Optimizing Breeding Strategies for Pekin Ducks Using Genomic Selection: Genetic Parameter Evaluation and Selection Progress Analysis in Reproductive Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Population and Phenotype Measurement
2.2. Genotyping and SNP Identification
2.3. BLUP, GBLUP, and ssGBLUP Models
2.4. Prediction Assessment
3. Results
3.1. Description of Phenotypic Data
3.2. Estimation of Heritability and Genetic Correlation
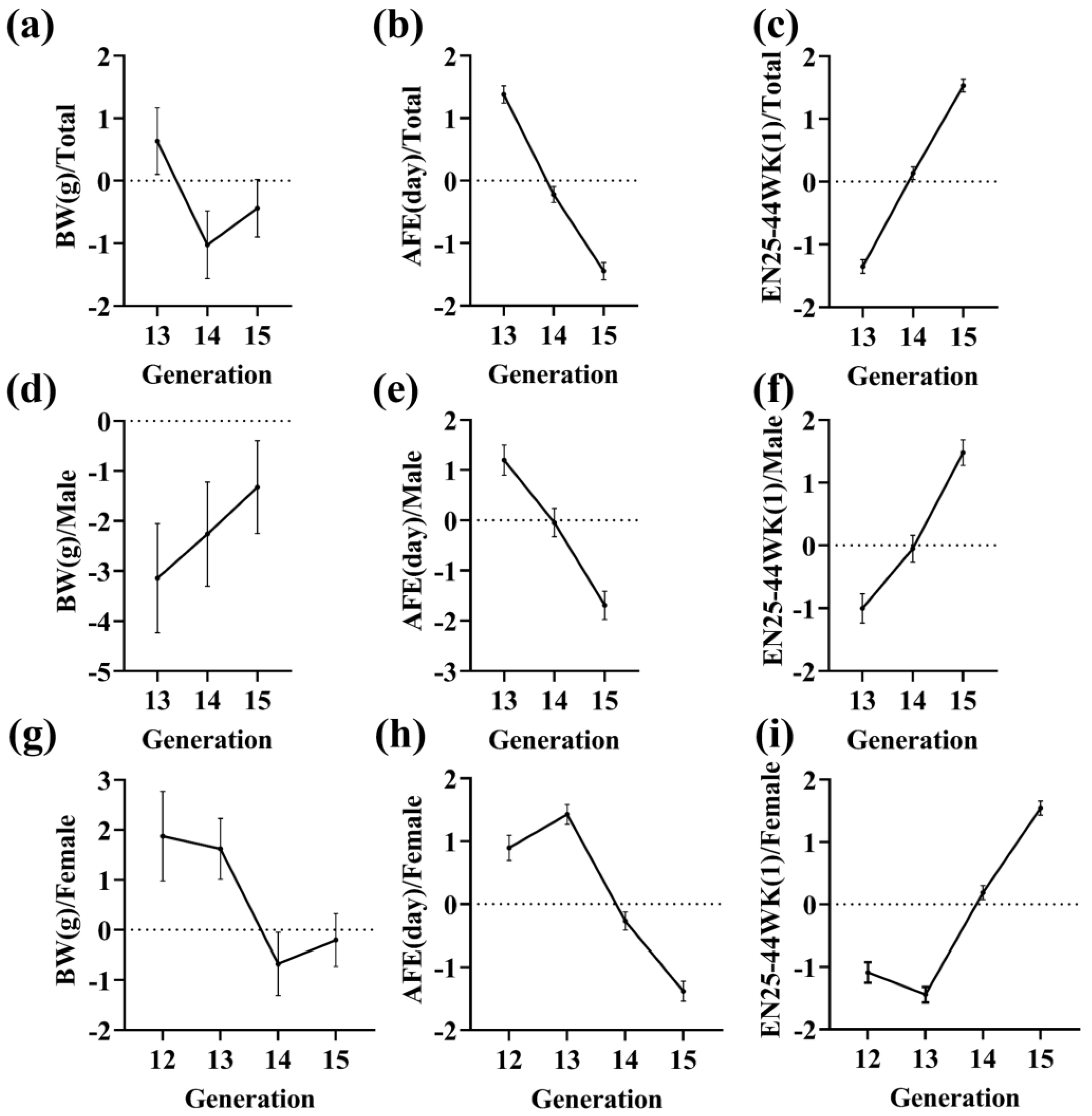
3.3. Selection Response
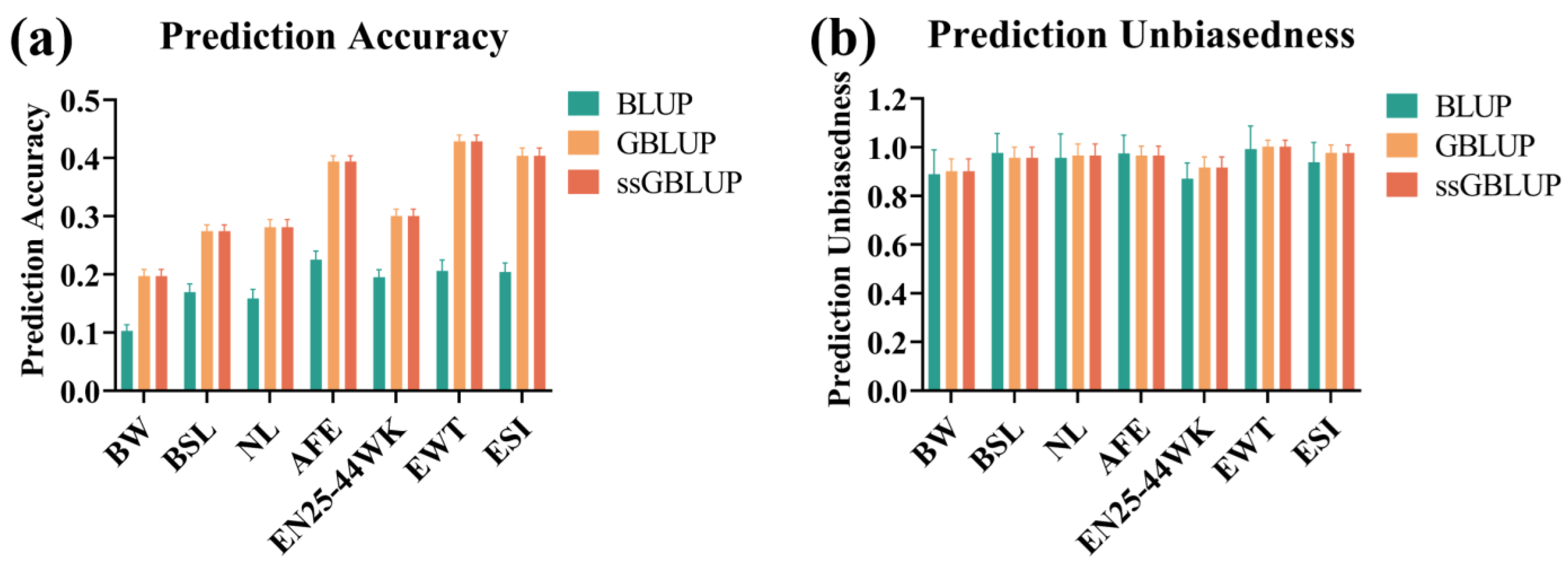
3.4. Genomic Prediction Using BLUP, GBLUP, and ssGBLUP Models
4. Discussion
4.1. Phenotypic and Genetic Parameters
4.2. Improvements in the Selection Process
4.3. Genomic Selection Predictive Performance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bello, S.F.; Adeola, A.C.; Nie, Q. The study of candidate genes in the improvement of egg production in ducks—A review. Poult. Sci. 2022, 101, 101850. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Zhou, X.; Yuan, X.; Zhao, S.; Li, X.; Liu, X. KAML: Improving genomic prediction accuracy of complex traits using machine learning determined parameters. Genome Biol. 2020, 21, 146. [Google Scholar] [CrossRef] [PubMed]
- Dreisigacker, S.; Pérez-Rodríguez, P.; Crespo-Herrera, L.; Bentley, A.R.; Crossa, J. Results from rapid-cycle recurrent genomic selection in spring bread wheat. G3 Genes Genom. Genet. 2023, 13, jkad025. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Sabadin, F.; DoVale, J.C.; Platten, J.D.; Fritsche-Neto, R. Optimizing self-pollinated crop breeding employing genomic selection: From schemes to updating training sets. Front. Plant Sci. 2022, 13, 935885. [Google Scholar] [CrossRef]
- Cai, W.; Hu, J.; Fan, W.; Xu, Y.; Tang, J.; Xie, M.; Zhang, Y.; Guo, Z.; Zhou, Z.; Hou, S. Genetic parameters and genomic prediction of growth and breast morphological traits in a crossbreed duck population. Evol. Appl. 2024, 17, e13638. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.A.; Hickey, J.M.; Daetwyler, H.D.; van der Werf, J.H. The importance of information on relatives for the prediction of genomic breeding values and the implications for the makeup of reference data sets in livestock breeding schemes. Genet. Sel. Evol. 2012, 44, 4. [Google Scholar] [CrossRef]
- Wolc, A.; Stricker, C.; Arango, J.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Preisinger, R.; Habier, D.; Fernando, R.; Garrick, D.J.; et al. Breeding value prediction for production traits in layer chickens using pedigree or genomic relationships in a reduced animal model. Genet. Sel. Evol. 2011, 43, 5. [Google Scholar] [CrossRef]
- D’Ambrosio, J.; Morvezen, R.; Brard-Fudulea, S.; Bestin, A.; Acin Perez, A.; Guéméné, D.; Poncet, C.; Haffray, P.; Dupont-Nivet, M.; Phocas, F. Genetic architecture and genomic selection of female reproduction traits in rainbow trout. BMC Genom. 2020, 21, 558. [Google Scholar] [CrossRef] [PubMed]
- Bolormaa, S.; Brown, D.J.; Swan, A.A.; van der Werf, J.H.J.; Hayes, B.J.; Daetwyler, H.D. Genomic prediction of reproduction traits for Merino sheep. Anim. Genet. 2017, 48, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Veronese, A.; Marques, O.; Moreira, R.; Belli, A.L.; Bisinotto, R.S.; Bilby, T.R.; Peñagaricano, F.; Chebel, R.C. Genomic merit for reproductive traits. I: Estrous characteristics and fertility in Holstein heifers. J. Dairy Sci. 2019, 102, 6624–6638. [Google Scholar] [CrossRef]
- Schaeffer, L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef]
- Goddard, M.E.; Hayes, B.J. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009, 10, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhu, F.; Yang, F.X.; Hao, J.P.; Hou, Z.C. Genomic selection for meat quality traits in Pekin duck. Anim. Genet. 2022, 53, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Hu, J.; Fan, W.; Xu, Y.; Tang, J.; Xie, M.; Zhang, Y.; Guo, Z.; Zhou, Z.; Hou, S. Strategies to improve genomic predictions for 35 duck carcass traits in an F(2) population. J. Anim. Sci. Biotechnol. 2023, 14, 74. [Google Scholar] [CrossRef]
- Zhu, F.; Yin, Z.T.; Wang, Z.; Smith, J.; Zhang, F.; Martin, F.; Ogeh, D.; Hincke, M.; Lin, F.B.; Burt, D.W.; et al. Three chromosome-level duck genome assemblies provide insights into genomic variation during domestication. Nat. Commun. 2021, 12, 5932. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.W.; Flint, J.; Myers, S.; Mott, R. Rapid genotype imputation from sequence without reference panels. Nat. Genet. 2016, 48, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.R. Best linear unbiased estimation and prediction under a selection model. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef] [PubMed]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef]
- Santana, L.G.; Flores-Mir, C.; Iglesias-Linares, A.; Pithon, M.M.; Marques, L.S. Influence of heritability on occlusal traits: A systematic review of studies in twins. Prog. Orthod. 2020, 21, 29. [Google Scholar] [CrossRef]
- Overholser, B.R.; Sowinski, K.M. Biostatistics primer: Part 2. Nutr. Clin. Pract. 2008, 23, 76–84. [Google Scholar] [CrossRef]
- VanRaden, P.M.; Van Tassell, C.P.; Wiggans, G.R.; Sonstegard, T.S.; Schnabel, R.D.; Taylor, J.F.; Schenkel, F.S. Invited review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009, 92, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Butler, D.G.; Cullis, B.R.; Gilmour, A.R.; Gogel, B.G.; Thompson, R. ASReml-R Reference Manual, v. 4.2; VSN International Ltd.: Hemel Hempstead, UK, 2023. [Google Scholar]
- Haldar, A.; Pal, P.; Datta, M.; Paul, R.; Pal, S.K.; Majumdar, D.; Biswas, C.K.; Pan, S. Prolificacy and Its Relationship with Age, Body Weight, Parity, Previous Litter Size and Body Linear Type Traits in Meat-type Goats. Asian-Australas. J. Anim. Sci. 2014, 27, 628–634. [Google Scholar] [CrossRef]
- Ablondi, M.; Summer, A.; Vasini, M.; Simoni, M.; Sabbioni, A. Genetic parameters estimation in an Italian horse native breed to support the conversion from agricultural uses to riding purposes. J. Anim. Breed Genet. 2020, 137, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Kanlisi, R.A.; Amuzu-Aweh, E.N.; Naazie, A.; Otsyina, H.R.; Kelly, T.R.; Gallardo, R.A.; Lamont, S.J.; Zhou, H.; Dekkers, J.; Kayang, B.B. Genetic architecture of body weight, carcass, and internal organs traits of Ghanaian local chickens. Front. Genet. 2024, 15, 1297034. [Google Scholar] [CrossRef]
- Walugembe, M.; Amuzu-Aweh, E.N.; Botchway, P.K.; Naazie, A.; Aning, G.; Wang, Y.; Saelao, P.; Kelly, T.; Gallardo, R.A.; Zhou, H.; et al. Genetic Basis of Response of Ghanaian Local Chickens to Infection With a Lentogenic Newcastle Disease Virus. Front. Genet. 2020, 11, 739. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, J.Z.; Zhu, M.Y.; Yang, F.X.; Hao, J.P.; He, Y.; Zhu, X.L.; Hou, Z.C.; Zhu, F. Genome-Wide Association Analysis and Genetic Parameters for Egg Production Traits in Peking Ducks. Animals 2024, 14, 1891. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, X.; Zheng, W.; Wang, P.; Duan, Z.; Xu, G. Dynamic Changes in Egg Quality, Heritability and Correlation of These Traits and Yolk Nutrient throughout the Entire Laying Cycle. Foods 2023, 12, 4472. [Google Scholar] [CrossRef]
- Yuan, J.; Sun, C.; Dou, T.; Yi, G.; Qu, L.; Qu, L.; Wang, K.; Yang, N. Identification of Promising Mutants Associated with Egg Production Traits Revealed by Genome-Wide Association Study. PLoS ONE 2015, 10, e0140615. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Z.; Qu, Y.; Li, Q.; Tian, Y.; Chen, L.; Tang, J.; Li, C.; Li, G.; Shen, J.; et al. Genome-Wide Association Studies and Haplotype-Sharing Analysis Targeting the Egg Production Traits in Shaoxing Duck. Front. Genet. 2022, 13, 828884. [Google Scholar] [CrossRef]
- Ni, A.; Calus, M.P.L.; Bovenhuis, H.; Yuan, J.; Wang, Y.; Sun, Y.; Chen, J. Genetic parameters, reciprocal cross differences, and age-related heterosis of egg-laying performance in chickens. Genet. Sel. Evol. 2023, 55, 87. [Google Scholar] [CrossRef]
- Çelik, Ş.; Eyduran, E.; Şengül, A.Y.; Şengül, T. Relationship among egg quality traits in Japanese quails and prediction of egg weight and color using data mining algorithms. Trop Anim. Health Prod. 2021, 53, 382. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.G.; Xu, X.L.; Cao, H.Y.; Zhou, W.; Yin, Z.Z. Effect of age at first egg on reproduction performance and characterization of the hypothalamo-pituitary-gonadal axis in chickens. Poult. Sci. 2021, 100, 101325. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tan, L.; Zhi, Y.; Bu, L.; Wang, Y.; Wang, Z.; Guo, Y.; Tian, W.; Xu, C.; Li, D.; et al. Genome-wide variation study and inter-tissue communication analysis unveil regulatory mechanisms of egg-laying performance in chickens. Nat. Commun. 2024, 15, 7069. [Google Scholar] [CrossRef]
- Savegnago, R.P.; Caetano, S.L.; Ramos, S.B.; Nascimento, G.B.; Schmidt, G.S.; Ledur, M.C.; Munari, D.P. Estimates of genetic parameters, and cluster and principal components analyses of breeding values related to egg production traits in a White Leghorn population. Poult. Sci. 2011, 90, 2174–2188. [Google Scholar] [CrossRef]
- Shi, L.; Li, Y.; Isa, A.M.; Ma, H.; Yuan, J.; Wang, P.; Ge, P.; Gong, Y.; Chen, J.; Sun, Y. Characterization of clutch traits and egg production in six chicken breeds. Anim. Biosci. 2023, 36, 899–907. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, N.; Yan, Y.; Li, G.; Liu, A.; Wu, G.; Sun, C. Genome-wide association analysis of egg production performance in chickens across the whole laying period. BMC Genet. 2019, 20, 67. [Google Scholar] [CrossRef]
- Chomchuen, K.; Tuntiyasawasdikul, V.; Chankitisakul, V.; Boonkum, W. Genetic Evaluation of Body Weights and Egg Production Traits Using a Multi-Trait Animal Model and Selection Index in Thai Native Synthetic Chickens (Kaimook e-san2). Animals 2022, 12, 335. [Google Scholar] [CrossRef]
- Li, G.S.; Zhu, F.; Yang, F.X.; Hao, J.P.; Hou, Z.C. Selection response and genetic parameter estimation of feeding behavior traits in Pekin ducks. Poult. Sci. 2020, 99, 2375–2384. [Google Scholar] [CrossRef]
- da Silva, É.D.B.; Xavier, A.; Faria, M.V. Impact of Genomic Prediction Model, Selection Intensity, and Breeding Strategy on the Long-Term Genetic Gain and Genetic Erosion in Soybean Breeding. Front. Genet. 2021, 12, 637133. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Singh, V.K.; Bohra, A.; Kumar, A.; Reif, J.C.; Varshney, R.K. Genomics and breeding innovations for enhancing genetic gain for climate resilience and nutrition traits. Theor. Appl. Genet. 2021, 134, 1829–1843. [Google Scholar] [CrossRef]
- Mehrban, H.; Naserkheil, M.; Lee, D.; Ibáñez-Escriche, N. Multi-Trait Single-Step GBLUP Improves Accuracy of Genomic Prediction for Carcass Traits Using Yearling Weight and Ultrasound Traits in Hanwoo. Front. Genet. 2021, 12, 692356. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Xu, Z.; Wang, Q.; Zhu, D.; Bian, C.; Ren, J.; Huang, Z.; Zhu, X.; Tian, Z.; Wang, Y.; et al. Genome-wide association study and genomic prediction for growth traits in yellow-plumage chicken using genotyping-by-sequencing. Genet. Sel. Evol. 2021, 53, 82. [Google Scholar] [CrossRef]
| Traits 1 | Generation | Num 2 | Mean | SD 3 | CV (%) 4 | Min | Max | Num (M) 5 | Mean (M) | Num (F) 6 | Mean (F) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BW (g) | 12 | 709 | 2878 | 105 | 3.66 | 2682 | 3098 | \ | \ | 709 | 2878 (105) |
| 13 | 1638 | 2818 | 108 | 3.85 | 2630 | 3090 | 339 | 2970 (55) | 1299 | 2778 (80) | |
| 14 | 1878 | 2815 | 130 | 4.60 | 2555 | 3150 | 409 | 2968 (92) | 1469 | 2772 (104) | |
| 15 | 1820 | 2768 | 107 | 3.87 | 2565 | 3015 | 384 | 2923 (48) | 1436 | 2726 (76) | |
| BSL (cm) | 12 | 708 | 25.31 | 0.87 | 3.44 | 22.9 | 27.2 | \ | \ | 708 | 25.31 (0.87) |
| 13 | 1629 | 25.59 | 0.92 | 3.61 | 22.7 | 28.19 | 331 | 26.28 (0.79) | 1298 | 25.41 (0.87) | |
| 14 | 1878 | 24.34 | 0.80 | 3.27 | 22.1 | 26.5 | 409 | 25.28 (0.58) | 1469 | 24.07 (0.63) | |
| 15 | 1817 | 24.38 | 0.76 | 3.11 | 22.5 | 26.5 | 384 | 25.38 (0.51) | 1433 | 24.11 (0.57) | |
| NL (cm) | 12 | 709 | 20.25 | 0.65 | 3.19 | 18.1 | 21.8 | \ | \ | 709 | 20.25 (0.65) |
| 13 | 1626 | 20.39 | 0.95 | 4.66 | 17.34 | 23.3 | 331 | 21.57 (0.86) | 1295 | 20.09 (0.71) | |
| 14 | 1878 | 20.92 | 0.84 | 4.03 | 18.7 | 23.6 | 409 | 22.04 (0.66) | 1469 | 20.61 (0.59) | |
| 15 | 1819 | 20.74 | 0.82 | 3.95 | 18.8 | 23.1 | 384 | 21.85 (0.59) | 1435 | 20.45 (0.59) |
| Traits 1 | Generation | Num 2 | Mean | SD 3 | CV (%) 4 | Min | Max |
|---|---|---|---|---|---|---|---|
| AFE | 12 | 704 | 176.04 | 11.57 | 6.57 | 148 | 211 |
| 13 | 1217 | 174.20 | 12.26 | 7.04 | 146 | 210 | |
| 14 | 1395 | 173.51 | 11.22 | 6.47 | 147 | 206 | |
| 15 | 1362 | 165.89 | 14.09 | 8.49 | 134 | 206 | |
| EN25-44WK | 12 | 691 | 119.55 | 11.96 | 10.00 | 73 | 140 |
| 13 | 1199 | 120.03 | 12.38 | 10.32 | 76 | 140 | |
| 14 | 1378 | 120.53 | 12.31 | 10.22 | 77 | 141 | |
| 15 | 1342 | 124.05 | 11.91 | 9.60 | 78 | 140 |
| Traits 1 | Generation | Num 2 | Mean | SD 3 | CV (%) 4 | Min | Max |
|---|---|---|---|---|---|---|---|
| EWT | 12 | 681 | 89.53 | 5.89 | 6.58 | 72.6 | 105.3 |
| 13 | 1216 | 90.24 | 5.32 | 5.89 | 74.5 | 106.5 | |
| 14 | 1354 | 90.68 | 5.45 | 6.01 | 75.8 | 107 | |
| 15 | 1361 | 88.28 | 5.09 | 5.77 | 73.7 | 104 | |
| ESI | 12 | 680 | 1.36 | 0.04 | 3.14 | 1.24 | 1.48 |
| 13 | 1211 | 1.36 | 0.04 | 3.28 | 1.24 | 1.49 | |
| 14 | 1352 | 1.36 | 0.04 | 3.11 | 1.23 | 1.49 | |
| 15 | 1362 | 1.36 | 0.04 | 3.08 | 1.23 | 1.48 |
| Traits 2 | BW | BSL | NL | AFE | EN25-44WK | EWT | ESI |
|---|---|---|---|---|---|---|---|
| BW | 0.19 (0.02) 0.11 (0.02) | 0.52 (0.05) ** | 0.12 (0.07) | 0.11 (0.06) | –0.14 (0.07) * | 0.19 (0.06) ** | 0.03 (0.06) |
| BSL | 0.39 (0.09) ** | 0.27 (0.02) 0.20 (0.02) | 0.39 (0.06) ** | –0.01 (0.06) | 0.06 (0.06) | 0.24 (0.05) ** | 0.07 (0.06) |
| NL | 0.27 (0.11) ** | 0.25 (0.09) ** | 0.23 (0.02) 0.16 (0.02) | –0.04 (0.06) | 0.12 (0.06) * | 0.09 (0.06) | –0.01 (0.06) |
| AFE | 0 (0.1) | –0.22 (0.08) ** | –0.08 (0.09) | 0.37 (0.02) 0.26 (0.03) | –0.91 (0.02) ** | 0.06 (0.05) | –0.07 (0.05) |
| EN25-44WK | –0.04 (0.11) | 0.22 (0.08) ** | 0.14 (0.09) | –0.89 (0.04) ** | 0.27 (0.02) 0.24 (0.03) | –0.03 (0.06) | 0 (0.06) |
| EWT | 0.32 (0.10) ** | 0.24 (0.08) ** | 0.08 (0.09) | –0.06 (0.08) | 0.16 (0.09) * | 0.39 (0.02) 0.24 (0.03) | –0.03 (0.05) |
| ESI | –0.01 (0.11) | 0.14 (0.08) * | –0.09 (0.09) | –0.17 (0.08) * | 0.03 (0.09) | 0 (0.09) | 0.36 (0.02) 0.23 (0.03) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Yu, J.-Z.; Zhu, M.-Y.; Yang, F.-X.; Hao, J.-P.; He, Y.; Zhu, X.-L.; Hou, Z.-C.; Zhu, F. Optimizing Breeding Strategies for Pekin Ducks Using Genomic Selection: Genetic Parameter Evaluation and Selection Progress Analysis in Reproductive Traits. Appl. Sci. 2025, 15, 194. https://doi.org/10.3390/app15010194
Zhou J, Yu J-Z, Zhu M-Y, Yang F-X, Hao J-P, He Y, Zhu X-L, Hou Z-C, Zhu F. Optimizing Breeding Strategies for Pekin Ducks Using Genomic Selection: Genetic Parameter Evaluation and Selection Progress Analysis in Reproductive Traits. Applied Sciences. 2025; 15(1):194. https://doi.org/10.3390/app15010194
Chicago/Turabian StyleZhou, Jun, Jiang-Zhou Yu, Mei-Yi Zhu, Fang-Xi Yang, Jin-Ping Hao, Yong He, Xiao-Liang Zhu, Zhuo-Cheng Hou, and Feng Zhu. 2025. "Optimizing Breeding Strategies for Pekin Ducks Using Genomic Selection: Genetic Parameter Evaluation and Selection Progress Analysis in Reproductive Traits" Applied Sciences 15, no. 1: 194. https://doi.org/10.3390/app15010194
APA StyleZhou, J., Yu, J.-Z., Zhu, M.-Y., Yang, F.-X., Hao, J.-P., He, Y., Zhu, X.-L., Hou, Z.-C., & Zhu, F. (2025). Optimizing Breeding Strategies for Pekin Ducks Using Genomic Selection: Genetic Parameter Evaluation and Selection Progress Analysis in Reproductive Traits. Applied Sciences, 15(1), 194. https://doi.org/10.3390/app15010194







