential of PGPR, mainly LAB, and to reveal their mechanisms of action via which they impact agroecosystems by securing their sustainability.
2. Functional Attributes of PGPR and Their Mechanisms
2.1. Phytostimulation
Bacterial phytohormone production is one of the important studied traits in the plant–microorganism relationship because plant growth and development are directly dependent on phytohormone levels. To date, nine categories of plant hormones have been recognized: auxins (mainly in the form of 3-indol acetic acid (IAA)), gibberellins (GAs), cytokinins (CKs), ethylene (ETH), abscisic acid (ABA), brassinosteroids (BRs), salicylic acid (SA), jasmonic acid (JA), and strigolactones (SLs) [46].
2.2. Biofertilization
Atmospheric nitrogen can be converted to plant-absorbable forms (NH4+) by PGPR with biological nitrogen-fixing (diazotrophy) ability [18]. Nitrogen-fixing bacteria (NFB) are categorized into two main groups including (i) symbiotic bacteria associated with leguminous plants (e.g., Rhizobium) and nonleguminous plants (e.g., Frankia genus and Azospirillum species associated with some dicotyledonous species and cereal grasses, respectively) and (ii) nonsymbiotic free-living bacteria (e.g., cyanobacteria and some genera, including Azotobacter, Arthrobacter, Beijerinckia, Pseudomonas, and Diazotrophicus) [47].
Phosphate solubilizing bacteria (PSB) are among the known PGPR, with a notable capability in solubilizing insoluble complexes of P in soil and making them available to plants using various mechanisms [48,49]. Typically, PSB affect a soil’s biological and physicochemical characteristics, particularly through the release of various organic acids that lead to the chelation of mineral ions and decrease the environmental pH, providing soluble forms of P into the soil [5]. Moreover, the secretion of some enzymes (e.g., phytases and phosphatases) by PSB into the soil can lead to the breaking down of complex organic P forms by catalyzing the mineralization process [49].
The application of potassium solubilizing bacteria (KSB) was proposed as one of the sustainable efficient practices in plant production by transforming insoluble K from feldspar and aluminosilicate minerals into available K and improving the K uptake by plants [24,50]. Various mechanisms are used by KSB, such as the synthesis of organic acids (e.g., oxalic acid, citric acid, succinic acid, tartaric acid, and α-ketogluconic acid), which can affect the dissolution of K-containing minerals by decreasing the pH of the environment as well as by attaching the polysaccharides to the mineral surface [51,52]. The complexation of metal ions (e.g., Fe2+, Al3+, and Ca2+) and proton supply are other mechanisms of KSB to enhance the dissolution of K compounds [52].
It has been reported that zinc (Zn) deficiency can be addressed by applying a type of PGPR known as zinc solubilizing bacteria (ZSB), which can mobilize Zn complexes and solubilize insoluble Zn forms in the soil, including ZnO, ZnCO3, and Zn3(PO4)2, through various mechanisms [27]. These PGPR exude organic acids and phenolic and flavonoid compounds in the rhizosphere, resulting in the sequestration of Zn cations, lowering the pH of the rhizosphere and, consequently, increasing the soluble form of Zn and the ratio of Zn2+ to organic Zn ligands [53]. In fact, Zn absorption is mainly affected by soil pH, in which Zn easily adsorbs on cation exchange places at high pH levels while being replaced by CaCl2 at low pH levels [27].
On the other hand, it has also been suggested that the high Zn mobilization in soils in the presence of high levels of low molecular weight organic acids, phenolics, siderophores, and other bacterial metabolites mostly depends on the complexing capacity of these metabolites compared to their ability to acidify the rhizosphere [53,54]. Such complexing capacity is raised at high levels of soil pH due to the high concentration of deprotonated carboxylic and phenolic moieties, which are more potent Lewis bases in reacting with metal cations. Moreover, some organic molecules possessing more than one acidic moiety (e.g., citric acid) show a greater complexing capacity at high pH levels and, therefore, can form polydentate complexes with cations possessing more than one positive charge, such as Fe3+ and Zn2+, when all the acidic functional groups are consecutively deprotonated [17,54].
Several studies have clearly shown the favorable effects of PGPR on nutrient uptake and plant production. For instance, Bakhshandeh et al. [52] reported an increment in P and K uptake by rice plants, influenced by three PGPR strains (Pantoea ananatis, Rahnella aquatilis, and Enterobacter sp.), of up to 35–77% in leaves, 17–53% in stems, and 25–75% in roots, as well as plant height (+11–15%) and biomass (+27–65%), depending on the PGPR strain. Moreover, some researchers have documented that the application of PGPR treatment promoted plant growth by improving leaf photosynthetic efficiency by up to 19, 12, 12, 16, and 20% in durum wheat [55], rice [56], eggplant [57], cucumber, and pepper [58], respectively, compared to nontreated control plants, which could lead to higher CO2 assimilation [59] and enhanced grain yield [56].
2.3. Bioprotection
Several enzymes synthesized by PGPR have a critical function in protecting a plant from stress and pathogens [60]. Some of them, including chitinases, cellulases, and glucanases, could be labeled as biopesticides since they hinder plant pathogen growth by hydrolyzing polysaccharides and fibrillar materials of the cell wall of pathogenic fungi [60,61]. In this regard, Saraf et al. [62] reported that enzyme synthesis (e.g., proteases, chitinase, and β-1,3-glucanase) by PGPR strains can be considered an important strategy to control soil-borne pathogens through enzymatic degradation or deformation of their cell wall.
Another effective mechanism of PGPR is the synthesis of volatile organic compounds (VOCs), which makes them able to interact with plants and other soil microorganisms by causing systemic resistance to disease and pathogens and promoting plant development [63,64]. In fact, some characteristics of these secondary metabolites, such as low molecular weight (<300 g mol−1), high vapor pressure (>0.01 kPa), and low boiling point, enable them to volatilize and act as the agents of cell signaling [63,65]. Nearly 846 different potential VOCs produced by soil bacteria have been identified [58], the most important of which is N,N-dimethylhexadecylamine (DMHDA), a plant protector against pathogens (e.g., Botrytis cinerea and Phytophthora cinnamomi), and dimethyl disulfide (DMSD), an elicitor of plant defense as well as a plant growth stimulator by increasing the status of sulfur nutrition in plants [49,63].
Plants can usually produce enough vitamins (e.g., biotin, riboflavin, niacin, thiamin, and pantothenate) for their development and provide them to soil microorganisms through root exudates as main nutritive compounds for their survival and development; on the other hand, unhealthy and stressed plants may suffer from vitamin deficiency [5,66]. In this context, some PGPR, especially Bacillus sp. and Rhizobium sp., have great potential to synthesize vitamins, such as pantothenic acid, thiamine, riboflavin, pyrroloquinoline quinone, and biotin, and can contribute to their supply to plants [66,67]. The main functions of vitamins are (i) to act as cofactors in various metabolic pathways, (ii) to facilitate the synthesis of vital metabolites for plants and microbes, (iii) to induce resistance to pathogens, (iv) to promote plant growth and productivity, and (v) to participate in energy transformation in the plant from reserved compounds [67].
Harnessing the antagonistic activity of PGPR has already been suggested as an effective approach to control plant pathogens and inhibit the metabolic activities of various microorganisms through antibiotics [66,68]. In addition to the direct antipathogenic potential of PGPR, they also act as the determinative agents to trigger induced systemic resistance (ISR) in plants and promote plant growth through antifeedant, anthelminthic, phytotoxic, antioxidant, cytotoxic, and antitumor activities in insects and mammals [69]. Among them, diacetylphloroglucinol (2,4-DAPG) synthesized by Pseudomonas sp. [70,71], phenazine by Pseudomonas sp. [72], lipopeptides (e.g., iturin, fengycin, and bacillomycin) and polyketide by Bacillus sp. [73], phenazione-1-carboxylic acid (PCA) by Pseudomonas fluorescens [74], and circulin and colistin by Bacillus subtilis [75] are the most efficient low molecular weight extracellular metabolites that have been extensively studied.
One of the potent biological approaches of PGPR strains is the ability to synthesize 1-aminocyclopropane-1-carboxylate deaminase (ACCD), which can regulate plant growth and induce stress tolerance by decreasing ethylene levels [76]. ACCD, as one of the major enzymes in the intermediate precursor of ethylene production in plants, is responsible for the conversion of ACC to a-ketobutyrate and ammonium [77,78]. The interactions between plants and ACCD-producing PGPR can modify plant defense reactions to a wide range of environmental stresses (e.g., salinity, flooding, high temperature, drought, phytopathogens, and heavy metal contamination) by the degradation of ACC enzymes and decreasing ACC levels in root and leaf tissues [77,79].
2.4. Soil Bioremediation
Recently, PGPR have come to the forefront because of their environmental cleanup ability (bioremediation) as a substitute approach to chemical and physical traditional techniques in eliminating (or controlling) pollutants in soils [80]. Although some organic compounds can endure in soil for a long time, they can be degraded by aerobic PGPR or even dechlorinated and mineralized by anaerobic bacteria [81,82]. For instance, polychlorinated biphenyls (PCBs), as a group of well-known organic contaminants, can be oxidized in aerobic bioremediation processes, where some genera of PGPR (e.g., Bacillus, Pseudomonas, Rhodococcus, and Achromobacter) can utilize the biphenyl (a vital primary substrate that supports PCB cometabolism) using several enzymes, such as dehydrogenases, dioxygenases, hydrolases, hydratases, and aldolases [82,83].
In anaerobic degradation processes, organic compounds are broken down by anaerobic bacterial strains to release the energy required for their metabolic processes. In this process, reductive dechlorination or dehalorespiration in contaminated soils replaces the normal bacterial respiration using aryl halides as electron acceptors for their respiration, resulting in the formation of less toxic and more biodegradable compounds [84].
Soil microbes serve various mechanisms for reducing the toxicity for plants and themselves, including intracellular retention, extracellular sequestration, biosorption, biotransformation, bioaccumulation, complexation reaction, precipitation, and enzymatic detoxification (oxidation and reduction) of toxic metals [85,86,87], whose efficiency depends on the great variability among the toxicity levels of heavy metals. Among them, the most effective mechanism of PGPR strains is the decline in ROS production through the production of some specific enzymatic and nonenzymatic antioxidants (e.g., hydrolases, dioxygenases, hydratases, dehydrogenases, and aldolases), which can preserve plants from ROS-induced oxidative damage [49,60].
3. Lactic Acid Bacteria (LAB): An Emerging Group of PGPR
3.1. Soil- and Plant-Associated LAB
Diverse genera of beneficial rhizobacteria have already been proposed as PGPR, with Bacillus and Pseudomonas being the predominant genera [88]. Nevertheless, metagenomic analyses of plant and rhizosphere microbiomes have resulted in the identification of an emerging group of PGPR, namely, lactic acid bacteria (LAB), which are barely detectable in the plant–soil ecosystem due to their low abundance [18,89]. LAB are known as microaerophilic, Gram-positive, cytochrome-deficient, and nonsporulating bacteria that are also involved in food and silage fermentation as well as soil health; however, some of them are recognized as human pathogens [90,91]. Despite intensive investigations into the conventional function of LAB in the food processing industry, too little attention has been given to their other functions, such as acting as biofertilizers, biocontrol agents, and biostimulants in plant growth. Furthermore, little is known about LAB due to the difficulty of isolating them by plating serial dilutions of rhizospheric soil samples since enrichment methods using selective culture media have been largely ignored [18,92,93]. Despite their low relative abundances, LAB have been isolated from the rhizosphere in some studies, which has consequently led to their introduction as a crucial component of sustainable agricultural approaches as environmentally sustainable and efficient strategies to control pests and diseases and enhance crop yield [93,94,95].
It has been stated that root exudates, including amino acids, carbohydrates, enzymes, organic acids, phenols, and flavonoids, account for a considerable proportion (5–21%) of photosynthetically fixed carbon in plants, which can change the soil environment and, consequently, shape microbial communities [96]. Although such a carbohydrate-rich rhizosphere is ideal for LAB, a quick breakdown of organic acids in the rhizosphere has been proposed as a limiting factor in the capability of LAB to acidify soil to their benefit, thus preventing LAB from being the predominant taxon in agricultural soils [97]. Moreover, recent research on the efficient transfer of LAB from the rhizosphere and phyllosphere to the plant endosphere [98] provided an interesting strategy to assess their roles in plant growth and production. In fact, LAB also constitute a small fraction of the epiphyte [99,100] and endophyte populations of plant microbiota [100,101]. Among the plant-associated LAB, there are some well-known generalist taxa, including Lactiplantibacillus plantarum, Lactococcus lactis, Leuconostoc spp., Weissella spp., and Enterococcus spp., and some specialist taxa, such as Fructilactobacillus florum and Fructobacillus spp., that have been discovered relatively recently [102]. However, the consequences of LAB on plant physiology still need to be fully deciphered. Overall, the genomic diversity in LAB is mainly due to the particular pressure applied by each plant niche [103,104].
3.2. Biofertilization and Bioremediation Effects of LAB
The ability of LAB to synthesize metabolites, including organic acids, phenolic acids and their flavonoid derivatives, phytohormones, and antimicrobial substrates, has already been reported [19,105,106,107]. LAB have been reported to have a high capacity to solubilize insoluble forms of phosphate [19,108,109] and potassium [18], to biologically fix nitrogen [110], and to produce iron-chelating compounds [111] and siderophores [112]. They are also involved in soil biochemical cycles through regulating soil organic matter content and detoxifying hazardous chemicals [111]. Heavy metal biosorption mechanisms of LAB have been previously reported, involving bacterial surface-associated functional groups, including carboxyl, hydroxyl, and phosphate [111,113]. Previous studies on food technology outlined the critical role of LAB in breaking down organic macromolecules and indigestible polysaccharides and converting disfavored flavor compounds [114].
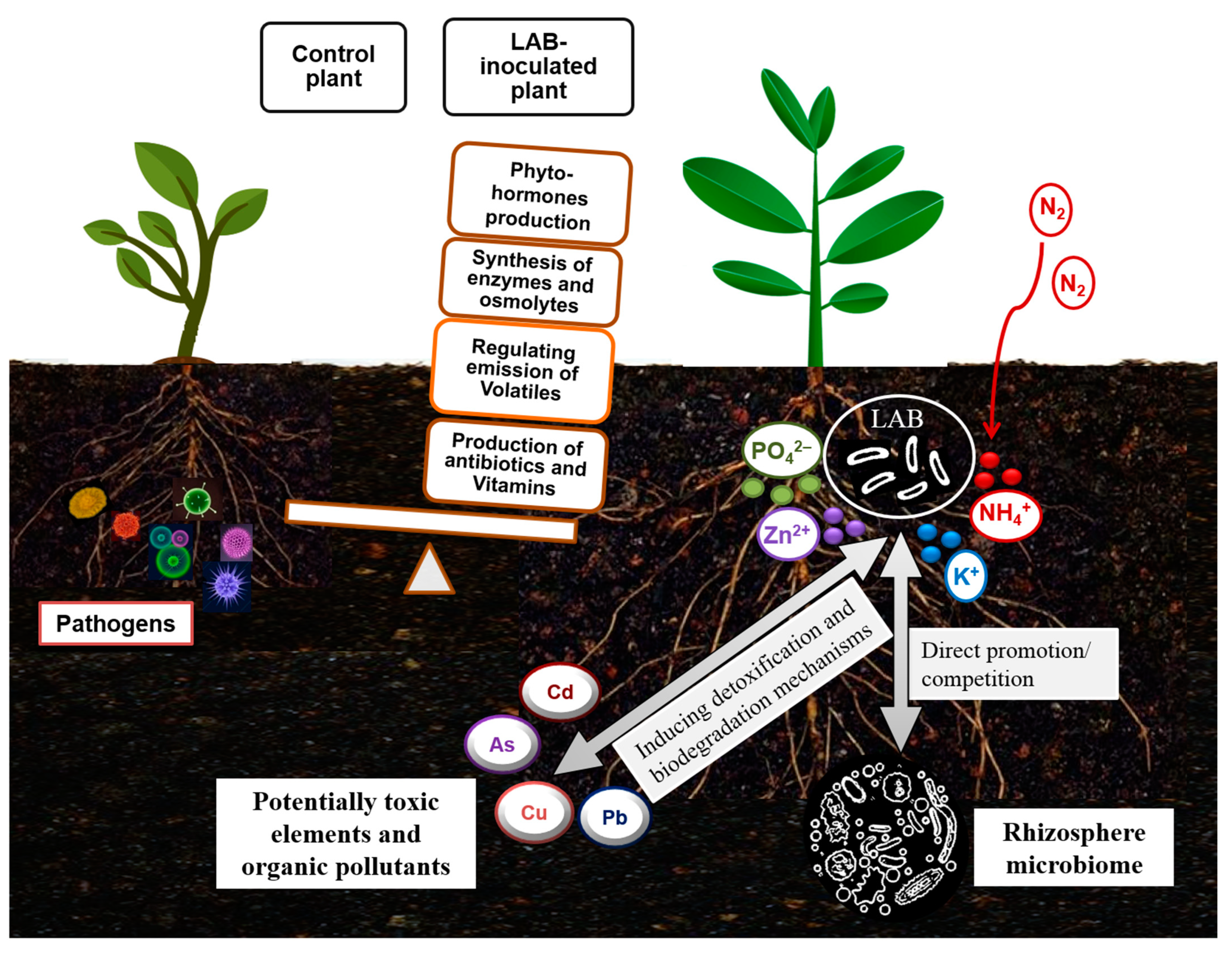
In addition, it has already been suggested that shifts in the microbiome in response to environmental changes may imply the plasticity of the available microbial genetic pool in aiding plant adaptation to environmental stress [115,116]. Accordingly, the finding of a rich diversity of LAB in the rhizosphere of plants grown in deserts [93,97] can confirm the role of LAB in improving the tolerance of associated plants. It can also be assumed that LAB conferring a specific stress tolerance can be derived from holobionts thriving under similar stress conditions. Improved tolerance of LAB-treated plants to abiotic stresses has been correlated with changes in plant metabolic responses related to proline content, phenolic acids, and antioxidant enzymes [97,117]. Such reported findings can support the assumption that LAB are effective as biofertilizers by increasing nutrient bioavailability and as biostimulants to stimulate plant growth or seed germination by alleviating diverse environmental stresses [97,118,119]. The beneficial outcomes of the application of LAB treatment in several plant species have been summarized in Table 2. A scheme of the biofertilization, bioprotection, and biodegradation potential of LAB is shown in Figure 2.

Table 2.
Beneficial effects of lactic acid bacteria in agroecosystems.
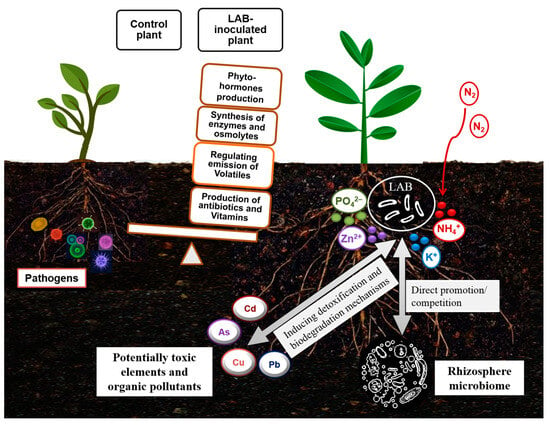
Figure 2.
Schematic portrayal of the biofertilization, bioprotection, and biodegradation potential of lactic acid bacteria.
3.3. Bioprotection Effects of LAB
LAB have also received considerable attention for their capability to synthesize antifungal metabolites (e.g., diketopiperazines, hydroxy derivatives of unsaturated fatty acids, and 3-phenyllactic acid), antibacterial (e.g., bacteriocins and bacteriocin-like substances), and general antimicrobial metabolites (e.g., hydrogen peroxide, organic acids, pyrrolidone-5-carboxylic acid, diacetyl, and reuterin) [133,134,135]. In addition to direct antagonism against pathogens, LAB can affect the plant response to pathogens by causing systemic acquired resistance (SAR) and enhancing plant innate immunity [97]. Mao et al. [136] observed that the antibacterial activity of Lacti plantarum DY-6 was dependent on the production of acetic acid, lactic acid, caprylic acid, propionic acid, and decyl acid. On the other hand, Magnusson et al. [137] observed that the ability to synthesize lactic acid in the bacterial strains without antimicrobial activity was in the same range or even higher than those possessing antimicrobial activity, while the amount of acetic acid corresponded to that normally detected in the culture medium used for assessment tests. Therefore, they concluded that the antimicrobial activity of Lacti. plantarum, Latilactobacillus sakei, Loigolactobacillus coryniformis, and Pediococcus. pentosaceus against Aspergillus sp., Fusarium sp., and Penicillium sp. was due to the synthesis of other metabolites [137]. Through an HPLC analysis of antagonistic bacteria supernatants, they detected two antifungal cyclic dipeptides, cyclo (Phe-Pro) and cyclo (Phe-4-OH-Pro), whose structures were similar to those found in Lacti. plantarum by Ström et al. [138]. Axel et al. [139] found that chemical acidification has no effect on mold inhibition in food, so it is more plausible that the antagonistic activity of LAB depends on the synergistic action between organic acids and other active compounds [140]. The production of bacteriocins by soil- and plant-associated LAB is rare but not excluded, as it was observed that the treatments of cell-free supernatants with organic solvents, surfactants, H2O2, high temperature, and different pH do not affect their antimicrobial activity [132]. Yanagida et al. [141] were the first to report the production of bacteriocins by Ligilactobacillus animalis C060203 and Enterococcus durans C102901, which exhibited strong antibacterial activity against Lati. sakei JCM 1157T. The defeat of antibacterial potential in response to proteinase K treatment confirmed the proteinaceous nature of antimicrobial compounds. A comparative genomic analysis between LAB isolated from plant/soil ecosystems and those isolated from dairy products, nondomestic animals, and human isolates revealed that plant/soil LAB are enriched in genes involved in bacteriocin synthesis, suggesting a probable role in plant fitness [18]. This finding confirms that LAB are a natural farm of antimicrobial metabolites [133] and can be used in agronomic fields to prevent or relieve disease sustainably.
4. Concluding Remarks and Future Perspectives
This review updates the eco-friendly approaches of PGPR application in sustainable agroecosystems as well as provides new insights into the direct or indirect mechanisms of action of these beneficial rhizobacteria involved in biological nitrogen fixation, the solubilization of insoluble minerals, biological control of soil-borne pathogens, stimulation of phytohormone synthesis (e.g., auxins, cytokinins, gibberellins, etc.) in plants, the promotion of enzyme activity involved in reactive oxygen species (ROS)-scavenging, and the biosynthesis of 1-amino cyclopropane-1-carboxylate deaminase (ACC deaminase), hydrogen cyanide, antibiotics, siderophore, and volatile organic compounds. Consequently, these mechanisms provide a bridge between PGPR, mainly LAB, and environmentally friendly crop productivity, which leads to sustainable production systems by reducing agrochemical use, improving soil quality, and minimizing environmental pollution. All these beneficial aspects of LAB need to be addressed in future research to plan methodologies to utilize them and/or to combine the use of these PGPR along with other organic or inorganic inputs in sustainable production systems.
Further work is needed to investigate the environmental sensitivity of LAB to determine how limiting they can be in widespread use. Moreover, a research question that could be asked includes how the competition of LAB with indigenous microorganisms can affect their survival in soils after inoculation. Satisfactory results can be achieved by keeping the bacterial load of the inoculum constant over time. Therefore, future research should focus on the development of efficient microbial formulations that are compatible with conventional techniques, including seed disinfection and pesticide use, to be efficient under various field conditions and soil types and be safe for humans, animals, and plants. Solid and liquid carriers that support the growth and longer viability of microorganisms as alternatives to expensive lyophilization processes have been identified, but each microorganism requires specific growth conditions, and the path to a solution that suits each of them is still a long one.
Author Contributions
Conceptualization, C.C., F.M. and M.Y.K.; writing—original draft preparation, M.Y.K. and S.S.; writing—review and editing, C.C., F.M. and P.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study was carried out within the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022). This manuscript reflects only the authors’ views and opinions; neither the European Union nor the European Commission can be considered responsible for them.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cerdà, A.; Hooke, J.; Romero-Diaz, A.; Montanarella, L.; Lavee, H. Soilerosion on Mediterraneantype-ecosystems. Land Degrad Dev. 2010, 21, 71–74. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Murgese, P.; Strafella, S.; Crecchio, C. Soil Biological Fertility and Bacterial Community Response to Land Use Intensity: A Case Study in the Mediterranean Area. Diversity 2019, 11, 211. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Cucci, G.; Lacolla, G.; Lanzellotti, L.; Crecchio, C. Soil fertility and bacterial community composition in a semiarid Mediterranean agricultural soil under long-term tillage management. Soil Use Manage 2020, 36, 604–615. [Google Scholar] [CrossRef]
- Racioppo, A.; d’Amelio, A.; De Santis, A.; Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. Potential Use of Plant Growth-Promoting Bacteria to Enhance Growth and Soil Fertility in Marginal Areas: Focus on the Apulia Region, Italy. Agronomy 2023, 13, 2983. [Google Scholar] [CrossRef]
- Meena, V.S.; Meena, S.K.; Verma, J.P.; Kumar, A.; Aeron, A.; Mishra, P.K.; Bisht, J.K.; Pattanayak, A.; Naveed, M.; Dotaniya, M.L. Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: A review. Ecol. Eng. 2017, 107, 8–32. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M. Nutrient use efficiency and nutrient uptake promoting of rice by potassium solubilizing bacteria (KSB). Cereal Res. Commun. 2018, 46, 739–750. [Google Scholar] [CrossRef]
- Nishimoto, R. Global trends in the crop protection industry. J. Pestic. Sci. 2019, 44, 141–147. [Google Scholar] [CrossRef]
- EUR-Lex—32019R1009—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2019/1009/oj (accessed on 10 December 2023).
- Hendriksen, N.B. Microbial Biostimulants—The Need for Clarification in EU Regulation. Trends Microbiol. 2022, 30, 311–313. [Google Scholar] [CrossRef]
- Ferioun, M.; Bouhraoua, S.; Srhiouar, N.; Tirry, N.; Belahcen, D.; Siang, T.C.; Louahlia, S.; El Ghachtouli, N. Optimized Drought Tolerance in Barley (Hordeum vulgare L.) Using Plant Growth-Promoting Rhizobacteria (PGPR). Biocatal. Agric. Biotechnol. 2023, 50, 102691. [Google Scholar] [CrossRef]
- Naqqash, T.; Malik, K.A.; Imran, A.; Hameed, S.; Shahid, M.; Hanif, M.K.; Majeed, A.; Iqbal, M.J.; Qaisrani, M.M.; Van Elsas, J.D. Inoculation with Azospirillum spp. Acts as the Liming Source for Improving Growth and Nitrogen Use Efficiency of Potato. Front. Plant Sci. 2022, 13, 929114. [Google Scholar] [CrossRef]
- Liu, F.; Ma, H.; Liu, B.; Du, Z.; Ma, B.; Jing, D. Effects of plant growth-promoting rhizobacteria on the physioecological characteristics and growth of walnut seedlings under drought stress. Agronomy 2023, 13, 290. [Google Scholar] [CrossRef]
- Kouam, I.D.; Mabah, J.; Germain Ntsoli, P.; Tchamani, L.; Yaouba, A.; Katte, B.; Bitom, D. Growth Promotion Potential of Bacillus spp. Isolates on Two Tomato (Solanum lycopersicum L.) Varieties in the West Region of Cameroon. Open Agric. 2023, 8, 20220154. [Google Scholar] [CrossRef]
- Hungria, M.; Barbosa, J.Z.; Rondina, A.B.L.; Nogueira, M.A. Improving Maize Sustainability with Partial Replacement of N Fertilizers by Inoculation with Azospirillum brasilense. Agron. J. 2022, 114, 2969–2980. [Google Scholar] [CrossRef]
- Conde-Avila, V.; Ortega-Martínez, L.D.; Loera, O.; Pérez-Armendáriz, B.; Martínez Valenzuela, C. Encapsulation of Azotobacter Vinelandii ATCC 12837 in Alginate-Na Beads as a Tomato Seedling Inoculant. Curr. Microbiol. 2022, 79, 112. [Google Scholar] [CrossRef]
- Deshwal, V.K.; Kumar, P. Production of Plant growth promoting substance by Pseudomonas. J. Acad. Indus. Res. JAIR 2013, 2, 221–225. [Google Scholar]
- Yaghoubi Khanghahi, M.; Strafella, S.; Allegretta, I.; Crecchio, C. Isolation of bacteria with potential plant-promoting traits and optimization of their growth conditions. Curr. Microbiol. 2021, 78, 464–478. [Google Scholar] [CrossRef]
- Strafella, S.; Simpson, D.J.; Yaghoubi Khanghahi, M.; De Angelis, M.; Gänzle, M.; Minervini, F.; Crecchio, C. Comparative Genomics and In Vitro Plant Growth Promotion and Biocontrol Traits of Lactic Acid Bacteria from the Wheat Rhizosphere. Microorganisms 2021, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Murgese, P.; Santamaria, P.; Leoni, B.; Crecchio, C. Ameliorative effects of PGPB on yield, physiological parameters, and nutrient transporter genes expression in barattiere (Cucumis melo L.). J. Soil Sci. Plant Nutr. 2020, 20, 784–793. [Google Scholar] [CrossRef]
- Scagliola, M.; Pii, Y.; Mimmo, T.; Cesco, S.; Ricciuti, P.; Crecchio, C. Characterization of plant growth promoting traits of bacterial isolates from the rhizosphere of barley (Hordeum vulgare L.) and tomato (Solanum lycopersicon L.) grown under Fe sufficiency and deficiency. Plant Physiol Biochem. 2016, 107, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Wang, Z.; Wang, F.Y.; Li, C.N.; Lan, T.J.; Singh, R.K.; Singh, P.; Yang, L.T.; Li, Y.R. Intercropping in sugarcane cultivation influenced the soil properties and enhanced the diversity of vital diazotrophic bacteria. Sugar Tech. 2017, 19, 136–147. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Luo, Y.; Freitas, H. Inoculation of endophytic bacteriaon host and non-host plants-effects on plant growth and Ni uptake. J Hazard. Mater. 2011, 195, 230–237. [Google Scholar] [CrossRef]
- Bakhshandeh, E.; Gholamhosseini, M.; Yaghoubian, Y.; Pirdashti, H. Plant growth promoting microorganisms can improve germination, seedling growth and potassium uptake of soybean under drought and salt stress. Plant Growth Regul. 2020, 90, 123–136. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M. Potassium solubilising bacteria (KSB) isolated from rice paddy soil: From isolation, identification to K use efficiency. Symbiosis 2018, 76, 13–23. [Google Scholar] [CrossRef]
- Saha, M.; Maurya, B.R.; Meena, V.S.; Bahadur, I.; Kumar, A. Identification and characterization of potassium solubilizing bacteria (KSB) from Indo-Gangetic Plains of India. Biocatal. Agric. Biotechnol. 2016, 7, 202–209. [Google Scholar] [CrossRef]
- Liu, C.; Mou, L.; Yi, J.; Wang, J.; Liu, A.; Yu, J. The Eno Gene of Burkholderia cenocepacia Strain 71-2 is involved in phosphate solubilization. Curr. Microbiol. 2019, 76, 495–502. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Ricciuti, P.; Allegretta, I.; Terzano, R.; Crecchio, C. Solubilization of insoluble zinc compounds by zinc solubilizing bacteria (ZSB) and optimization of their growth conditions. Environ. Sci. Pollut. Res. 2018, 25, 25862–25868. [Google Scholar] [CrossRef]
- Scagliola, M.; Valentinuzzi, F.; Mimmo, T.; Cesco, S.; Crecchio, C.; Pii, Y. Bioinoculants as promising complement of chemical fertilizers for a more sustainable agricultural practice. Front. Sustain. Food Syst. 2021, 4, 622169. [Google Scholar] [CrossRef]
- Sharma, A.; Shankhdhar, D.; Sharma, A.; Shankhdhar, S.C. Growth promotion of the rice genotypes by pgprs isolated from rice rhizosphere. J. Soil Sci. Plant Nutr. 2014, 14, 505–517. [Google Scholar] [CrossRef]
- Purwanto, P.; Yuwariah, Y.; Sumadi, S.; Simarmata, T. Nitrogenase Activity and IAA Production of Indigenous Diazotroph and Its Effect on Rice Seedling Growth. J. Agric. Sci. 2016, 39, 31–37. [Google Scholar] [CrossRef][Green Version]
- Ghavami, N.; Alikhani, H.A.; Pourbabaei, A.A.; Besharati, H. Effects of two new siderophore-producing rhizobacteria on growth and iron content of maize and canola plants. J. Plant Nutr. 2017, 40, 736–746. [Google Scholar] [CrossRef]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Asaf, S.; Khan, M.A.; Kang, S.M.; Yun, B.W.; Lee, I.J. Seed-borne endophytic Bacillus amyloliquefaciens RWL-1 produces gibberellins and regulates endogenous phytohormones of Oryza sativa. Plant Physiol. Biochem. 2016, 106, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Mohite, B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Tabatabaei, F.S.; Saeedizadeh, A. Rhizobacteria cooperative effect against Meloidogyne javanica in rhizosphere of legume seedlings. Hell. Plant Prot. J. 2017, 10, 25–34. [Google Scholar] [CrossRef][Green Version]
- Myresiotis, C.K.; Vryzas, Z.; Papadopoulou-Mourkidou, E. Biodegradation of soil-applied pesticides by selected strains of plant growth-promoting rhizobacteria (PGPR) and their effects on bacterial growth. Biodegradation 2012, 23, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Parewa, H.P.; Meena, V.S.; Jain, L.K.; Choudhary, A. Sustainable Crop Production and Soil Health Management Through Plant Growth-Promoting Rhizobacteria. In Role of Rhizospheric Microbes in Soil; Meena, V., Ed.; Springer: Singapore, 2018. [Google Scholar] [CrossRef]
- Aka, R.J.A.; Babalola, O.O. Effect of bacterial inoculation of strains of pseudomonas aeruginosa, alcaligenes feacalis and bacillus subtilis on germination, growth and heavy metal (Cd, Cr, and Ni) uptake of brassica juncea. Int. J. Phytoremediat. 2016, 18, 200–209. [Google Scholar]
- Rakian, T.C.; Karimuna, L.; Taufik, M.; Sutariati, G.A.K.; Fermin, U. The effectiveness of various Rhizobacteria carriers to improve the shelf life and the stability of Rhizobacteria as Bioherbicide. IOP Conf. Ser. Earth Environ. Sci. 2018, 122, 012032. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S.; Behrendt, U.; Lentzsch, P.; Müller, M.E.H. Antagonistic potential of fluorescent Pseudomonads colonizing wheat heads against mycotoxin producing alternaria and fusaria. Front. Microbiol. 2018, 10, 2124. [Google Scholar] [CrossRef]
- Jog, R.; Nareshkumar, G.; Rajkumar, S. Plant growth promoting potential and soil enzyme production of the most abundant Streptomyces spp. from wheat rhizosphere. J. Appl. Microbiol. 2012, 113, 1154–1164. [Google Scholar] [CrossRef]
- Porcel, R.; Zamarreño, Á.M.; García-Mina, J.M.; Aroca, R. Involvement of plant endogenous ABA in Bacillus megaterium PGPR activity in tomato plants. BMC Plant Biol. 2014, 14, 36. [Google Scholar] [CrossRef]
- Thilagar, G.; Bagyaraj, D.J.; Podile, A.R.; Vaikuntapu, P.R. Bacillus sonorensis, a novel plant growth promoting rhizobacterium in improving growth, nutrition and yield of chilly (Capsicum annuum L.). Proc. Natl. Acad. Sci. USA India Sect. B Biol. Sci. 2018, 88, 813–818. [Google Scholar] [CrossRef]
- Abraham, J.; Silambarasan, S. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol using a novel bacterium Ochrobactrum sp. JAS2: A proposal of its metabolic pathway. Pestic. Biochem. Phys. 2016, 126, 13–21. [Google Scholar] [CrossRef]
- Hofmann, K.; Heinz, E.B.; Charles, T.C.; Hoppert, M.; Liebl, W.; Streit, W.R. Sinorhizobiummeliloti strain 1021 bioS and bdhA gene transcriptions are both affected by biotin available in defined medium. FEMS Microbiol Lett. 2000, 182, 41–44. [Google Scholar] [CrossRef]
- Phillips, D.A.; Joseph, C.M.; Yang, G.-P.; Martínez-Romero, E.; Sanborn, J.R.; Volpin, H. Identification of lumichrome as a Sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl. Acad. Sci. USA 1999, 96, 12275–12280. [Google Scholar] [CrossRef]
- Su, Y.; Xia, S.; Wang, R.; Xiao, L. Phytohormonal quantification based on biological principles. In Hormone Metabolism and Signaling in Plants; Li, J., Li, C., Smith, S.M., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 431–470. [Google Scholar]
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia—The roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Bakhshandeh, E.; Pirdashti, H.; Lendeh, K.S.; Gilani, Z.; Yaghoubi Khanghahi, M.; Crecchio, C. Effects of plant growth promoting microorganisms inoculums on mineral nutrition, growth and productivity of rice (Oryza sativa L.). J. Plant Nutr. 2020, 43, 1643–1660. [Google Scholar] [CrossRef]
- Khatoon, Z.; Huang, S.; Rafique, M.; Fakhar, A.; Kamran, M.A.; Santoyo, G. Unlocking the potential of plant growth-promoting rhizobacteria on soil health and the sustainability of agricultural systems. J. Environ. Manag. 2020, 273, 111118. [Google Scholar] [CrossRef] [PubMed]
- Yaghoubi Khanghahi, M.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M. The role of potassium solubilizing bacteria (KSB) inoculations on grain yield, dry matter remobilization and translocation in rice (Oryza sativa L.). J. Plant Nutr. 2019, 42, 1165–1179. [Google Scholar] [CrossRef]
- Liu, W.; Xu, X.; Wu, S.; Yang, Q.; Luo, Y.; Christie, P. Decomposition of silicate minerals by Bacillus mucilaginosus in liquid culture. Environ. Geochem. Health 2006, 28, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Bakhshandeh, E.; Pirdashti, H.; Lendeh, K.S. Phosphate and potassium-solubilizing bacteria effect on the growth of rice. Ecol. Eng. 2017, 103, 164–169. [Google Scholar] [CrossRef]
- Kamran, S.; Shahid, I.; Baig, D.N.; Rizwan, M.; Malik, K.A.; Mehnaz, S. Contribution of zinc solubilizing bacteria in growth promotion and zinc content of wheat. Front. Microbiol. 2017, 8, 2593. [Google Scholar] [CrossRef]
- Mimmo, T.; Del Buono, D.; Terzano, R.; Tomasi, N.; Vigani, G.; Crecchio, C.; Pinton, R.; Zocchi, G.; Cesco, S. Rhizospheric organic compounds in the soil-microorganism-plant system: Their role in iron availability. Eur. J. Soil Sci. 2014, 65, 629–642. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Strafella, S.; Crecchio, C. Changes in photo-protective energy dissipation of photosystem II in response to beneficial bacteria consortium in durum wheat under drought and salinity stresses. Appl. Sci. 2020, 10, 5031. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Pirdashti, H.; Rahimian, H.; Nematzadeh, G.A.; Ghajar Sepanlou, M.; Salvatori, E.; Crecchio, C. Evaluation of leaf photosynthetic characteristics and photosystem II photochemistry of rice (Oryza sativa L.) under potassium soloubilizing bacteria (KSB) inoculation. Photosynthetica 2019, 57, 500–511. [Google Scholar] [CrossRef]
- Han, H.; Lee, K. Phosphate and potassium solubilizing bacteria effect on mineral uptake, soil availability and growth of eggplant. Res. J. Biol. Sci. 2005, 1, 176–180. [Google Scholar]
- Han, H.S.; Supanjani, E.; Lee, K.D. Effect of co-inoculation with phosphate and potassium solubilizing bacteria on mineral uptake and growth of pepper and cucumber. Plant Soil Environ. 2006, 52, 130–136. [Google Scholar] [CrossRef]
- Kramer, D.M.; Johnson, G.; Kiirats, O.; Edwards, G.E. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth Res. 2004, 79, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Manoj, S.R.; Karthik, C.; Kadirvelu, K.; Arulselvi, P.I.; Shanmugasundaram, T.; Bruno, B.; Rajkumar, M. Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: A review. J. Environ. Manag. 2020, 254, 109779. [Google Scholar] [CrossRef]
- Beauvais, A.; Latgé, J.-P. Special Issue: Fungal Cell Wall. J. Fungi 2018, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Saraf, M.; Pandya, U.; Thakkar, A. Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol. Res. 2014, 169, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Cappellari, L.; Giordano, W.; Banchio, E. Production of Volatile OrganicCompounds in PGPR. In Handbook for Azospirillum; Cassán, F., Okon, Y., Creus, C., Eds.; Springer: Cham, Germany, 2015; pp. 307–317. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Fincheira, P.; Quiroz, A. Microbial volatiles as plant growth inducers. Microbiol. Res. 2018, 208, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Shameer, S.; Prasad, T.N.V.K.V. Plant growth promoting rhizobacteria for sustainable agricultural practices with special reference to biotic and abiotic stresses. Plant Growth Regul. 2018, 84, 603–615. [Google Scholar] [CrossRef]
- Palacios, O.A.; Bashan, Y.; de-Bashan, L.E. Proven and potential involvement of vitamins in interactions of plants with plant growth-promoting bacteria—An overview. Biol. Fertil. Soils 2014, 50, 415–432. [Google Scholar] [CrossRef]
- Tariq, M.; Noman, M.; Ahmed, T.; Hameed, A.; Manzoor, N.; Zafar, M. Antagonistic features displayed by Plant Growth Promoting Rhizobacteria (PGPR): A Review. J. Plant Sci. Phytopathol. 2017, 1, 38–43. [Google Scholar] [CrossRef]
- Fernando, W.G.D.; Ramarathnam, R.; Krishnamoorthy, A.S.; Savchuk, S.C. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 2005, 37, 955–964. [Google Scholar] [CrossRef]
- De Souza, J.T.; Weller, D.M.; Raaijmakers, J.M. Frequency, diversity and activity of 2, 4-diacetylphloroglucinol producing fluorescent Pseudomonas spp. in Dutch take-all decline soils. Phytopathology 2003, 93, 54–63. [Google Scholar] [CrossRef]
- Couillerot, O.; Prigent-Combaret, C.; Caballero-Mellado, J.; Moënne-Loccoz, Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 2009, 48, 505–512. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.; Bloemberg, G.V.; Lugtenberg, B.J. Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 2003, 157, 503–523. [Google Scholar] [CrossRef]
- Romero, D.; de Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef]
- Weller, D.M. Pseudomonas biocontrol agents of soil borne pathogens: Looking back over 30 years. Phytopathology 2007, 97, 250–256. [Google Scholar] [CrossRef]
- Maksimov, I.V.; Abizgil’Dina, R.R.; Pusenkova, L.I. Plant growth promoting rhizobacteria as alternative to chemical crop protectors from pathogens. Appl. Biochem. Microbiol. 2011, 47, 333–345. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.C.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef]
- Saraf, M.; Jha, C.K.; Patel, D. The Role of ACC Deaminase Producing PGPR in Sustainable Agriculture. In Plant Growth and Health Promoting Bacteria. Microbiology Monographs; Maheshwari, D., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; Volume 18. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-Aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef]
- Zhuang, X.; Chen, J.; Shim, H.; Bai, Z. New advances in plant growth-promoting rhizobacteria for bioremediation. Environ. Int. 2007, 33, 406–413. [Google Scholar] [CrossRef]
- Chien, S.W.C.; Chou, J.S.; Chen, S.W.; Chang, J.H.; Chen, S.H. Oxidative mineralization and dechlorination effects of micron/nanosize birnessite on pentachlorophenol in contaminated soil. Water Air Soil Pollut. 2019, 230, 97. [Google Scholar] [CrossRef]
- Vergani, L.; Mapelli, F.; Zanardini, E.; Terzaghi, E.; Di Guardo, A.; Morosini, C.; Raspa, G.; Borin, S. Phyto-rhizoremediation of polychlorinated biphenyl contaminated soils: An outlook on plant-microbe beneficial interactions. Sci. Total Environ. 2017, 575, 1395–1406. [Google Scholar] [CrossRef]
- Reddy, A.V.B.; Moniruzzaman, M.; Aminabhavi, T.M. Polychlorinated biphenyls (PCBs) in the environment: Recent updates on sampling, pretreatment, cleanup technologies and their analysis. Chem. Eng. J. 2019, 358, 1186–1207. [Google Scholar] [CrossRef]
- El-Sayed, W.; Al-Senani, S.R.; Elbahloul, Y. Diversity of dehalorespiring bacteria and selective enrichment of aryl halides-dechlorinating consortium from sedimentary environment near an oil refinery. J. Taibah Univ. Sci. 2018, 12, 711–722. [Google Scholar] [CrossRef]
- Terzano, R.; Rascio, I.; Allegretta, I.; Porfido, C.; Spagnuolo, M.; Yaghoubi Khanghahi, M.; Crecchio, C.; Sakellariadou, F.; Gattullo, C.E. Fire effects on the distribution and bioavailability of potentially toxic elements (PTE) in agricultural soils. Chemosphere 2021, 281, 130752. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Yin, K.; Wang, Q.; Lv, M.; Chen, L. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 2019, 360, 1553–1563. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Jaffar, N.S.; Jawan, R.; Chong, K.P. The potential of lactic acid bacteria in mediating the control of plant diseases and plant growth stimulation in crop production—A mini review. Front. Plant Sci. 2023, 13, 1047945. [Google Scholar] [CrossRef] [PubMed]
- Duar, R.M.; Lin, X.B.; Zheng, J.; Martino, M.E.; Grenier, T.; Pérez-Muñoz, M.E.; Leulier, F.; Gänzle, M.; Walter, J. Lifestyles in transition: Evolution and natural history of the genus Lactobacillus. FEMS Microbiol. Rev. 2017, 41, S27–S48. [Google Scholar] [CrossRef] [PubMed]
- Bintsis, T. Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiol. 2018, 4, 665–684. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Yanagida, F.; Shinohara, T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbial. 2005, 40, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Fhoula, I.; Najjari, A.; Turki, Y.; Jaballah, S.; Boudabous, A.; Ouzari, H. Diversity and antimicrobial properties of lactic acid bacteria isolated from rhizosphere of olive trees and desert truffles of Tunisia. BioMed Res. Int. 2013, 2013, 405708. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.P.; Michel, V.; Martinez, C.; Camps, C. Lactic acid bacteria as biocontrol agents of soil-borne pathogens. Bull. IOBC/WPRS 2012, 78, 285–288. [Google Scholar]
- Murindangabo, Y.T.; Kopecký, M.; Perná, K.; Nguyen, T.G.; Konvalina, P.; Kavková, M. Prominent use of lactic acid bacteria in soil-plant systems. Appl. Soil Ecol. 2023, 189, 104955. [Google Scholar] [CrossRef]
- Liao, Z.; Fan, J.; Lai, Z.; Bai, Z.; Wang, H.; Cheng, M.; Zhang, F.; Li, Z. Chapter Three—Response network and regulatory measures of plant-soil-rhizosphere environment to drought stress. Adv. Agron. 2023, 180, 93–196. [Google Scholar] [CrossRef]
- Lamont, J.R.; Wilkins, O.; Bywater-Ekegärd, M.; Smith, D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil Biol. Biochem. 2017, 111, 1–9. [Google Scholar] [CrossRef]
- Viscardi, S.; Marileo, L.; Barra, P.J.; Durán, P.; Inostroza-Blancheteau, C. From farm to fork: It could be the case of Lactic Acid Bacteria in the stimulation of folates biofortification in food crops. Curr. Opin. Food Sci. 2020, 34, 1–8. [Google Scholar] [CrossRef]
- Di Cagno, R.; Coda, R.; De Angelis, M.; Gobbetti, M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013, 33, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pontonio, E.; Di Cagno, R.; Tarraf, W.; Filannino, P.; De Mastro, G.; Gobbetti, M. Dynamic and assembly of epiphyte and endophyte lactic acid bacteria during the life cycle of Origanum vulgare L. Front. Microbiol. 2018, 9, 1372. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Celano, G.; Lattanzi, A.; Tedone, L.; De Mastro, G.; Gobbetti, M.; De Angelis, M. Lactic acid bacteria in durum wheat flour are endophytic components of the plant during its entire life cycle. Appl. Environ. Microbiol. 2015, 81, 6736–6748. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.O.; Leveau, J.H.; Marco, M.L. Abundance, diversity and plant-specific adaptations of plant-associated lactic acid bacteria. Environ. Microbiol. Rep. 2020, 12, 16–29. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O. Symposium review: Lactococcus lactis from nondairy sources: Their genetic and metabolic diversity and potential applications in cheese1. J. Dairy Sci. 2018, 101, 3597–3610. [Google Scholar] [CrossRef]
- Golomb, B.L.; Marco, M.L. Lactococcus lactis metabolism and gene expression during growth on plant tissues. J. Bacteriol. 2015, 197, 371. [Google Scholar] [CrossRef]
- Dalié, D.K.D.; Deschamps, A.M.; Richard-Forget, F. Lactic acid bacteria—Potential for control of mould growth and mycotoxins: A review. Food Control. 2010, 21, 370–380. [Google Scholar] [CrossRef]
- Filannino, P.; Gobbetti, M.; De Angelis, M.; Di Cagno, R. Hydroxycinnamic acids used as external acceptors of electrons: An energetic advantage for strictly heterofermentative lactic acid bacteria. Appl. Environ. Microbiol. 2014, 80, 7574–7582. [Google Scholar] [CrossRef]
- Filannino, P.; Di Cagno, R.; Gobbetti, M. Metabolic and functional paths of lactic acid bacteria in plant foods: Get out of the labyrinth. Curr. Opin. Biotechnol. 2018, 49, 64–72. [Google Scholar] [CrossRef]
- De Lacerda, J.R.M.; Da Silva, T.F.; Vollú, R.E.; Marques, J.M.; Seldin, L. Generally recognized as safe (GRAS) Lactococcus lactis strains associated with Lippia sidoides Cham. are able to solubilize/mineralize phosphate. Springer Plus 2016, 5, 828. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Million, T.; Assefa, F. Rhizospheric bacterial isolates of grass pea (Lathyrus sativus L.) endowed with multiple plant growth promoting traits. J. Appl. Microbiol. 2018, 125, 1786–1801. [Google Scholar] [CrossRef] [PubMed]
- Giassi, V.; Kiritani, C.; Kupper, K.C. Bacteria as growth-promoting agents for citrus rootstocks. Microbiol. Res. 2016, 190, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Raman, J.; Kim, J.-S.; Choi, K.R.; Eun, H.; Yang, D.; Ko, Y.-J.; Kim, S.-J. Application of Lactic Acid Bacteria (LAB) in Sustainable Agriculture: Advantages and Limitations. Int. J. Mol. Sci. 2022, 23, 7784. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Kim, B.S.; Park, D.H. Biological control of bacterial spot disease and plant growth-promoting effects of lactic acid bacteria on pepper. Biocontrol Sci. Technol. 2014, 24, 763–779. [Google Scholar] [CrossRef]
- Ameen, F.A.; Hamdan, A.M.; El-Naggar, M.Y. Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci. Rep. 2020, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Mukherjee, A.; Singh, S.; Gaurav, A.K.; Chouhan, G.K.; Jaiswal, D.K.; de Araujo Pereira, A.P.; Passari, A.K.; Abdel-Azeem, A.M.; Verma, J.P. Harnessing of phytomicrobiome for developing potential biostimulant consortium for enhancing the productivity of chickpea and soil health under sustainable agriculture. Sci. Total Environ. 2022, 836, 155550. [Google Scholar] [CrossRef] [PubMed]
- Bandopadhyay, S.; Li, X.; Bowsher, A.W.; Last, R.L.; Shade, A. Disentangling plant- and environment-mediated drivers of active rhizosphere bacterial community dynamics during short-term drought. BioRxiv 2023, preprint. [Google Scholar] [CrossRef]
- Phoboo, S.; Sarkar, D.; Bhowmik, P.C.; Jha, P.K.; Shetty, K. Improving salinity resilience in Swertia chirayita clonal line with Lactobacillus plantarum. Can. J. Plant Sci. 2016, 96, 117–127. [Google Scholar] [CrossRef]
- Daranas, N.; Roselló, G.; Cabrefiga, J.; Donati, I.; Francés, J.; Badosa, E.; Spinelli, F.; Montesinos, E.; Bonaterra, A. Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol. 2019, 174, 92–105. [Google Scholar] [CrossRef]
- Afanador-Barajas, L.N.; Navarro-Noya, Y.E.; Luna-Guido, M.L. Impact of a bacterial consortium on the soil bacterial community structure and maize (Zea mays L.) cultivation. Sci. Rep. 2021, 11, 13092. [Google Scholar] [CrossRef] [PubMed]
- Steglińska, A.; Kołtuniak, A.; Motyl, I.; Berłowska, J.; Czyżowska, A.; Cieciura-Włoch, W. Lactic acid bacteria as biocontrol agents against potato (Solanum tuberosum L.) pathogens. Appl. Sci. 2022, 12, 7763. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; You, Y.H.; Khan, A.L.; Park, J.M.; Lee, S.M.; Lee, I.J. Cucumber performance is improved by inoculation with plant growth promoting microorganisms. Acta Agric. Scand. Sect. B Soil 2015, 65, 36–44. [Google Scholar] [CrossRef]
- Baffoni, L.; Gaggia, F.; Dalanaj, N.; Prodi, A.; Nipoti, P.; Pisi, A.; Biavati, B.; Di Gioia, D. Microbial inoculants for the biocontrol of Fusarium spp. in durum wheat. BMC Microbiol. 2015, 30, 242. [Google Scholar] [CrossRef]
- Limanska, N.; Ivanytsia, T.; Basiul, O.; Krylova, K.; Biscola, V.; Chobert, J.M.; Ivanytsia, V.; Haertle, T. Effect of Lactobacillus plantarum on germination and growth of tomato seedlings. Acta Physiol. Plant. 2013, 35, 1587–1595. [Google Scholar] [CrossRef]
- Tsuda, K.; Tsuji, K.G.; Higashiyama, M.; Ogiyama, H.; Umemura, K.; Mitomi, M.; Kubo, Y.; Kosaka, Y. Biological control of bacterial soft rot in Chinese cabbage by Lactobacillus plantarum strain BY under field conditions. Biol. Control. 2016, 100, 63–69. [Google Scholar] [CrossRef]
- Konappa, N.M.; Maria, M.; Uzma, F.; Krishnamurthy, S.; Nayaka, S.C.; Niranjana, S.R.; Chowdapp, S. Lactic acid bacteria mediated induction of defense enzymes to enhance the resistance in tomato against Ralstonia solanacearum causing bacterial wilt. Sci. Hortic. 2016, 207, 183–192. [Google Scholar] [CrossRef]
- Ghosh, R.; Barman, S.; Mukhopadhyay, A.; Mandal, N.C. Biological control of fruit-rot of jackfruit by rhizobacteria and food grade lactic acid bacteria. Biol. Control. 2015, 83, 29–36. [Google Scholar] [CrossRef]
- Byrne, M.B.; Thapa, G.; Doohan, F.M.; Burke, J.I. Lactic Acid Bacteria as Potential Biocontrol Agents for Fusarium Head Blight Disease of Spring Barley. Front. Microbiol. 2022, 13, 912632. [Google Scholar] [CrossRef]
- Ma, J.; Hong, Y.; Deng, L.; Yi, L.; Zeng, K. Screening and characterization of lactic acid bacteria with antifungal activity against Penicillium digitatum on citrus. Biol. Control. 2019, 138, 104044. [Google Scholar] [CrossRef]
- Hamed, H.A.; Moustafa, Y.A.; Abdel-Aziz, S.M. In vivo efficacy of lactic acid bacteria in biological control against Fusarium oxysporum for protection of tomato plant. Life Sci. J. 2011, 8, 462–468. [Google Scholar]
- Kharazian, Z.A.; Jouzani, G.S.; Aghdasi, M.; Khorvash, M.; Zamani, M.; Mohammadzadeh, H. Biocontrol potential of Lactobacillus strains isolated from corn silages against some plant pathogenic fungi. Biol Control. 2017, 110, 33–43. [Google Scholar] [CrossRef]
- Lynch, K.M.; Zannini, E.; Guo, J.; Axel, C.; Arendt, E.K.; Kildea, S.; Coffey, A. Control of Zymoseptoria tritici cause of Septoria tritici blotch of wheat using antifungal Lactobacillus strains. J. Appl. Microbiol. 2016, 121, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Tiwari, S.K. Isolation, identification and characterization of Pediococcus pentosaceus LB44 and Weissella confusa LM85 for the presence of bacteriocin-like inhibitory substances (BLIS). Microbiology 2016, 85, 540–547. [Google Scholar] [CrossRef]
- Stoyanova, L.G.; Ustiugova, E.A.; Netrusov, A.I. Antibacterial metabolites of lactic acid bacteria: Their diversity and properties. Prikl. Biokhim. Mikrobiol. 2012, 48, 259–275. [Google Scholar] [CrossRef]
- Benítez-Chao, D.F.; León-Buitimea, A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. Bacteriocins: An Overview of Antimicrobial, Toxicity, and Biosafety Assessment by in vivo Models. Front. Microbiol. 2021, 12, 630695. [Google Scholar] [CrossRef]
- Fiorino, G.M.; Tlais, A.Z.A.; Losito, I.; Filannino, P.; Gobbetti, M.; Di Cagno, R. Triacylglycerols hydrolysis and hydroxy-and epoxy-fatty acids release during lactic fermentation of plant matrices: An extensive study showing inter-and intra-species capabilities of lactic acid bacteria. Food Chem. 2023, 412, 135552. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, X.; Xu, Z. Identification of Antibacterial Substances of Lactobacillus plantarum DY-6 for Bacteriostatic Action. Food Sci. Nutr. 2020, 8, 2854–2863. [Google Scholar] [CrossRef] [PubMed]
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Ström, K.; Sjögren, J.; Bröberg, A.; Schnürer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and phenyllactic acid. Appl. Environ. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [PubMed]
- Axel, C.; Zannini, E.; Arendt, E.K.; Waters, D.M.; Czerny, M. Quantification of cyclic dipeptides from cultures of Lactobacillus brevis R2D by HRGC/MS using stable isotope dilution assay. Anal. Bioanal. Chem. 2014, 406, 2433–2444. [Google Scholar] [CrossRef]
- Axel, C.; Zannini, E.; Arendt, E.K. Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Crit. Rev. Food Sci. 2017, 57, 3528–3542. [Google Scholar] [CrossRef]
- Yanagida, F.; Chen, Y.S.; Shinohara, T. Searching for bacteriocin-producing lactic acid bacteria in soil. J. Gen. Appl. Microbiol. 2006, 52, 21–28. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).