A New Permutation-Based Method for Ranking and Selecting Group Features in Multiclass Classification
Abstract
1. Introduction
2. Related Work
2.1. Individual Feature Ranking and Selection
2.2. Group Feature Selection Method
3. Method
3.1. Removing Irrelevant Individual Features
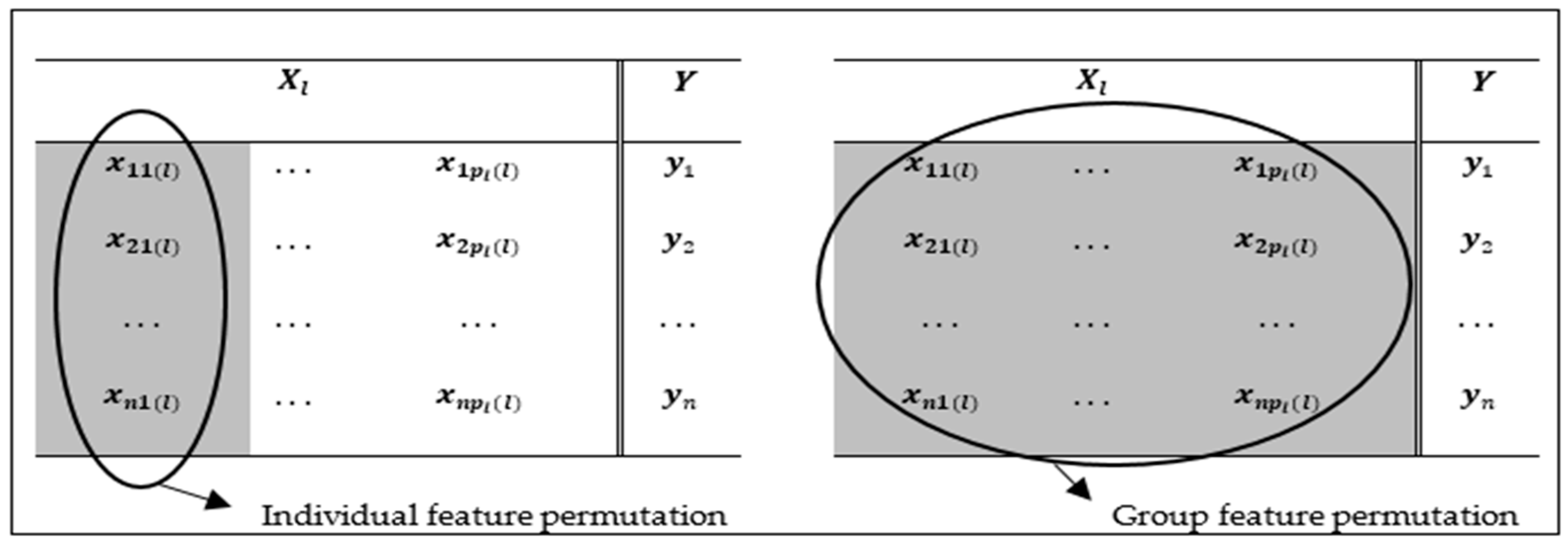
3.2. A Novel Permutation-Based Group Feature Importance Measure
| Algorithm 1: Pseudocode of permutation group importance based on random forest | ||
| Input: X matrix with d individual features, group features, and Y response variable | ||
| Output: vector W | ||
| 1 | procedure Split dataset X into training and testing datasets | |
| 2 | Train the model on the training dataset and compute the baseline out-of-bag by using a random forest classifier | |
| 3 | Initialize all group feature score w[] = [ ] | |
| 4 | For l = 1 to L do | |
| 5 | Compute the permutation importance of in tree t | |
| 6 | Calculate the overall permutation importance of | |
| 7 | end | |
| 8 | return vector W of group features score | |
| 9 | end procedure | |
3.3. Ranking and Selection of GF
4. Results
4.1. Data Description
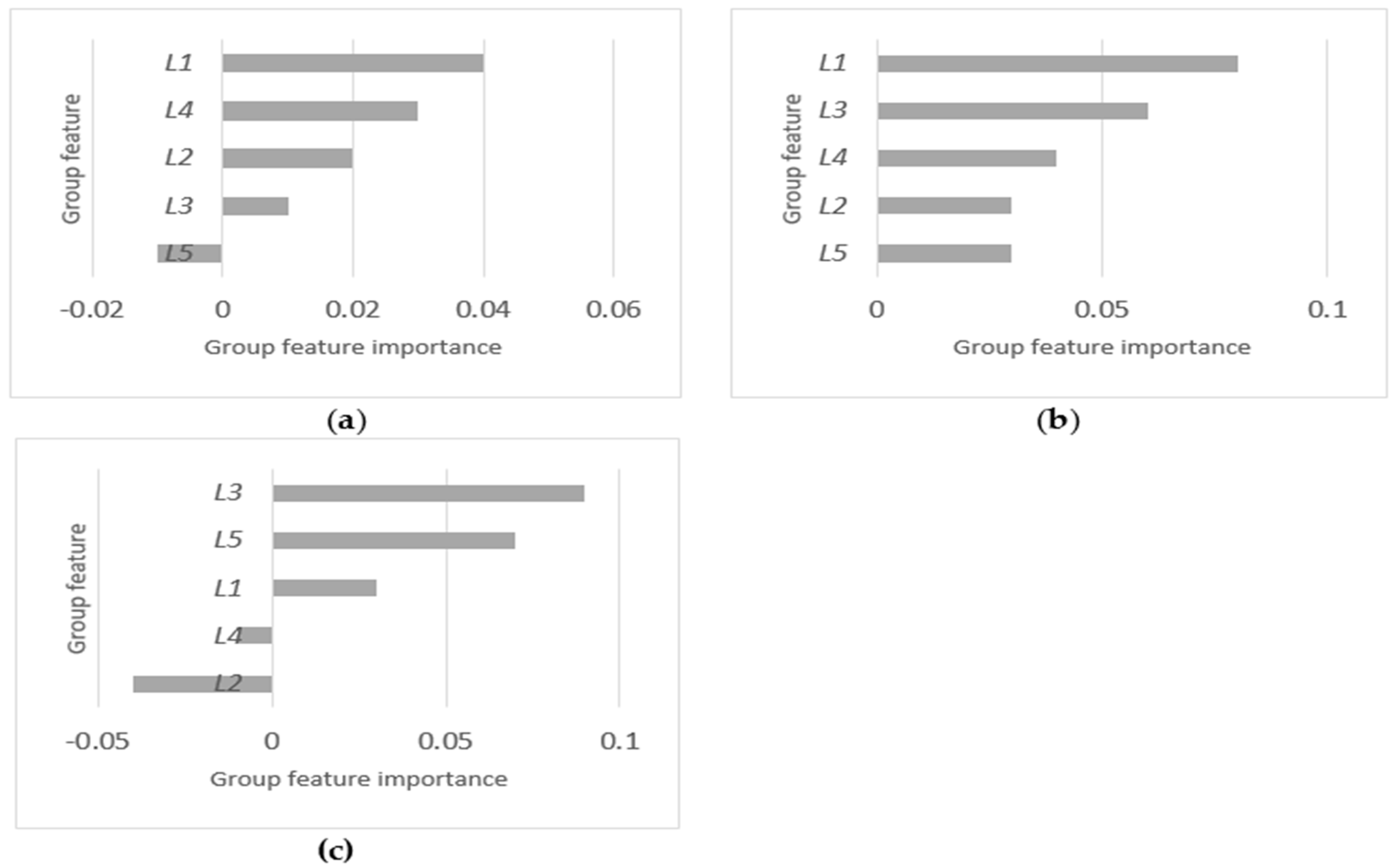
4.2. Relative Importance and Selection of Groups
4.3. Comparison of the Classification Results
5. Discussion and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cavalheiro, L.P.; Bernard, S.; Barddal, J.P.; Heutte, L. Random forest kernel for high-dimension low sample size classification. Stat. Comput. 2024, 34, 9. [Google Scholar] [CrossRef]
- Jiménez, F.; Sánchez, G.; Palma, J.; Miralles-Pechuán, L.; Botía, J.A. Multivariate feature ranking with high-dimensional data for classification tasks. IEEE Access 2022, 10, 60421–60437. [Google Scholar] [CrossRef]
- Bolón-Canedo, V.; Sánchez-Marono, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A review of microarray datasets and applied feature selection methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Tang, F.; Adam, L.; Si, B. Group feature selection with multiclass support vector machine. Neurocomputing 2018, 317, 42–49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Ruiz, R. Weighted general group lasso for gene selection in cancer classification. IEEE Trans. Cybern. 2018, 49, 2860–2873. [Google Scholar] [CrossRef] [PubMed]
- Bakin, S. Adaptive Regression and Model Selection in Data Mining Problems; The Australian National University: Canberra, Australia, 1999. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Yuan, M.; Lin, Y. Model selection and estimation in regression with grouped variables. J. R. Stat. Soc. Ser. B Stat. Methodol. 2006, 68, 49–67. [Google Scholar] [CrossRef]
- Meier, L.; Van De Geer, S.; Bühlmann, P. The group lasso for logistic regression. J. R. Stat. Soc. Ser. B Stat. Methodol. 2008, 70, 53–71. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.; Hastie, T.; Tibshirani, R. A sparse-group lasso. J. Comput. Graph. Stat. 2013, 22, 231–245. [Google Scholar] [CrossRef]
- Fang, K.; Wang, X.; Zhang, S.; Zhu, J.; Ma, S. Bi-level variable selection via adaptive sparse group Lasso. J. Stat. Comput. Simul. 2015, 85, 2750–2760. [Google Scholar] [CrossRef]
- Vincent, M.; Hansen, N.R. Sparse group lasso and high dimensional multinomial classification. Comput. Stat. Data Anal. 2014, 71, 771–786. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, J.; Sun, Z.; Zurada, J.M.; Pal, N.R. Feature selection for neural networks using group lasso regularization. IEEE Trans. Knowl. Data Eng. 2019, 32, 659–673. [Google Scholar] [CrossRef]
- Zubair, I.M.; Kim, B. A Group Feature Ranking and Selection Method Based on Dimension Reduction Technique in High-Dimensional Data. IEEE Access 2022, 10, 125136–125147. [Google Scholar] [CrossRef]
- Theng, D.; Bhoyar, K.K. Feature selection techniques for machine learning: A survey of more than two decades of research. Knowl. Inf. Syst. 2024, 66, 1575–1637. [Google Scholar] [CrossRef]
- Egozi, O.; Gabrilovich, E.; Markovitch, S. Concept-Based Feature Generation and Selection for Information Retrieval. In Proceedings of the AAAI, Chicago, IL, USA, 13–17 July 2008; pp. 1132–1137. [Google Scholar]
- Chen, J.; Huang, H.; Tian, S.; Qu, Y. Feature selection for text classification with Naïve Bayes. Expert Syst. Appl. 2009, 36, 5432–5435. [Google Scholar] [CrossRef]
- Vajda, S.; Karargyris, A.; Jaeger, S.; Santosh, K.; Candemir, S.; Xue, Z.; Antani, S.; Thoma, G. Feature selection for automatic tuberculosis screening in frontal chest radiographs. J. Med. Syst. 2018, 42, 146. [Google Scholar] [CrossRef] [PubMed]
- Dy, J.G.; Brodley, C.E.; Kak, A.; Broderick, L.S.; Aisen, A.M. Unsupervised feature selection applied to content-based retrieval of lung images. IEEE Trans. Pattern Anal. Mach. Intell. 2003, 25, 373–378. [Google Scholar] [CrossRef]
- Lazar, C.; Taminau, J.; Meganck, S.; Steenhoff, D.; Coletta, A.; Molter, C.; de Schaetzen, V.; Duque, R.; Bersini, H.; Nowe, A. A survey on filter techniques for feature selection in gene expression microarray analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2012, 9, 1106–1119. [Google Scholar] [CrossRef]
- Haq, A.U.; Zhang, D.; Peng, H.; Rahman, S.U. Combining multiple feature-ranking techniques and clustering of variables for feature selection. IEEE Access 2019, 7, 151482–151492. [Google Scholar] [CrossRef]
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Hsu, H.-H.; Hsieh, C.-W.; Lu, M.-D. Hybrid feature selection by combining filters and wrappers. Expert Syst. Appl. 2011, 38, 8144–8150. [Google Scholar] [CrossRef]
- Abeel, T.; Helleputte, T.; Van de Peer, Y.; Dupont, P.; Saeys, Y. Robust biomarker identification for cancer diagnosis with ensemble feature selection methods. Bioinformatics 2010, 26, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Setiono, R. Chi2: Feature selection and discretization of numeric attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; pp. 388–391. [Google Scholar]
- Chuang, L.-Y.; Ke, C.-H.; Chang, H.-W.; Yang, C.-H. A two-stage feature selection method for gene expression data. OMICS A J. Integr. Biol. 2009, 13, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Göcs, L.; Johanyák, Z.C. Feature Selection with Weighted Ensemble Ranking for Improved Classification Performance on the CSE-CIC-IDS2018 Dataset. Computers 2023, 12, 147. [Google Scholar] [CrossRef]
- Cheng, Y.; Shi, Q. PCMIgr: A fast packet classification method based on information gain ratio. J. Supercomput. 2023, 79, 7414–7437. [Google Scholar] [CrossRef]
- Kira, K.; Rendell, L.A. A practical approach to feature selection. In Machine Learning Proceedings 1992; Elsevier: Amsterdam, The Netherlands, 1992; pp. 249–256. [Google Scholar]
- Kononenko, I.; Šimec, E.; Robnik-Šikonja, M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Appl. Intell. 1997, 7, 39–55. [Google Scholar] [CrossRef]
- Eppstein, M.J.; Haake, P. Very large scale ReliefF for genome-wide association analysis. In Proceedings of the 2008 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, Sun Valley, ID, USA, 15–17 September 2008; pp. 112–119. [Google Scholar]
- Greene, C.S.; Penrod, N.M.; Kiralis, J.; Moore, J.H. Spatially uniform relieff (SURF) for computationally-efficient filtering of gene-gene interactions. BioData Min. 2009, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.S.; Himmelstein, D.S.; Kiralis, J.; Moore, J.H. The informative extremes: Using both nearest and farthest individuals can improve relief algorithms in the domain of human genetics. In Proceedings of the European Conference on Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics, Istanbul, Turkey, 7–9 April 2010; pp. 182–193. [Google Scholar]
- Urbanowicz, R.J.; Meeker, M.; La Cava, W.; Olson, R.S.; Moore, J.H. Relief-based feature selection: Introduction and review. J. Biomed. Inform. 2018, 85, 189–203. [Google Scholar] [CrossRef]
- Granizo-Mackenzie, D.; Moore, J.H. Multiple threshold spatially uniform relieff for the genetic analysis of complex human diseases. In Proceedings of the Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics: 11th European Conference, EvoBIO 2013, Vienna, Austria, 3–5 April 2013; pp. 1–10. [Google Scholar]
- Stokes, M.E.; Visweswaran, S. Application of a spatially-weighted Relief algorithm for ranking genetic predictors of disease. BioData Min. 2012, 5, 20. [Google Scholar] [CrossRef]
- Le, T.T.; Urbanowicz, R.J.; Moore, J.H.; McKinney, B.A. Statistical inference relief (STIR) feature selection. Bioinformatics 2019, 35, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Breitling, R.; Armengaud, P.; Amtmann, A.; Herzyk, P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004, 573, 83–92. [Google Scholar] [CrossRef]
- Ye, J.; Xiong, T.; Madigan, D. Computational and Theoretical Analysis of Null Space and Orthogonal Linear Discriminant Analysis. J. Mach. Learn. Res. 2006, 7, 1183–1204. [Google Scholar]
- Dudoit, S.; Yang, Y.H.; Callow, M.J.; Speed, T.P. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sin. 2002, 12, 111–139. [Google Scholar]
- Bolón-Canedo, V.; Sánchez-Maroño, N.; Alonso-Betanzos, A. A review of feature selection methods on synthetic data. Knowl. Inf. Syst. 2013, 34, 483–519. [Google Scholar] [CrossRef]
- Huang, J.; Breheny, P.; Ma, S. A selective review of group selection in high-dimensional models. Stat. Sci. A Rev. J. Inst. Math. Stat. 2012, 27, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Huang, J. Concave 1-norm group selection. Biostatistics 2015, 16, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Konietschke, F.; Marmolejo-Ramos, F.; Pauly, M. Permutation tests are robust and powerful at 0.5% and 5% significance levels. Behav. Res. Methods 2021, 53, 2712–2724. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]


| Category | Dataset | No. of Samples | No. of Features | No. of Classes |
|---|---|---|---|---|
| Microarray | GLA-BRA-180 | 180 | 49151 | 4 |
| Microarray | CLL-SUB-111 | 111 | 11340 | 3 |
| Microarray | TOX-171 | 171 | 5749 | 4 |
| Group | GLA-BRA-180 | CLL-SUB-111 | TOX-171 | |||
|---|---|---|---|---|---|---|
| No. of Features | Selected Groups | No. of Features | Selected Groups | No. of Features | Selected Groups | |
| L1 | 14116 | √ | 4411 | √ | 8605 | |
| L2 | 14044 | √ | 2113 | 5156 | ||
| L3 | 5089 | 1451 | √ | 2774 | √ | |
| L4 | 3494 | √ | 1766 | 4941 | ||
| L5 | 12412 | 1598 | 1768 | √ | ||
| Predicted Class | |||
|---|---|---|---|
| Positive | Negative | ||
| Actual class | Positive | tp | fn |
| Negative | fp | tn | |
| Dataset | Classifier | No-FS | Group-Lasso | Proposed Method | |||
|---|---|---|---|---|---|---|---|
| Accuracy | F1 Score | Accuracy | F1 Score | Accuracy | F1 Score | ||
| GLA-BRA-180 | LR | 0.74 | 0.68 | 0.81 | 0.79 | 0.83 | 0.82 |
| SVM | 0.73 | 0.67 | 0.88 | 0.83 | 0.89 | 0.89 | |
| RF | 0.7 | 0.57 | 0.71 | 0.63 | 0.71 | 0.65 | |
| CLL-SUB-111 | LR | 0.82 | 0.83 | 0.87 | 0.91 | 0.95 | 0.97 |
| SVM | 0.85 | 0.88 | 0.9 | 0.93 | 0.97 | 0.98 | |
| RF | 0.71 | 0.75 | 0.83 | 0.87 | 0.88 | 0.91 | |
| TOX-171 | LR | 0.86 | 0.85 | 0.87 | 0.88 | 0.9 | 0.89 |
| SVM | 0.9 | 0.89 | 0.97 | 0.97 | 0.98 | 0.98 | |
| RF | 0.72 | 0.72 | 0.71 | 0.72 | 0.74 | 0.73 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zubair, I.M.; Lee, Y.-S.; Kim, B. A New Permutation-Based Method for Ranking and Selecting Group Features in Multiclass Classification. Appl. Sci. 2024, 14, 3156. https://doi.org/10.3390/app14083156
Zubair IM, Lee Y-S, Kim B. A New Permutation-Based Method for Ranking and Selecting Group Features in Multiclass Classification. Applied Sciences. 2024; 14(8):3156. https://doi.org/10.3390/app14083156
Chicago/Turabian StyleZubair, Iqbal Muhammad, Yung-Seop Lee, and Byunghoon Kim. 2024. "A New Permutation-Based Method for Ranking and Selecting Group Features in Multiclass Classification" Applied Sciences 14, no. 8: 3156. https://doi.org/10.3390/app14083156
APA StyleZubair, I. M., Lee, Y.-S., & Kim, B. (2024). A New Permutation-Based Method for Ranking and Selecting Group Features in Multiclass Classification. Applied Sciences, 14(8), 3156. https://doi.org/10.3390/app14083156






