Abstract
Neurodegenerative diseases (NDDs) are marked by progressive degeneration of neurons within the central nervous system. A notable rise in the prevalence of NDDs has been noticed in the recent past. There is an undeniable requirement for the discovery of innovative therapies aimed at treating NDDs, as current medications predominantly address symptoms rather than provide cures. Approved therapies often experience a decline in therapeutic efficacy over time and are associated with significant side effects. The current investigation explores the potential of spilanthol, the major bioactive compound isolated from Acmella paniculata, in attenuating NDDs through a multi-faceted approach combining in silico, and in vitro methodologies. In silico pharmacokinetic and toxicity screening of spilanthol indicated favorable characteristics for oral delivery, blood–brain barrier permeability, and minimal toxicity. Network pharmacology predicts that spilanthol attenuates neuroinflammation in NDDs by suppressing the toll-like receptor signaling pathway. Molecular docking and dynamics simulations demonstrate robust binding affinities between spilanthol and key proteins in the TLR4 pathway. In vitro experiments conducted using BV-2 microglial cells demonstrate the potential of spilanthol to reduce the production of proinflammatory cytokines and mediators such as NO, TNF-α, and IL-6 induced by lipopolysaccharide. The cumulative findings of the present study indicate that spilanthol mitigates neurodegeneration by alleviating neuroinflammation.
1. Introduction
Neurodegenerative diseases (NDDs) and multifactorial deliberation diseases caused by structural and functional neuronal anomalies in the central nervous system (CNS). NDDs are typified by persistent degeneration of neurons in the CNS, which may lead to physical disabilities and psychobehavioral manifestations including ataxia and dementia [1]. Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), frontotemporal dementia, and Huntington’s disease (HD) are examples of classic NDDs. Although partially attributable to the expansion of life, a discernible increase in the prevalence of NDDs has been noticed during the past ten years [2,3]. Importantly, many of the presently approved drug regimens alleviate symptoms but neither cure nor prevent the propagation of neurodegenerative diseases [4]. Additionally, the prognosis is poor for most NDDs due to the lack of effective therapies. Moreover, the complications of current therapies and the deterioration of the therapeutic efficacy over time have further increased the demand for discovering safe and efficacious treatment modalities for NDDs.
Although there are key pathophysiological differences, microglial activation and chronic neuroinflammation are pathological hallmarks in most NDDs [5,6]. Pathogenesis of AD is not restricted to the neuronal compartment and is strongly related to the innate immune response triggered by microglia. The identification of amyloid-β (Aβ) species by pattern recognition receptors (PRRs) on microglia enhances the expression of cytokines and mediators that are responsible for neuroinflammation, including nitric oxide (NO), tumor necrosis factor-alpha (TNF-α), and interleukin-6 (IL-6), through activation of nuclear factor-κB (NF-κB) via toll-like receptor (TLR) signaling pathways. The chronic overabundant production of pro-inflammatory cytokines further enhances neuronal damage and induces the upregulation of β-secretase, which is responsible for the production of pathogenic Aβ species [7]. In Parkinsonism, chronic release of proinflammatory cytokines from activated microglia is considered a key contributor to the deterioration of dopaminergic neurons in the substantia nigra pars compacta [8]. Therefore, proinflammatory cytokines released from activated microglia have a crucial involvement in the initiation and perpetuation of neurodegeneration and attenuation of chronic neuroinflammation, and proinflammatory cytokine expression is considered to produce a therapeutic benefit in NDDs [9,10].
Phytochemicals have captured significant interest as neuroprotective agents. The therapeutic efficacy of phytochemicals is mainly attributed to their antioxidant, anti-inflammatory, and anticholinesterase activities [2]. Acmella paniculata (Wall ex DC.) R. K. Jansen, from the plant family Asteraceae, is a versatile medicinal plant with a plethora of ethnopharmacological applications. A. paniculata is known to possess a variety of bioactivities including local anesthetic, antimicrobial, antiulcer, antipyretic, antioxidant, anti-inflammatory, insecticidal, anticancer, and larvicidal activity [11,12,13]. Floral inflorescence of A. paniculata is a rich source of N-alkylamides, and the fresh floral inflorescence has been widely used in many traditional medical systems to alleviate pain and inflammation and to induce local anesthesia. Spilanthol [(2E,6Z,8E)-N-Isobutyldeca-2,6,8-trienamide] is one of the most prominent biologically active N-alkylamides in the inflorescence of A. paniculate [14]. Spilanthol is renowned for its bioactivities, including anti-fungal, anti-inflammatory, anti-nociceptive, local anesthetic, and aphrodisiac activity [15]. Although a few studies have been conducted to evaluate the anti-neuroinflammatory effectiveness of spilanthol, the specific mechanism through which it confers neuroprotection remains unclear.
The advancement of computer-aided drug discovery tools has markedly improved natural product-related drug discovery, effectively reducing the historically high attrition rates. Therefore, structure-based virtual screening (SBVS) tools including molecular docking and molecular dynamic (MD) simulation, together with in silico absorption, distribution, metabolism, excretion, and toxicity (ADMET) screening and network pharmacology analysis, have been frequently utilized by both academia and industry to accelerate the drug discovery process. Network pharmacology, a holistic approach, leverages the integration of systems biology, pharmacology, bioinformatics, and network analysis to predict the interactions of compounds with multiple biochemical pathways involved in a specific disease condition [16]. Accordingly, network pharmacology analysis allows the opportunity to comprehensively understand the mechanism of action of a therapeutic candidate and provides the opportunity to repurpose existing bioactive compounds for novel therapeutic applications [17]. Molecular docking and MD simulation studies have become major elements in the SBVS [18]. Molecular docking analysis elucidates the binding interactions between drug candidates and biological targets, offering valuable insights into their binding affinity, intermolecular interactions, and optimal binding orientations [19]. Moreover, protein-based drug targets possess dynamic structures and are frequently susceptible to conformational changes. Hence, MD simulation studies are necessary for the investigation of the time-dependent dynamic interactions between therapeutic candidates and respective targets, as they allow the investigation of time-dependent dynamic interactions. A combination of both molecular docking and MD simulation approaches provides a more holistic view of biomolecular interaction [20]. In addition to efficacy, for a compound to be considered a viable therapeutic candidate, it should also demonstrate a favorable pharmacokinetic and toxicity profile. Therefore, we conducted a comprehensive assessment of the pharmacokinetics and toxicity of spilanthol through in silico ADMET screening tools. Subsequently, we employed network pharmacology to assess the potential of spilanthol in attenuating NDDs. Thereafter, we utilized molecular docking and MD simulation studies to examine the biomolecular interactions of spilanthol with key target proteins identified through the network pharmacology results. Finally, the in silico findings were validated using a lipopolysaccharides (LPS)-induced BV-2 microglia cell model.
2. Results and Discussion
2.1. In Silico ADMET Prediction
Early determination of the ADMET properties of a therapeutic candidate is crucial, as they significantly reduce the attrition rates in drug discovery. Conventional approaches in the determination of ADMET parameters are often time-consuming, expensive, and involve ethical concerns [21]. Hence, computer-aided ADMET prediction has captured significant popularity owing to its feasibility and reliability. In the current study, we utilized the pkCSM web tool and the SwissADME web tool to determine pharmacokinetic descriptors and drug-likeness, respectively. In terms of pharmacokinetics, spilanthol has gained a favorable result in various pharmacokinetic descriptors. Importantly, spilanthol has demonstrated substantial blood–brain barrier (BBB) permeability, favoring its potential therapeutic application in NDDs. Additionally, spilanthol has accounted for high intestinal absorption, thereby favoring oral administration (Supplementary Table S1).
Drug-likeness entails a qualitative assessment aimed at evaluating the likelihood of a compound to serve as an oral drug through scrutiny of its structural and physicochemical attributes. Lipinski’s rule of five is a widespread guideline to evaluate drug-likeness compounds, and it consists of five descriptors including molecular weight (MW) ≤ 500 g/mol, Log p ≤ 5 (n-octanol/water distribution coefficient), nHA ≤ 10 (number of hydrogen bond acceptors), and nHD ≤ 5 (number of hydrogen bond donors) [22]. Interestingly, spilanthol exhibited promising drug-likeness with zero violation in the descriptors coming under Lipinski’s rule. Additionally, spilanthol did not violate any descriptors of the Ghose, Veber, Egan, and Muegge rules, which are also frequently utilized to evaluate the drug-likeness of compounds. Notably, spilanthol exhibited substantial oral bioavailability indicated by an Abbot bioavailability score of 0.55, predicting the probability of spilanthol achieving at least 10% oral bioavailability in rats (Supplementary Table S2 and Supplementary Figure S1) [23]. Protox-II is a widespread web server used to predict the rodent oral toxicity of compounds, and we leveraged Protox-II to evaluate the toxicity of spilanthol. Interestingly, spilanthol demonstrated relatively low toxicity with predicted LD50: 4378 mg/kg, and it was classified under predicted toxicity class 5 with both average similarity and average accuracy of 100%, indicating the safe therapeutic applicability of spilanthol (Supplementary Table S3).
Despite the extensive ethnopharmacological uses of spilanthol-rich herbal extracts and herbal preparations, a very limited number of safety studies have been conducted regarding the safety and toxicity of spilanthol. Stein et al. evaluated the impact of ethanolic extracts of A. oleracea flowers and leaves, as well as pure spilanthol in male Wistar rats. Intraperitoneal administration of crude extracts (10–100 mg/kg) and spilanthol (6.2 mg/kg) have not induced nephrotoxicity and hepatotoxicity [24]. Similarly, intraperitoneal treatment of A. oleracea floral extract for 7 days up to 500 mg/kg has not produced significant toxic outcomes [25]. Chronic toxicity evaluation in male Wistar mice using 100 mg/kg/day of A. oleracea floral extract orally for 60 days showed no signs of toxicity [26]. However, oral and immersion treatment with spilanthol and spilanthol-rich extracts has induced identifiable toxicity characterized by histopathological changes in vital organs in zebrafish models [27]. Similarly, substantial maternal toxicity and teratogenicity have been observed in zebrafish preclinical models upon treatment with spilanthol and spilanthol-rich crude extracts. Additionally, few metabolites of spilanthol with mutagenic potential have been identified during in silico evaluations [28,29]. Despite the available literature, due to the unavailability of human safety data as well as dose escalation studies regarding toxicity, it is not rational to definitively conclude the safety of spilanthol. Moreover, it is essential to evaluate the safety of spilanthol by comprehensively targeting vital physiological systems using in vivo models before considering spilanthol for clinical use.
2.2. Network Pharmacology Analysis
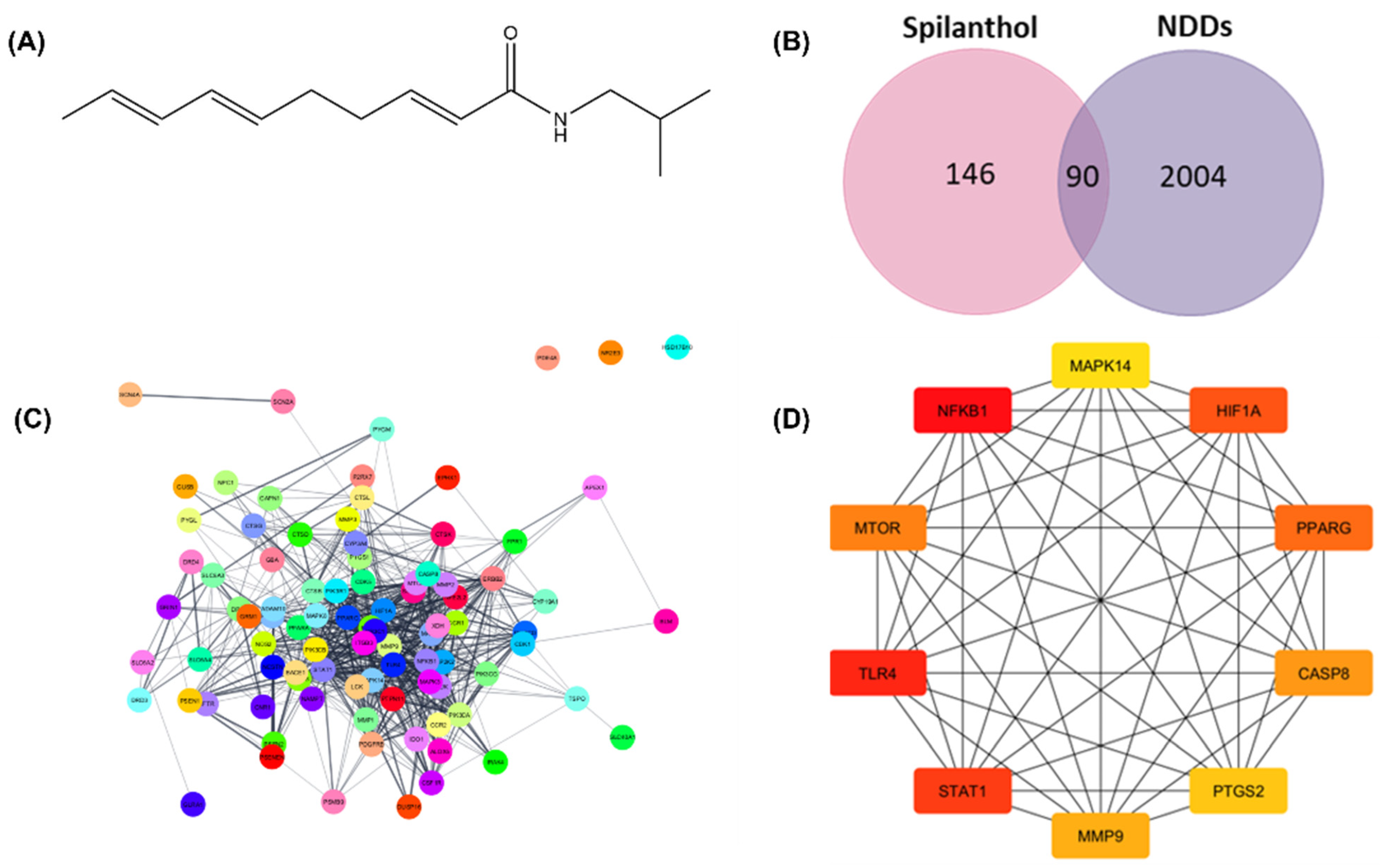
The current study employs network pharmacology analysis to predict the anti-neurodegenerative efficacy of spilanthol. Network pharmacology is a multidisciplinary approach that merges the fields of pharmacology and systems biology, aiming to investigate the complex interactions between bioactive compounds and multiple targets implicated in complex pathophysiologies [16]. The SMILES notation of spilanthol retrieved from the PubChem database was used for structure-based target fishing. The chemical structure of spilanthol is shown in Figure 1A. Plausible targets of the spilanthol were determined using SwissTargetPrediction, SEA SearchServer, and Super-PRED 3.0. As shown in Figure 1B, spilanthol accounted for 236 potential targets. Moreover, 2094 targets linked with NDDs were obtained from three databases: GeneCard, DisGeNET, and OMIM.
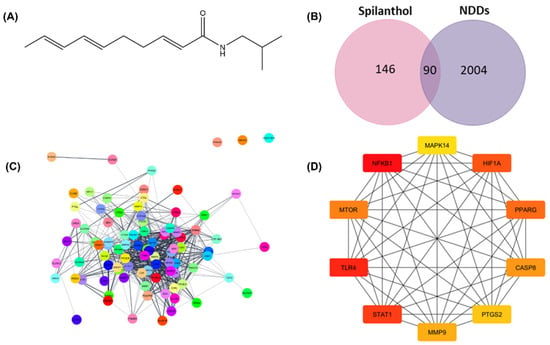
Figure 1.
Network pharmacology analysis for prediction of the underlying mechanism of spilanthol in the attenuation of neurodegenerative diseases. (A) Chemical structure of spilanthol, MW = 221.339 g/mol. (B) Intersection between spilanthol target genes and differentially expressed genes in neurodegenerative diseases. (C) Protein–protein interactions of 90 common targets. (D) Network of the top 10 hub genes using Maximal Clique Centrality analysis. Red to yellow colors signify varying degrees of scores, with red typically indicating a higher score and yellow indicating a lower score. SP, spilanthol; NDDs, Neurodegenerative diseases.
The common targets of spilanthol and NDDs are identified from the intersection of a Venn diagram (Figure 1B). The STRING database was employed for the assessment of protein–protein interactions (PPI), revealing intricate relationships among the involved target proteins. Moreover, there were 690 edges and 90 nodes in the network (Figure 1C). To determine the top 10 interacting proteins, the PPI network was also investigated further for Maximal Clique Centrality (MCC) using the CytoHubba. TLR4, NF-κB1, PTGS2, MMP9, PPARG, MAPK3, HIF1A, GSK3B, STAT1, and MTOR are among the top 10 interacting proteins (Figure 1D). The anti-neurodegenerative effectiveness of spilanthol could potentially be achieved by the modulation of these targets. To gain insight into the pathways mediated by spilanthol in neurodegenerative disorders, GO and KEGG pathway enrichment studies were subsequently executed. A sum of 1732 biological processes, 124 cellular components, and 182 molecular functions were enriched in the GO enrichment analysis. Moreover, 159 pathways were significantly enriched in the KEGG enrichment analysis. Figure 2 summarizes the most significantly enriched KEGG pathways and GO terms.
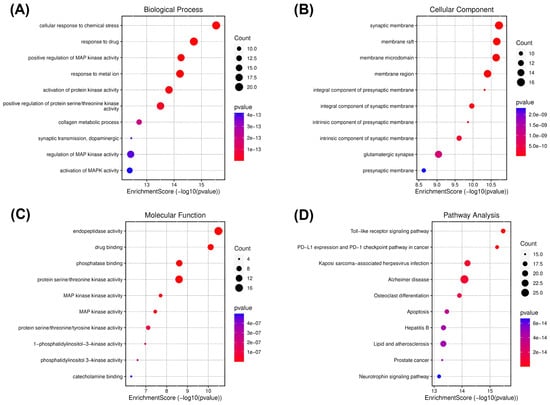
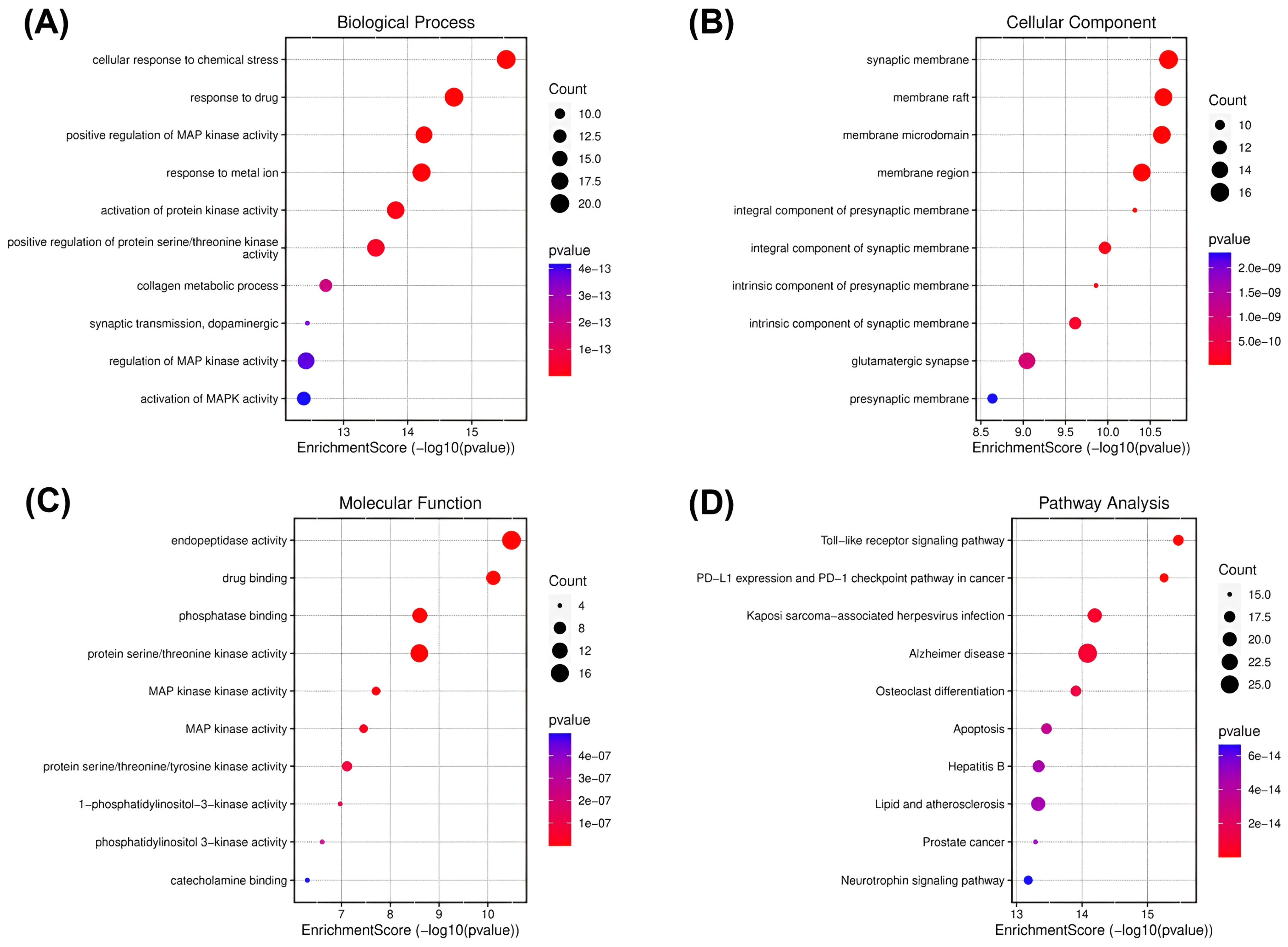
Figure 2.
Gene ontology (GO) and KEGG enrichment analysis of spilanthol- and neurodegenerative disease-related targets. Illustrations of enriched biological processes (A), cellular components (B), molecular functions (C), and KEGG (D) enrichment pathway analyses. The color of each GO and KEGG term indicates the p-value, and the area of the circle indicates the number of enriched genes.
Cellular response to chemical stress was significantly enriched under the biological processes (Figure 2A). This biological process plays a major role in neuroinflammation and subsequently neurodegeneration, indicating the potential of the spilanthol to attenuate neuroinflammation in NDDs [30]. To gather insight into the plausible pathways of spilanthol in the attenuating NDDs, the probable targets were analyzed for KEGG pathway enrichment analysis. The TLR signaling pathway emerged as the most significantly enriched pathway. Meanwhile, the AD pathway was among the top 5 significant pathways (Figure 2D).
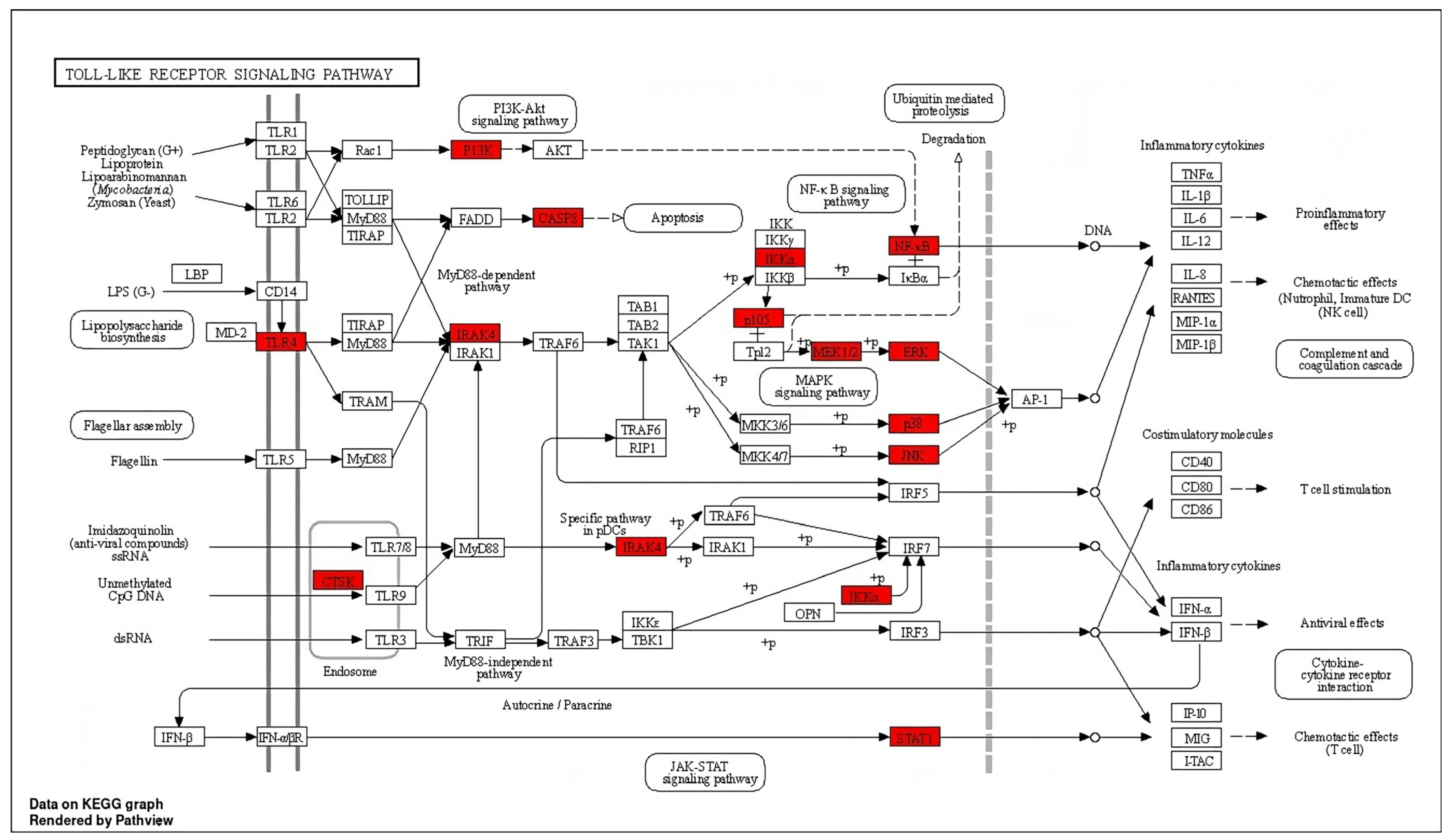
Chronic neuroinflammation and microglial activation are pathological hallmarks in the majority of NDDs. A TLR4 signaling pathway is a predominant pathway involved in microglial activation and neuroinflammation. As illustrated in Figure 3, spilanthol shows interactions with multiple targets in the TLR4 signaling pathway. Microglia, which play a crucial role in immune responses within the central nervous system (CNS), contain toll-like receptor 4 (TLR-4) as a major pattern-recognition receptor. Activation of TLR4 promotes the polarization of microglia toward the M1 phenotype which is associated with the pro-inflammatory response. M1 phenotype releases pro-inflammatory cytokines and chemokines including IL-1, IL-6, IL-12, TNF-α, and NO. The activation of TLR4 triggers microglial downstream signaling via TLR adapters, namely MyD88 and TRIF. Among these, the MyD88-dependent pathway is crucial for the polarization of microglia to M1 phenotype [31]. MyD88 functions as a bridge to employ and activate downstream signaling molecules. MyD88 induces autophosphorylation of interleukin-1 receptor-associated kinases (IRAK4). The phosphorylated IRAK4 then induces tumor necrosis factor receptor-associated factor 6 (TRAF6) to encounter self-ubiquitination and conjugate ubiquitin chains onto TAK1. Activated TAK1 is responsible for the activation of the IKK complex via phosphorylation. Upon activation, the IKK complex phosphorylates the NF-κB inhibitor IκBα, leading to NF-κB activation [32,33]. The translocation of NF-κB induces the transcription of proinflammatory genes and ultimately induces the release of proinflammatory cytokines, chemokines, and enzymes involved in inflammation. As indicated in Figure 3, spilanthol interacts with multiple proteins in the TLR-4 pathway, including TLR4, IRAK4, IKK-α, and NF-κB, in favor of attenuating NDDs.
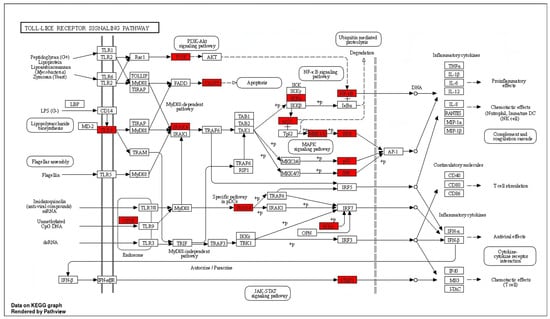
Figure 3.
The distribution of genes in the TLR signaling pathway. ⟶ represents the activation effect, T arrows represent the inhibition effect, and dashed lines represent activation or inhibition effects. The intersection genes are highlighted in red.
Suppression of the TLR4 pathway has been proven to produce favorable outcomes against many NDDs, including AD, PD, ALS, MS, and HD [34,35]. Interestingly, suppression of TLR4 signaling in AD has been proven to improve cognitive dysfunction in streptozotocin-induced murine models of AD [36]. Similarly, inhibition of TLR4-induced microglial polarization through the administration of TLR4-specific inhibitor TAK-242 has significantly improved neurological function, reduced the level of M1 microglia, and inhibited neuronal apoptosis [37]. Although, TLR4 signaling produces a protective effect in acute phases of PD, by inducing the clearance of α-synuclein, chronic activation of the TLR4 signaling pathway was found to exacerbate neurodegeneration by inducing chronic neuroinflammation, and suppression of TLR4 signaling was found to alleviate the features of α-synucleinopathies, excessive inflammation, and progress of neurodegeneration [38]. Importantly, compounds capable of suppressing TLR4/NF-κB signaling have significantly reduced the loss of dopaminergic neurons and have improved locomotor impairments in LPS-induced murine models of Parkinson’s disease [39,40]. As per the findings of previous studies, suppression of TLR signaling has produced therapeutic benefits in NDDs and, interestingly, spilanthol has interacted with multiple proteins in the TLR pathway in favor of attenuating neurodegenerative diseases.
Considering the AD pathway, spilanthol demonstrated substantial potential to modulate with multiple pathophysiological pathways by interacting with 27 target proteins involved in AD pathophysiology, including NMDAR, CASP8, BACE, JNK, and mTOR, in addition to targets in the inflammatory pathway (Supplementary Figure S2) [41,42]. Considering the complex pathophysiology and potential of spilanthol to interact with multiple targets involved in different pathological pathways, future studies are warranted to comprehensively evaluate the potential of spilanthol in attenuating AD focusing on different pathways by evaluating different biomarkers [43,44].
2.3. Molecular Docking and Molecular Dynamics Simulation Analysis
To elucidate the biomolecular interactions between spilanthol and pivotal proteins involved in the TLR4 signaling pathway, molecular docking and MD simulations were undertaken. The Protein Data Bank (PDB) serves as a vital source, containing three-dimensional structural data on numerous biologically important macromolecules, including proteins and ligands. This extensive resource serves as a cornerstone for researchers seeking profound insights into molecular biology. In our study, we focused on specific macromolecular constituents involved in the TLR pathway. Accordingly, three-dimensional structural representations of the target proteins were retrieved from the PDB. Subsequently, we conducted docking experiments and assessed the interaction energy using the GOLD program. The relevant PDB identifiers together with the GOLD fitness score for spilanthol and specific inhibitors are tabulated (Table 1).

Table 1.
GOLD fitness scores for spilanthol and specific inhibitors of pivotal proteins in the TLR4 signaling pathway.
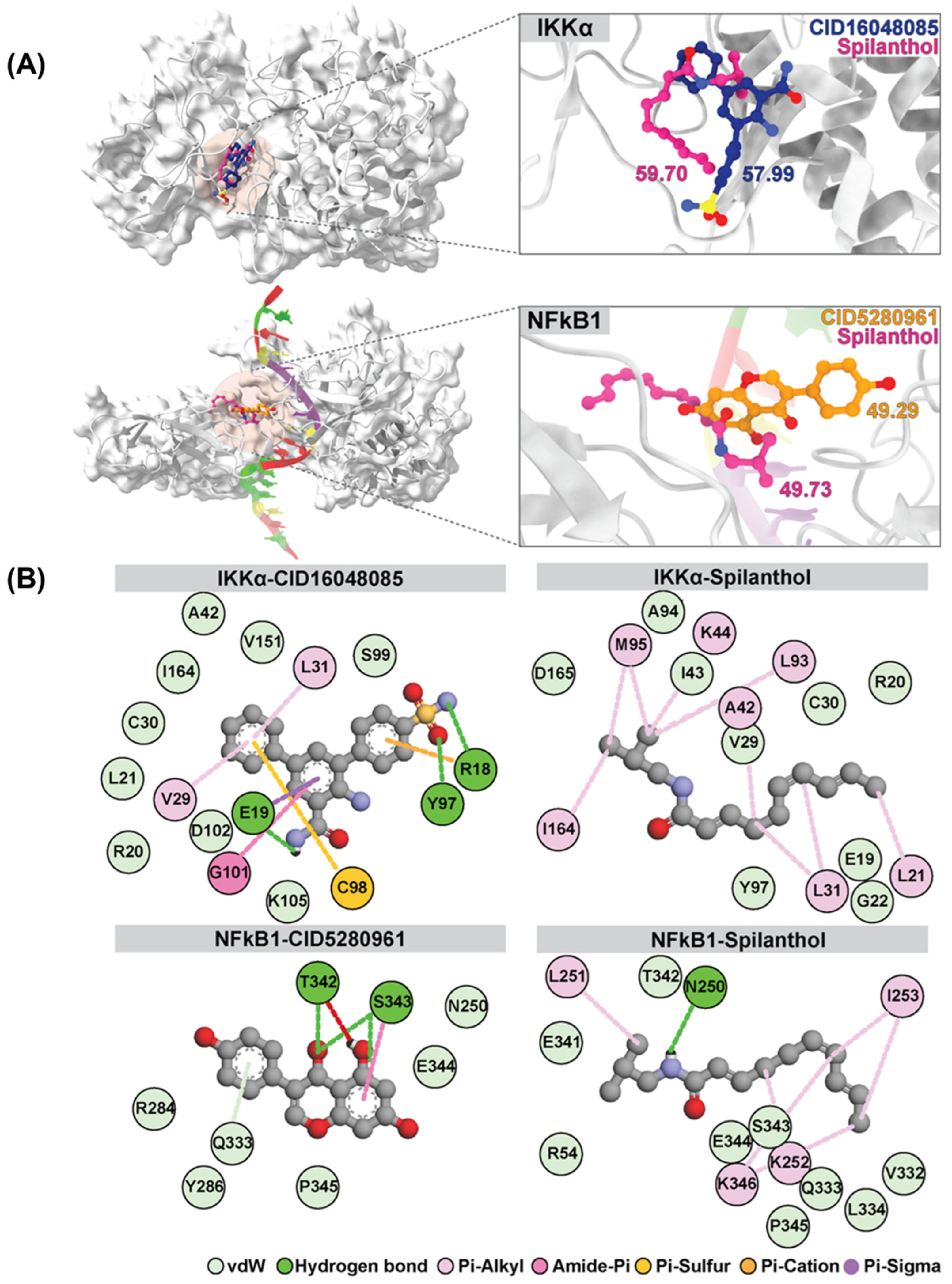
Interestingly, spilanthol demonstrated moderate binding affinity to multiple targets, and notably, spilanthol exhibited a substantial affinity for the active sites of IKKα and NF-kB1 proteins, as indicated by GOLD fitness scores of 59.70 and 49.73, respectively. Through analysis of the superimposed three-dimensional interactions, we observed that spilanthol fit favorably within the active site that is comparable to the inhibitors CID16048085 and CID5280961 (Figure 4A).
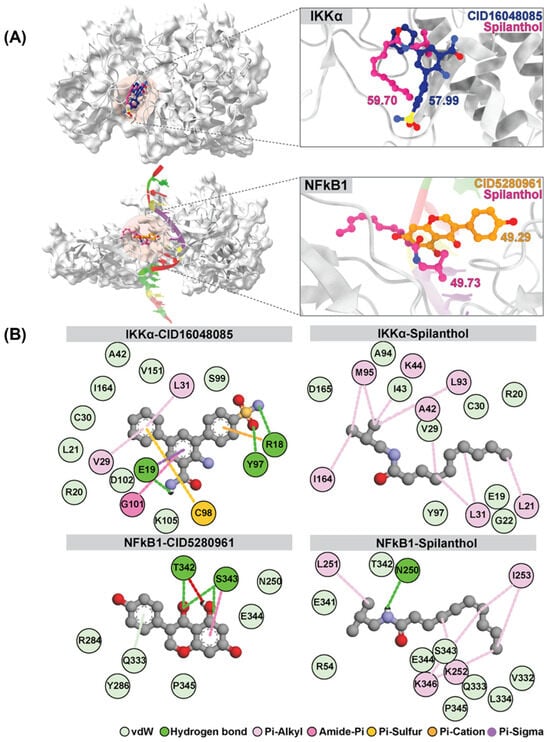
Figure 4.
Superimposition and GOLD docking score comparison of reference ligands CID16048085 and CID5280961 for IKKα and NFkB1, respectively, with spilanthol from molecular docking results. (A) Depicts the superimposition for IKKα and NFkB1. (B) The two-dimensional interaction map illustrates the average structure derived from the last 20 ns of MD simulations involving IKKα, NFkB1, and all ligands.
To delve deeper into the matter, MD simulation of the protein–ligand association complexes was conducted for 100 ns. MD simulations of the complexes formed between spilanthol-IKKα and NF-kB1 proteins were all located within the active sites. We calculated the root-mean-square displacement (RMSD) throughout the simulation by tracking the geometric coordinates of all atoms in the complex (Supplementary Figure S3). In the IKKα-CID16048085 and IKKα-spilanthol, the RMSD values exhibited an initial increase to approximately 4.00–4.50 Å, persisting until the end of the simulation. In the same way as IKKα-spilanthol, the RMSD values rose to 5.00–5.50 Å throughout the entire 100 ns simulation. In the case of NF-kB1-CID5280961 and NF-kB1-spilanthol complex, the RMSD values initially increased, followed by a subsequent rise to 6.00–6.50 Å throughout the entire 100 ns simulation. Given that the active site of NF-kB1 resides within the loop region, the RMSD values of genistein and spilanthol displayed some fluctuations; however, during the final 20 ns, the RMSD values remained stable until the completion of the simulation.
The binding affinity values of the protein–ligand complexes were determined using the SIE method, with data extracted from the analysis of 1000 snapshots obtained during the final 20 ns of the simulations (Table 2). The average binding free energy values (ΔGbind) for different complexes involving IKKα and various ligands are as follows: IKKα-CID5280961 and IKKα-spilanthol exhibit values of −3.81 ± 1.74 and −6.65 ± 0.37 kcal/mol, respectively. Similarly, for NF-kB1 complexes, the values for NF-kB1-CID16048085 and NF-kB1-spilanthol are −3.05 ± 0.62 and −5.06 ± 1.79 kcal/mol, respectively. The SIE method exposes a remarkable and robust binding affinity between spilanthol and both IKKα and NF-kB1. This affinity is notably stronger than that observed with their inhibitors. The map of protein–ligand interactions shows the frequency of interactions over the last 20 ns, as determined by the average structure. (Figure 4B) Molecular docking and dynamics collectively demonstrate the inherent capability of spilanthol to bind with NF-kB1 and IKKα, thereby reinforcing the findings of the network pharmacology analysis.

Table 2.
Binding free energies (kcal/mol), including SD, of IKKα and NF-kB1, complexed with reference ligand and spilanthol and evaluated using the SIE method (n = 100, avg, average; and SD, standard deviation).
2.4. Assessment of Cytotoxicity of Spilanthol in BV-2 Microglial Cells
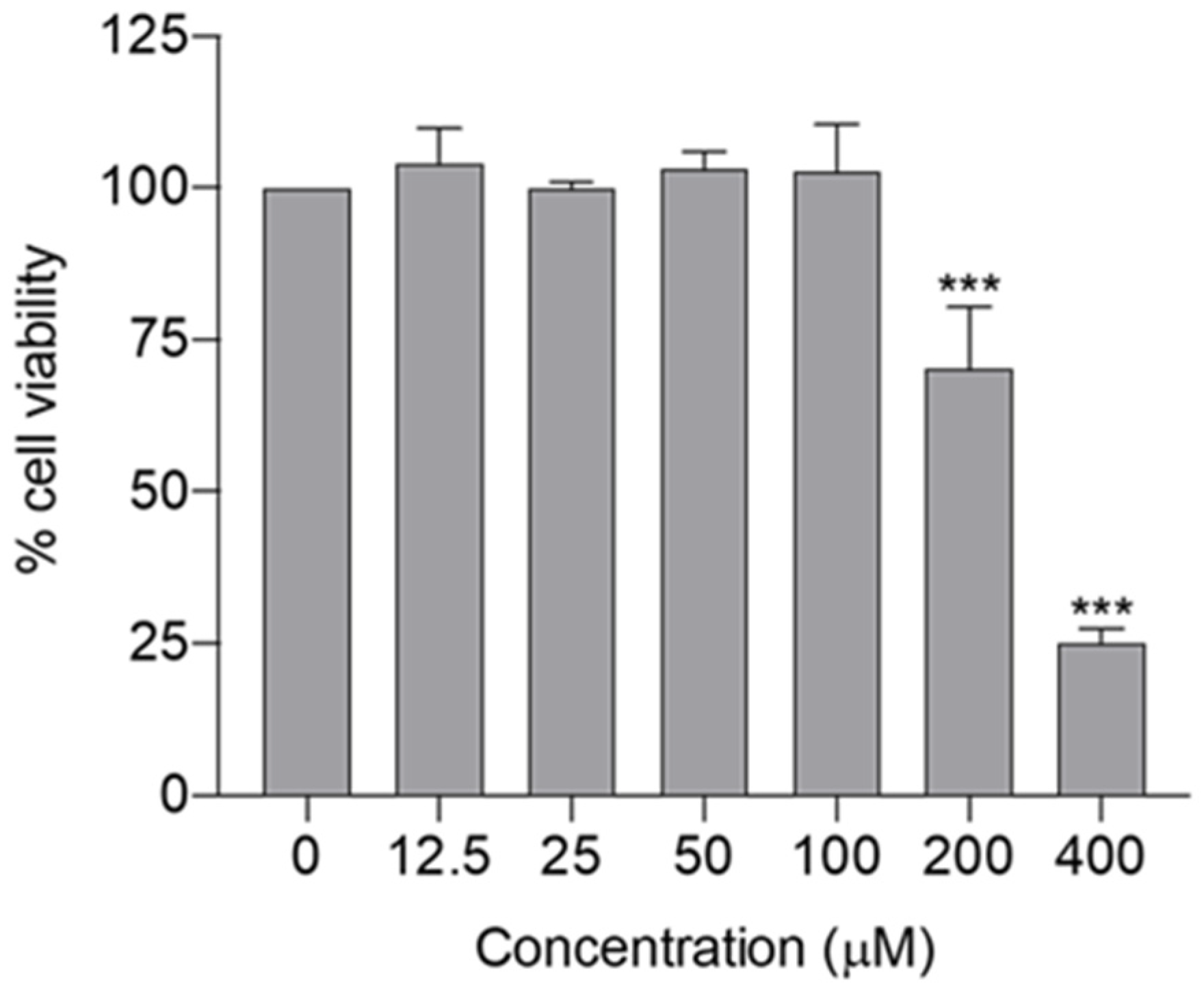
In silico findings suggest that spilanthol attenuates neurodegeneration mainly by attenuating neuroinflammation via suppression of the TLR4 signaling pathway. Subsequently, we conducted in vitro studies to validate the in silico findings. The BV-2 cell line was employed for in vitro evaluation, as microglia predominantly contribute to the chronic inflammation linked with neurodegenerative diseases [5,6]. Initially, the maximum non-cytotoxic concentration of spilanthol was determined, as non-cytotoxic concentrations had to be employed to evaluate the anti-neuroinflammatory efficacy. The cytotoxicity profile of spilanthol on BV-2 cells was assessed using the MTT assay, revealing that concentrations of spilanthol up to 100 µM did not exhibit a significant impact on the viability of BV-2 cells (Figure 5). Hence, 100 µM was determined as the maximum non-cytotoxic concentration. Spilanthol concentrations ranging from 25 to 100 µM were used in subsequent experiments to evaluate its anti-inflammatory properties while eliminating the confounding factors of cytotoxicity.
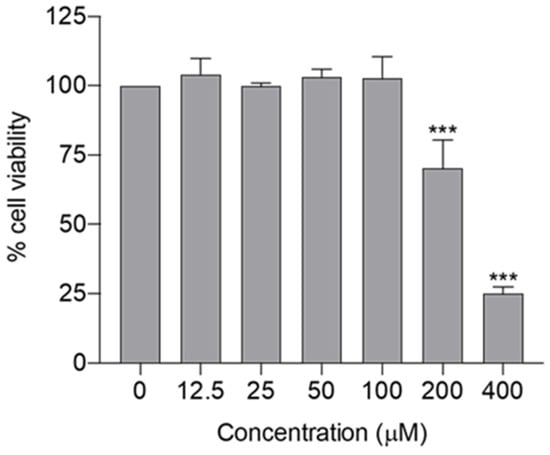
Figure 5.
Cytotoxicity profile of spilanthol on BV-2 cells; cell viability was determined by MTT assay. Data show the mean ± SD, n = 3 independent experiments. *** p < 0.001 compared with the negative control group; ANOVA followed by Dunnett’s post hoc test.
2.5. Evaluation of the Anti-Neuroinflammatory Efficacy of Spilanthol
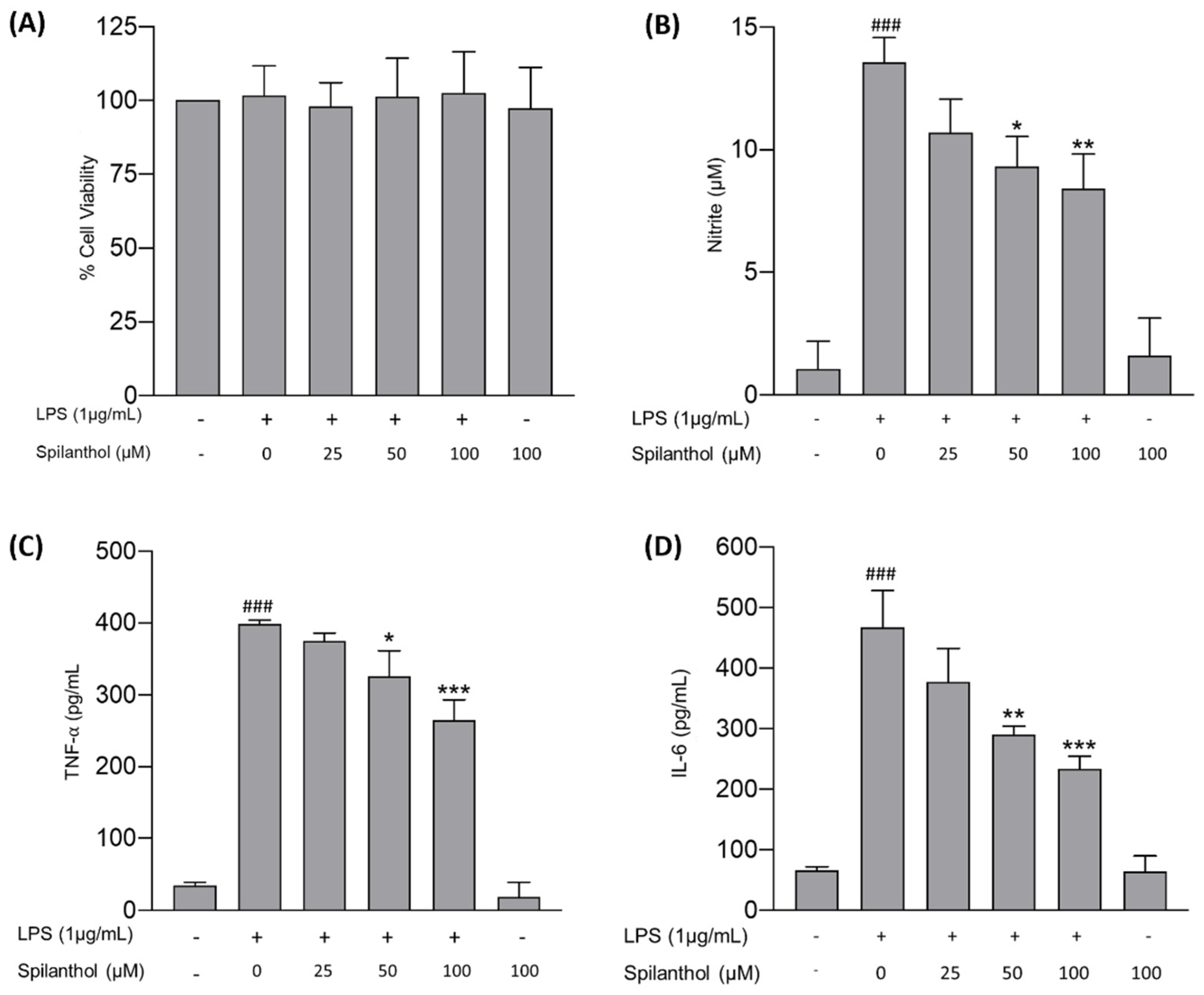
Chronic neuroinflammation is a pathogenic hallmark in NDDs and affects neuronal plasticity, impairs memory, induces secondary neurotoxicity, and sustains neurodegeneration [45]. Microglia in the CNS are predominantly responsible for sustained neuroinflammation. LPS is a well-known endotoxin produced by gram-negative bacteria that elicits a strong inflammatory response in microglia via the TLR4/NF-κB signaling pathway [46]. Therefore, the potential of spilanthol to attenuate neuroinflammation was assessed in LPS-induced BV-2 cells by measuring NO, TNF-α, and IL-6 concentration in the culture supernatant. Briefly, the cells were pretreated with spilanthol (25, 50, and 100 μM) for 2 h, followed by induction with LPS (1 μg/mL) for 22 h. The NO expression in the cell culture supernatant was then measured using the Griess reaction. The results demonstrated that the LPS-induced cells accounted for significantly high NO levels (13.58 ± 0.99 μM) in comparison with the untreated cells (1.07 ± 1.13 μM). Pretreatment with spilanthol produced a concentration-dependent decline in NO levels. In comparison with the LPS-only group, treatment with spilanthol at 100 and 50 μM significantly decreased the concentrations of nitrite in the cell culture supernatants by 38.59% (8.43 ± 1.39 μM) and 31.3% (9.33 ± 1.22 μM), respectively. however, treatment with spilanthol alone had no significant effects on NO production in BV-2 cells (Figure 6B). In addition, treatment with spilanthol exhibited no noticeable effect on cell viability (Figure 6A), indicating that the reduction of NO expression is not due to the decrease in cell viability or proliferation.
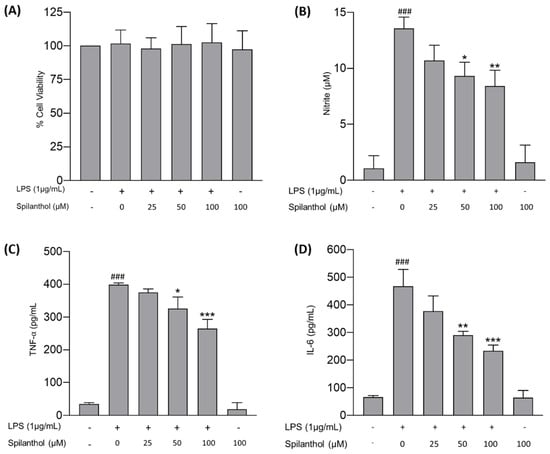
Figure 6.
Anti-neuroinflammatory efficacy of spilanthol on LPS-treated BV-2 cells. (A) Impact of the treatment on cell viability. (B) Impact of spilanthol on NO release. Influence of spilanthol on proinflammatory cytokine expression: (C) TNF-α and (D) IL-6 levels. Data show the mean ± SD values of three independent experiments. ### p < 0.001 compared with the control group, * p < 0.05, ** p < 0.01, and *** p < 0.001 compared with the LPS-control group, ANOVA followed by Tukey’s post hoc test.
ELISA was utilized to assess the potential of spilanthol to attenuate the release of proinflammatory cytokines, specifically TNF-α and IL-6, in LPS-induced BV-2 cells. The treatment of BV-2 cells with LPS has demonstrated significantly high levels of both TNF-α and IL-6, attaining concentrations of 399.5 ± 5.07 and 467.5 ± 60.58 pg/mL, respectively, compared with the control (34.36 ± 4.48 and 66.87 ± 6.11 pg/mL, respectively) (Figure 6C,D). However, treatment with spilanthol induced a concentration-dependent reduction in the LPS-induced production of TNF-α and IL-6, and the inhibition was statistically significant for the treatments using 50 μM and 100 μM. Treatment with spilanthol at 100 and 50 μM significantly reduces the release of TNF-α by 33.64% (265.1 ± 28.23 pg/mL) and 18.42% (325.9 ± 35.52 pg/mL), respectively, with respect to the LPS-induced group. Similarly, IL-6 expression was also significantly inhibited by the treatment of spilanthol. At 100 μM, IL-6 expression was reduced by 49.75% (234.9 ± 20.13 pg/mL), and at 50 μM, IL-6 expression was reduced by 37.82% (290 ± 14.00 pg/mL). Moreover, treatment of spilanthol alone did not elicit any significant alterations in the levels of proinflammatory cytokines.
Spinozzi and co-workers evaluated the anti-inflammatory effect of spilanthol in BV-2 cells and revealed that treatment of spilanthol at a concentration of 10 μM significantly inhibits the expression of IL-1β. However, expression of TNF-α, iNOS, and COX-2 is significantly inhibited at 10 μM concentration [47]. Importantly, the findings of the present study are in line with the previous study conducted by Wu and co-workers in 2008. They discovered the potential of spilanthol to suppress the expression of proinflammatory cytokines in RAW 264.7 murine macrophages at a concentration between 20 and 180 μM without inducing significant cytotoxicity by downregulating the NF-kB pathway [48]. These findings further validate the potential therapeutic application of spilanthol in NDDs by attenuating neuroinflammation.
3. Conclusions
In conclusion, the present study comprehensively explores the potential of spilanthol isolated from A. paniculata in attenuating NDDs through a combination of in silico and in vitro studies. Our research indicates that spilanthol demonstrates favorable pharmacokinetic properties and drug-like characteristics based on in silico ADMET prediction. Network pharmacology analysis reveals a complex interaction between spilanthol and key target proteins in the TLR4 signaling pathway involved in NDDs. Additionally, molecular docking and dynamics simulations demonstrate robust binding affinities between spilanthol and key proteins in the TLR4 pathway. Experimental validation using LPS-induced BV-2 microglial cells demonstrates the ability of spilanthol to reduce the expression of proinflammatory cytokines and mediators including NO, IL-6, and TNF-α without causing significant cytotoxicity. However, comprehensive investigations are necessary to assess the efficacy of spilanthol in in vivo models. Additionally, extensive toxicological studies are imperative to pave the way for its clinical application.
4. Materials and Methods
4.1. Materials and Chemicals
Silica gel (230–400 mesh ASTM), Silica gel 60 F254 Aluminum TLC plates, and chloroform-D1 were purchased from Merck KGaA. (Darmstadt, Germany). Sephadex LH-20, Lipolysaccharide (LPS), Cell culture regents including Fetal Bovine Serum (FBS), Dulbecco’s modified Eagle’s medium (DMEM), and Trypsin-EDTA were obtained from Invitrogen (Grand Island, NY, USA). ELISA kits for the qualification of proinflammatory cytokines were purchased from BioLegend (San Diego, CA, USA). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
4.2. In Silico ADMET Screening
The pkCSM web tool (https://biosig.lab.uq.edu.au/pkcsm/, accessed on 1 January 2024) and the SwissADME web tool (http://www.swissadme.ch/, accessed on 1 January 2024) were used to assess the pharmacokinetic descriptors of spilanthol and its drug-likeness, respectively. The Protox-II virtual toxicity lab (https://tox-new.charite.de/protox_II/, accessed 1 January 2024) was used to evaluate the toxicity of spilanthol.
4.3. Network Pharmacology Analysis
4.3.1. Acquisition of the Target Genes of Spilanthol
SMILES notation of spilanthol retrieved from the PubChem database was utilized for in silico structure-based-target fishing. Target genes of spilanthol were identified from three target fishing web servers, specifically SwissTargetPrediction (http://www.swisstargetprediction.ch/, accessed on 20 October 2023), SEA SearchServer (https://sea.bkslab.org/, accessed on 20 October 2023), and Super-PRED 3.0 (https://prediction.charite.de/subpages/target_prediction.php, accessed on 20 October 2023). Subsequently, gene symbols for potential targets were obtained and validated from the UniProt database (https://www.uniprot.org/, accessed on 20 October 2023). Subsequently, gene symbols retrieved from the three sources were combined, and duplicate entries were excluded prior to analysis.
4.3.2. Acquisition of the Target Genes of Neurodegenerative Diseases
Genes related to NDDs were identified from GeneCards (https://www.genecards.org/, accessed on 19 October 2023) (relevance score ≥ 10), OMIM (https://www.omim.org/, accessed on 19 October 2023), and the DisGeNET database (https://www.disgenet.org/, accessed on 19 October 2023) (fit score ≥ 0.1) using “Neurodegenerative disease” as the keyword. Retrieved target genes were subsequently validated and standardized using the UniProt database. Finally, the genes obtained from all sources were compiled, and duplicates were removed before the analysis.
4.3.3. Construction of Protein–Protein Interaction (PPI) Network
Initially, shared targets between spilanthol and targets related to NDDs were identified and depicted using Venny 2.1.0, an online tool used to generate Venn diagrams (https://bioinfogp.cnb.csic.es/tools/venny/, accessed on 19 October 2023). By using common targets, an interactive network integrating therapeutic targets of spilanthol and NDD-related targets was established, and the network was visualized using Cytoscape (version 3.9.1). The top 10 genes exhibiting the most significant degree of interaction were identified utilizing Cytoscape’s cytoHubba plugin v.0.1.
4.3.4. GO Function Enrichment and KEGG Pathway Analysis
Plausible mechanisms by which spilanthol attenuates neurodegeneration were assessed using the KEGG and GO pathway enrichment analyses using an online bioinformatics data analysis tool (http://www.bioinformatics.com.cn/, accessed on 20 October 2023) to determine the possible molecular mechanism of spilanthol in NDDs.
4.4. Dynamic Interactions: Insights from Docking and Molecular Dynamics
The process began with acquiring the protein structures of the IKKα and NF-kB1 proteins from the PDB database using the PDB IDs 5EBZ [49] and 1SVC [50], respectively. Visualization and removal of ligands and small molecules were facilitated using ChimeraX [51]. All ligands were constructed using the GaussView program, and both the ligands and receptors were saved in mol2 format. Subsequently, the ligand was docked with the receptor utilizing the GOLD Software (2022.1) [52]. It is important to note that docking was performed within a defined grid box of 8 × 8 × 8 dimensions, centered on specific XYZ coordinates for each model. For example, for the IKKα model (PDB ID: 5EBZ), the coordinates used were 88.11, 65.64, and 0.37. For the NF-kB1 model (PDB ID: 1SVC), the coordinates were 34.30, 9.39, and 40.52. From the ensemble of docked conformations, those achieving the highest GOLD fitness score were meticulously selected for further analysis. Following the docking phase, protonation states of the protein–ligand complexes were determined at a physiological pH of 7.4 using the PDB2PQR server [53]. Subsequently, all-atom Molecular Dynamics (MD) simulations were conducted on IKKα and NF-kB1 proteins, along with their respective inhibitors, CID16048085 and CID5280961. These simulations were carried out within a periodic boundary using AMBER22 [54]. For force field parameters, the ff19SB force field [55] was applied to the target proteins, encompassing both bonded and nonbonded interactions. Parameters for the inhibitors (CID16048085 and CID5280961) and spilanthol were generated using the leap module in conjunction with the general AMBER force field 2 (GAFF2) [56]. Furthermore, RESP charges for spilanthol were computed following established standard procedures [57]. To finalize the structural setup of each system, any missing hydrogen atoms were added using the LeaP module. Additionally, Na+ ions were introduced to neutralize charges. Subsequently, the newly introduced hydrogen atoms underwent an energy minimization process employing steepest descents (SD) and conjugate gradient (CG) algorithms. Finally, the entire protein–ligand complex underwent a complete energy minimization procedure to achieve an optimal state. The complex was then gradually heated from 10 to 310 K under 1 atm of pressure before initiating a 100 ns MD simulation. The MD trajectories were derived from the final 20 ns, and RMSD analysis was performed by CCPTRAJ module for assessment of the stability of the complex simulations [58]. Additionally, the average structure was computed based on data from the final 20 ns of simulation using the Chimera version 1.15 program [59]. Following this, 2D interaction diagrams were generated using the Discovery Studio program.
4.5. Extraction and Isolation
Acmella paniculata was collected and authenticated by Associate Professor Thatree Phadungcharoen. Then, fresh floral inflorescence was pulverized, and 828 g was used for Soxhlet extraction using 95% EtOH at 40–80 °C for 26 h. The extract was then filtered and concentrated by evaporating solvent using a rotatory evaporator at 40 °C to obtain the A. paniculata crude extract (75 g). For the isolation of spilanthol, the crude extract was initially partitioned between dichloromethane and water. The dichloromethane fraction was further fractionated using a series of Silica and Sephadex columns. Fractions from each column were collected and combined according to the TLC patterns. NMR analysis of the isolated compound was carried out using a Bruker Ascend 400 NMR spectrometer (Billerica, MA, USA) to obtain 1H-NMR, 13C-NMR, HSQC, and HMBC correlations. The compound’s identification was conducted by comparing its NMR spectrum with existing literature data [15,60].
4.6. In Vitro Evaluation
4.6.1. BV-2 Cell Culture
BV-2 cells, an immortalized murine microglia cell line, were obtained from AcceGen Biotechnology located in Fairfield, NJ, USA. The microglial cells were cultured and maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin within a controlled environment of 37 °C and 5% CO2 atmosphere in a humidified incubator.
4.6.2. Evaluation of the Cytotoxicity of Spilanthol
For determination of the maximum non-cytotoxic concentration, BV-2 cells were seeded at a density of 5 × 104 cells/well in a clear flat-bottomed multiwell cell culture plate with DMEM complete media (DMEM supplemented with 10% FBS containing 1% penicillin/streptomycin). Followed by 24 h of incubation, the culture supernatant was replaced with 200 µL of serum-free DMEM containing varying concentrations of spilanthol (0–400 µM). At 24 h after the treatments, the supernatants were carefully removed, and 100 µL of MTT solution (0.5 mg/mL in PBS) was added. Then, the multiwell cell culture plate was left to incubate in darkness for 3 h at 37 °C with 5% CO2 in a humidified incubator. After 3 h of incubation, the supernatant was discarded and 200 µL of DMSO was added, then the cell culture plate was shaken for 10 min to facilitate the dissolution of formazan crystals. Finally, the absorbance was measured at 570 nm using a multimode plate reader.
4.6.3. Anti-Neuroinflammatory Efficacy of Spilanthol
Twenty-four hours before the treatment, BV-2 cells were seeded at a density of 15 × 104 cells/well in 24-well plates with DMEM supplemented with 10% FBS. After an initial 24-h incubation, the cells were treated with spilanthol at concentrations of 0, 25, 50, and 100 µM for 2 h. Then followed by the treatment, the cells were treated with LPS at a final concentration of 1 µg/mL and incubated for 22 h. Finally, cell culture supernatant was collected to quantity inflammatory mediators and proinflammatory cytokines.
4.6.4. Nitrite Assay
The Griess reaction was utilized to evaluate nitrite concentration in the culture supernatant. Briefly, 100 µL of supernatant was added to a well of a 96-well plate, followed by 50 µL of 1% sulfanilamide in 5% phosphoric acid and 50 µL of 2.5% N-1-Napthylenediamine dihydrochloride. After 5 min of incubation in darkness, absorbance was measured at 520 nm using a microplate reader (CLARIOstar®, BMG Labtech, Ortenberg, Germany). Finally, the nitrite concentration was determined using a sodium nitrite standard curve.
4.6.5. Determination of IL-6 and TNF-α Levels Using ELISA
The impact of the spilanthol on the release of inflammatory cytokine in the LPS-induced murine microglia was assessed using ELISA. Followed by 24 h of respective treatment, the cytokine levels of the cell culture supernatant were assessed using commercial ELISA kits, as per the protocol given by the manufacturer. The absorbance was measured at 450 nm using a multimode plate reader (CLARIOstar®, BMG Labtech, Ortenberg, Germany). The concentrations of the cytokines were calculated from the respective calibration curves.
4.7. Statistical Analysis
Results of the cell culture experiments are demonstrated as means ± SD. The data underwent analysis utilizing one-way analysis of variance (ANOVA), followed by the Bonferroni post hoc test for multiple comparisons. Statistical significance was attained at its lowest when the p-value < 0.05.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app14093755/s1, Table S1: In silico ADMET prediction of spilanthol using pkCSM; Table S2: Drug-likeness prediction of spilanthol; Table S3: In silico toxicity prediction of spilanthol using protox-II. Figure S1. Bioavailability radar of the spilanthol; Figure S2. Interaction of spilanthol with multiple targets in the Alzheimer’s disease pathway. Figure S3. RMSD plots of all-atom residues for assessing structural stability and conformational changes in the molecular systems: IKKα and NFkB1; Figure S4. 1H-NMR (400 MHz, CDCl3) spectrum of spilanthol; Figure S5. 13C-NMR (125 MHz, CDCl3) spectrum of spilanthol. Video S1: MD simulation of NFKB-spilanthol complex.; Video S2: MD simulation of NFKB-CID5280961 complex; Video S3: MD simulation of IKKα-spilanthol complex, Video S4: MD simulation of IKKα- CID16048085 complex; https://drive.google.com/drive/folders/10dfFUUYuSWmOsIZhgiUW-XJ6BcCz74rF?usp=drive_link (accessed on 19 October 2023).
Author Contributions
Conceptualization, S.S.; Methodology, T.R.; Validation, P.W.D.W., W.T. and G.S.; Formal Analysis, S.S.J., S.Y.N. and N.D.; Investigation, S.S.J. and N.D.; Resources, P.T. and S.S.; Data Curation, W.T. and G.S.; Writing—Original Draft Preparation, S.S.J. and N.D.; Writing—Review and Editing, P.W.D.W., S.Y.N. and T.R.; Visualization, S.S.J., N.D. and G.S.; Supervision, S.S., P.T. and T.R.; Project Administration, S.S. and P.T.; Funding Acquisition, S.S. and P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary Materials.
Acknowledgments
This research project is supported by the Second Century Fund (C2F), Chulalongkorn University. The authors would also like to express their gratitude to the Faculty of Pharmaceutical Sciences, Chulalongkorn University, for providing facilities.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| TLR4 | Toll-like receptor 4 |
| NF-κB | Nuclear factor kappa B |
| IKKα | Inhibitory kappa B kinase α |
| IRAK4 | Interleukin-1 receptor-associated kinase 4 |
| MD2 | Myeloid differentiation factor 2 |
| MyD88 | Myeloid differentiation primary response protein 88 |
| IL-6 | Interleukin-6 |
| IL-1 | Interleukin-1 |
| TNF-α | Tumor necrosis factor-alpha |
| NO | Nitric oxide |
| iNOS | Inducible nitric oxide synthase |
| COX-2 | Cyclooxygenase-2 |
| LPS | Lipopolysaccharide |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| DMSO | Dimethyl sulfoxide |
| ELISA | Enzyme-linked immunosorbent assay |
| PPI | Protein–protein interactions |
| MCC | Maximal clique centrality |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate buffered saline |
| RMSD | Root-mean-square displacement |
| PDB | Protein data bank |
References
- Liu, S.; Zheng, G.; Chen, H.; Li, G.; Guo, X. Advances in integrative medicine for neurodegenerative diseases: From basic research to clinical practice. Front. Neurol. 2023, 14, 1197641. [Google Scholar] [CrossRef]
- Cai, C.; Fang, J.; Ke, H. Traditional medicine and phytochemicals for neurodegenerative diseases treatment: Application of interdisciplinary technologies in novel therapeutic target and drug discovery. Front. Neurosci. 2023, 17, 1268710. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.P.; Brown, R.H., Jr.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Yacoubian, T. Neurodegenerative disorders: Why do we need new therapies? In Drug Discovery Approaches for the Treatment of Neurodegenerative Disorders; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–16. [Google Scholar]
- Rubino, S.J.; Mayo, L.; Wimmer, I.; Siedler, V.; Brunner, F.; Hametner, S.; Madi, A.; Lanser, A.; Moreira, T.; Donnelly, D. Acute microglia ablation induces neurodegeneration in the somatosensory system. Nat. Commun. 2018, 9, 4578. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, D.; Mao, Q.; Xia, H. Role of neuroinflammation in neurodegeneration development. Signal Transduct. Target. Ther. 2023, 8, 267. [Google Scholar] [CrossRef]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zhou, J. Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl. Neurodegener. 2015, 4, 19. [Google Scholar] [CrossRef]
- Du, L.; Zhang, Y.; Chen, Y.; Zhu, J.; Yang, Y.; Zhang, H.-L. Role of microglia in neurological disorders and their potentials as a therapeutic target. Mol. Neurobiol. 2017, 54, 7567–7584. [Google Scholar] [CrossRef]
- Gao, Y.; Tu, D.; Yang, R.; Chu, C.-H.; Hong, J.-S.; Gao, H.-M. Through reducing ROS production, IL-10 suppresses caspase-1-dependent IL-1β maturation, thereby preventing chronic neuroinflammation and neurodegeneration. Int. J. Mol. Sci. 2020, 21, 465. [Google Scholar] [CrossRef]
- Abd Ghafar, S.A.; Salehuddin, N.S.; Abdul Rahman, N.Z.; Halib, N.; Mohamad Hanafiah, R. Transcriptomic profile analysis of Streptococcus mutans response to Acmella paniculata flower extracts. Evid. -Based Complement. Altern. Med. 2022, 2022, 7767940. [Google Scholar] [CrossRef]
- Meitei, L.R.; De, A.; Mao, A.A. An ethnobotanical study on the wild edible plants used by forest dwellers in Yangoupokpi Lokchao Wildlife Sanctuary, Manipur, India. Ethnobot. Res. Appl. 2022, 23, 5–18. [Google Scholar] [CrossRef]
- Panyadee, P.; Inta, A. Taxonomy and ethnobotany of Acmella (Asteraceae) in Thailand. Biodiversitas J. Biol. Divers. 2022, 23. [Google Scholar] [CrossRef]
- Patel, S.; Purohit, N.; Sapra, P.; Solanki, H.; Bishoyi, A.K. Genetic diversity analysis revealed the hot spot of Acmella paniculata (Wall ex DC.) RK Jansen existing in natural populations of Gujarat. Genet. Resour. Crop Evol. 2022, 69, 2249–2260. [Google Scholar] [CrossRef]
- Barbosa, A.F.; Carvalho, M.G.d.; Smith, R.E.; Sabaa-Srur, A.U. Spilanthol: Occurrence, extraction, chemistry and biological activities. Rev. Bras. Farmacogn. 2016, 26, 128–133. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Alegría-Arcos, M.; Barbosa, T.; Sepúlveda, F.; Combariza, G.; González, J.; Gil, C.; Martínez, A.; Ramírez, D. Network pharmacology reveals multitarget mechanism of action of drugs to be repurposed for COVID-19. Front. Pharmacol. 2022, 13, 952192. [Google Scholar] [CrossRef]
- Sadybekov, A.V.; Katritch, V. Computational approaches streamlining drug discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Agu, P.; Afiukwa, C.; Orji, O.; Ezeh, E.; Ofoke, I.; Ogbu, C.; Ugwuja, E.; Aja, P. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Sakhawat, A.; Khan, M.U.; Rehman, R.; Khan, S.; Shan, M.A.; Batool, A.; Javed, M.A.; Ali, Q. Natural compound targeting BDNF V66M variant: Insights from in silico docking and molecular analysis. AMB Express 2023, 13, 134. [Google Scholar] [CrossRef]
- Jia, C.-Y.; Li, J.-Y.; Hao, G.-F.; Yang, G.-F. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today 2020, 25, 248–258. [Google Scholar] [CrossRef]
- Abdullahi, M.; Uzairu, A.; Shallangwa, G.A.; Mamza, P.A.; Ibrahim, M.T.; Ahmad, I.; Patel, H. Structure-based drug design, molecular dynamics simulation, ADMET, and quantum chemical studies of some thiazolinones targeting influenza neuraminidase. J. Biomol. Struct. Dyn. 2023, 41, 13829–13843. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.; Berger, M.; Santana de Cecco, B.; Mallmann, L.P.; Terraciano, P.B.; Driemeier, D.; Rodrigues, E.; Beys-da-Silva, W.O.; Konrath, E.L. Chymase inhibition: A key factor in the anti-inflammatory activity of ethanolic extracts and spilanthol isolated from Acmella oleracea. J. Ethnopharmacol. 2021, 270, 113610. [Google Scholar] [CrossRef] [PubMed]
- Nomura, E.C.O.; Rodrigues, M.R.A.; da Silva, C.F.; Hamm, L.A.; Nascimento, A.M.; de Souza, L.M.; Cipriani, T.R.; Baggio, C.H.; Werner, M.F.d.P. Antinociceptive effects of ethanolic extract from the flowers of Acmella oleracea (L.) R.K. Jansen in mice. J. Ethnopharmacol. 2013, 150, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, E.T.; Peretti, P.; Bezerra, R.M.; Biancardi, M.F.; Sousa, F.F.; Mendes, E.P.; Dutra, J.B.; Silveira, C.C.; Castro, C.H.; Cruz, J.N. Pharmacological Characteristics of the Hydroethanolic Extract of Acmella oleracea (L) RK Jansen Flowers: ADME/Tox In Silico and In Vivo Antihypertensive and Chronic Toxicity Evaluation. Evid. -Based Complement. Altern. Med. 2023, 2023, 1278720. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.; Silva, I.; Viana, M.D.; Melo, N.; Sánchez-Ortiz, B.; Oliveira, M.; Barbosa, W.; Ferreira, I.M.; Carvalho, J. Acute Toxicity of the Hydroethanolic Extract of the Flowers of Acmella oleracea L. in Zebrafish (Danio rerio): Behavioral and Histopathological Studies. Pharmaceuticals 2019, 12, 173. [Google Scholar]
- De Souza, G.; Viana, M.; Goés, L.; Sanchez-Ortiz, B.; Silva, G.d.; Pinheiro, W.d.S.; Santos, C.R.D.; Carvalho, J.T. Reproductive toxicity of the hydroethanolic extract of the flowers of Acmella oleracea and spilanthol in zebrafish: In vivo and in silico evaluation. Hum. Exp. Toxicol. 2020, 39, 127–146. [Google Scholar] [CrossRef] [PubMed]
- de Souza, G.C.; Matias Pereira, A.C.; Viana, M.D.; Ferreira, A.M.; da Silva, I.D.R.; de Oliveira, M.M.R.; Barbosa, W.L.R.; Silva, L.B.; Ferreira, I.M.; Dos Santos, C.B.R. Acmella oleracea (L) RK Jansen reproductive toxicity in zebrafish: An in vivo and in silico assessment. Evid.-Based Complement. Altern. Med. 2019, 2019, 1237301. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhao, Y.; Yang, T.; Gao, X.; Meng, C. Duhuo Jisheng decoction alleviates neuroinflammation and neuropathic pain by suppressing microglial M 1 polarization: A network pharmacology research. J. Orthop. Surg. Res. 2023, 18, 629. [Google Scholar] [CrossRef]
- Guo, S.; Wang, H.; Yin, Y. Microglia polarization from M1 to M2 in neurodegenerative diseases. Front. Aging Neurosci. 2022, 14, 815347. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Xu, Y.-R.; Lei, C.-Q. TAK1-TABs complex: A central signalosome in inflammatory responses. Front. Immunol. 2021, 11, 608976. [Google Scholar] [CrossRef]
- Okun, E.; Griffioen, K.J.; Lathia, J.D.; Tang, S.-C.; Mattson, M.P.; Arumugam, T.V. Toll-like receptors in neurodegeneration. Brain Res. Rev. 2009, 59, 278–292. [Google Scholar] [CrossRef]
- Azam, S.; Jakaria, M.; Kim, I.-S.; Kim, J.; Haque, M.E.; Choi, D.-K. Regulation of toll-like receptor (TLR) signaling pathway by polyphenols in the treatment of age-linked neurodegenerative diseases: Focus on TLR4 signaling. Front. Immunol. 2019, 10, 1000. [Google Scholar] [CrossRef]
- Su, Y.; Wang, D.; Liu, N.; Yang, J.; Sun, R.; Zhang, Z. Clostridium butyricum improves cognitive dysfunction in ICV-STZ-induced Alzheimer’s disease mice via suppressing TLR4 signaling pathway through the gut-brain axis. PLoS ONE 2023, 18, e0286086. [Google Scholar] [CrossRef]
- Cui, W.; Sun, C.; Ma, Y.; Wang, S.; Wang, X.; Zhang, Y. Inhibition of TLR4 induces M2 microglial polarization and provides neuroprotection via the NLRP3 inflammasome in Alzheimer’s disease. Front. Neurosci. 2020, 14, 444. [Google Scholar] [CrossRef]
- Kouli, A.; Horne, C.; Williams-Gray, C. Toll-like receptors and their therapeutic potential in Parkinson’s disease and α-synucleinopathies. BrainBehav. Immun. 2019, 81, 41–51. [Google Scholar] [CrossRef]
- Heidari, A.; Yazdanpanah, N.; Rezaei, N. The role of Toll-like receptors and neuroinflammation in Parkinson’s disease. J. Neuroinflammation 2022, 19, 135. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, W.-M.; Tang, P.-C.; Zhang, X.; Zhang, X.-Y.; Yu, B.-C.; Fan, Y.-Y.; Ge, X.-Q.; Zhang, X.-L. Neuroprotective effects of natural cordycepin on LPS-induced Parkinson’s disease through suppressing TLR4/NF-κB/NLRP3-mediated pyroptosis. J. Funct. Foods 2020, 75, 104274. [Google Scholar] [CrossRef]
- Sharma, V.K.; Singh, T.G.; Singh, S.; Garg, N.; Dhiman, S. Apoptotic Pathways and Alzheimer’s Disease: Probing Therapeutic Potential. Neurochem. Res. 2021, 46, 3103–3122. [Google Scholar] [CrossRef]
- Mohamed Asik, R.; Suganthy, N.; Aarifa, M.A.; Kumar, A.; Szigeti, K.; Mathe, D.; Gulyás, B.; Archunan, G.; Padmanabhan, P. Alzheimer’s disease: A molecular view of β-amyloid induced morbific events. Biomedicines 2021, 9, 1126. [Google Scholar] [CrossRef]
- Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 2018, 19, 687–700. [Google Scholar] [CrossRef]
- Abubakar, M.B.; Sanusi, K.O.; Ugusman, A.; Mohamed, W.; Kamal, H.; Ibrahim, N.H.; Khoo, C.S.; Kumar, J. Alzheimer’s disease: An update and insights into pathophysiology. Front. Aging Neurosci. 2022, 14, 742408. [Google Scholar] [CrossRef] [PubMed]
- Muzio, L.; Viotti, A.; Martino, G. Microglia in neuroinflammation and neurodegeneration: From understanding to therapy. Front. Neurosci. 2021, 15, 742065. [Google Scholar] [CrossRef]
- Rahimifard, M.; Maqbool, F.; Moeini-Nodeh, S.; Niaz, K.; Abdollahi, M.; Braidy, N.; Nabavi, S.M.; Nabavi, S.F. Targeting the TLR4 signaling pathway by polyphenols: A novel therapeutic strategy for neuroinflammation. Ageing Res. Rev. 2017, 36, 11–19. [Google Scholar] [CrossRef]
- Spinozzi, E.; Pavela, R.; Bonacucina, G.; Perinelli, D.R.; Cespi, M.; Petrelli, R.; Cappellacci, L.; Fiorini, D.; Scortichini, S.; Garzoli, S. Spilanthol-rich essential oil obtained by microwave-assisted extraction from Acmella oleracea (L.) RK Jansen and its nanoemulsion: Insecticidal, cytotoxic and anti-inflammatory activities. Ind. Crops Prod. 2021, 172, 114027. [Google Scholar] [CrossRef]
- Wu, L.-C.; Fan, N.-C.; Lin, M.-H.; Chu, I.-R.; Huang, S.-J.; Hu, C.-Y.; Han, S.-Y. Anti-inflammatory effect of spilanthol from Spilanthes acmella on murine macrophage by down-regulating LPS-induced inflammatory mediators. J. Agric. Food Chem. 2008, 56, 2341–2349. [Google Scholar] [CrossRef] [PubMed]
- Polley, S.; Passos, D.O.; Huang, D.-B.; Mulero, M.C.; Mazumder, A.; Biswas, T.; Verma, I.M.; Lyumkis, D.; Ghosh, G. Structural Basis for the Activation of IKK1/α. Cell Rep. 2016, 17, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-κB p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Cerutti, D.S.; Cisneros, G.A.; Cruzeiro, V.W.D.; Forouzesh, N.; Giese, T.J.; Götz, A.W.; Gohlke, H.; et al. AmberTools. J. Chem. Inf. Model. 2023, 63, 6183–6191. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q. ff19SB: Amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 2019, 16, 528–552. [Google Scholar] [CrossRef]
- He, X.; Man, V.H.; Yang, W.; Lee, T.-S.; Wang, J. A fast and high-quality charge model for the next generation general AMBER force field. J. Chem. Phys. 2020, 153, 114502. [Google Scholar] [CrossRef]
- Wang, J.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- de Freitas Blanco, V.S.; Michalak, B.; Zelioli, Í.A.M.; de Oliveira, A.d.S.S.; Rodrigues, M.V.N.; Ferreira, A.G.; Garcia, V.L.; Cabral, F.A.; Kiss, A.K.; Rodrigues, R.A.F. Isolation of spilanthol from Acmella oleracea based on Green Chemistry and evaluation of its in vitro anti-inflammatory activity. J. Supercrit. Fluids 2018, 140, 372–379. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).