Enhancing Colony Detection of Microorganisms in Agar Dishes Using SAM-Based Synthetic Data Augmentation in Low-Data Scenarios
Abstract
Featured Application
Abstract
1. Introduction
2. Related Works
2.1. Colony Detection and Segmentation
2.2. Addressing Data Scarcity in Medical Deep Learning Applications
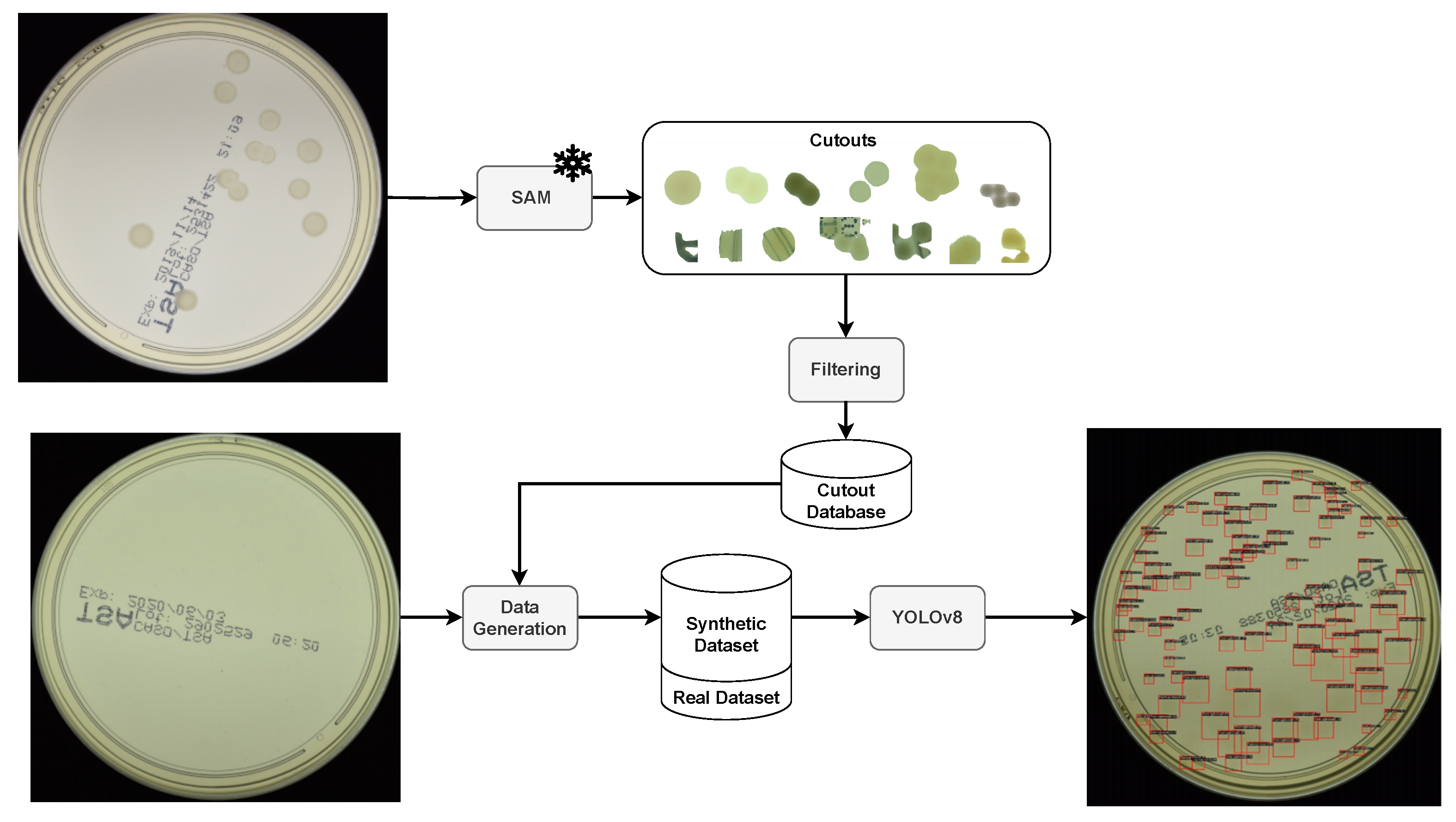
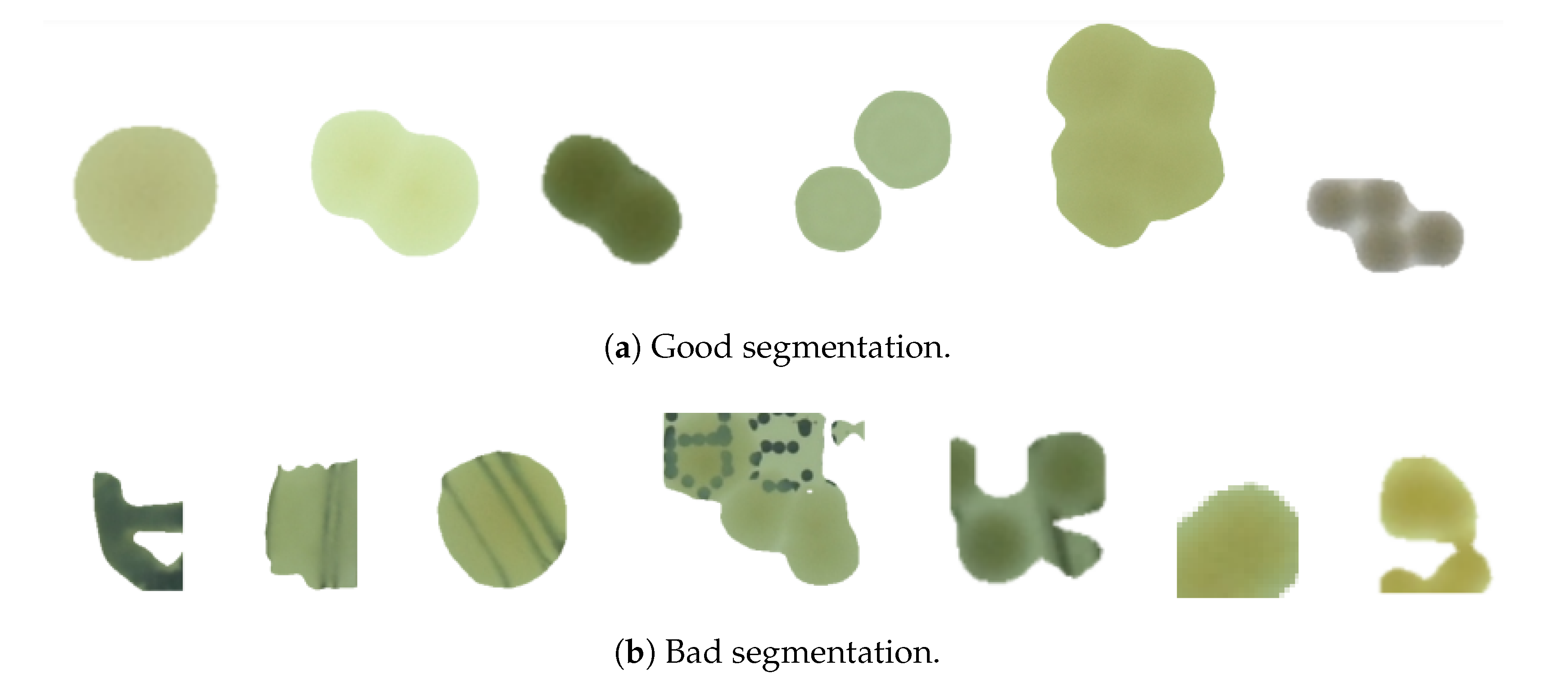
3. Method
| Algorithm 1 Synthetic Agar Dish Generation | |
| 1: | Initialization |
| 2: | B: List of different backgrounds |
| 3: | I: List of color intensities of the colonies |
| 4: | H: List of hues of the colonies |
| 5: | Cl: List of colony classes |
| 6: | |
| 7: | Input: |
| 8: | C: Pre-sorted colony cutouts |
| 9: | E: Images of empty agar dishes |
| 10: | |
| 11: | for each synthetic image do: |
| 12: | = Randomly selected from [B, I, H, Cl] |
| 13: | e = Randomly selected form E based on |
| 14: | N = Randomly select from range(1,100) |
| 15: | for n in range(N) do: |
| 16: | = randomly seceded from C based on |
| 17: | Find viable regions in e without overlap |
| 18: | Create annotation (bounding box) for |
| 19: | Save the final synthetic image |
| 20: | Save the corresponding annotation file with bounding box information |
| 21: | |
| 22: | Output: |
| 23: | Synthetic dataset with annotations |
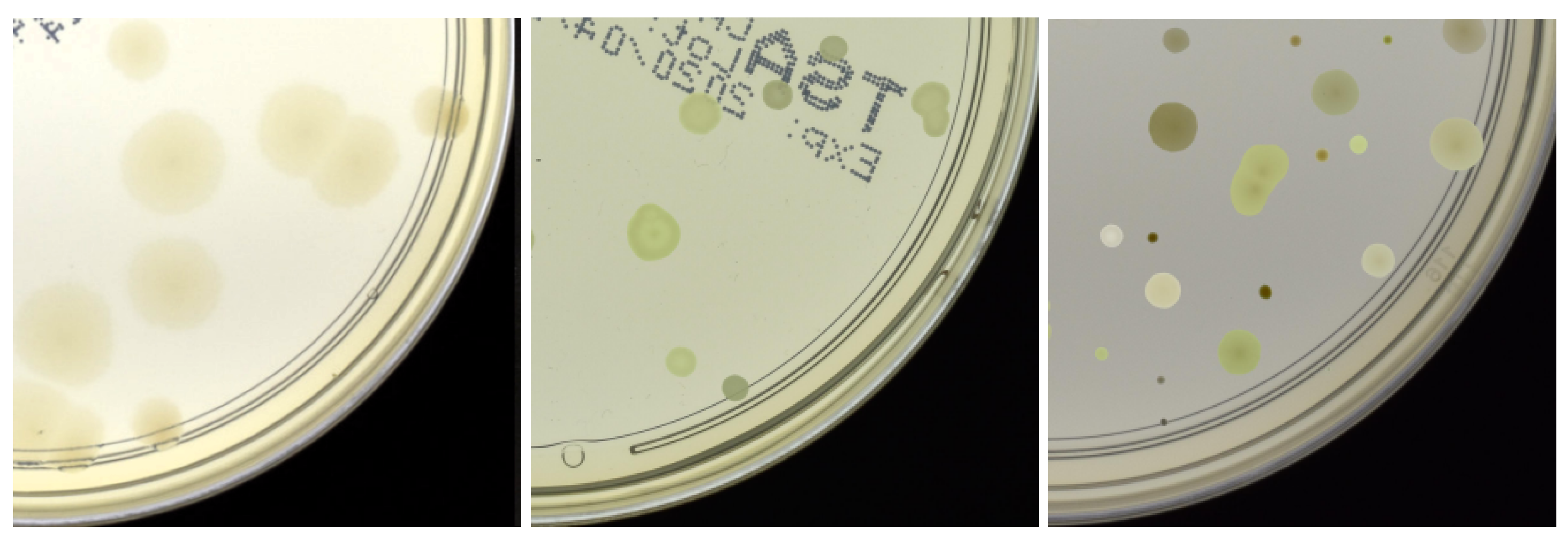
4. Results and Discussion
4.1. Dataset and Metrics
4.2. Data Generation Configuration
4.2.1. Ablation Study
4.2.2. Domain Adaption
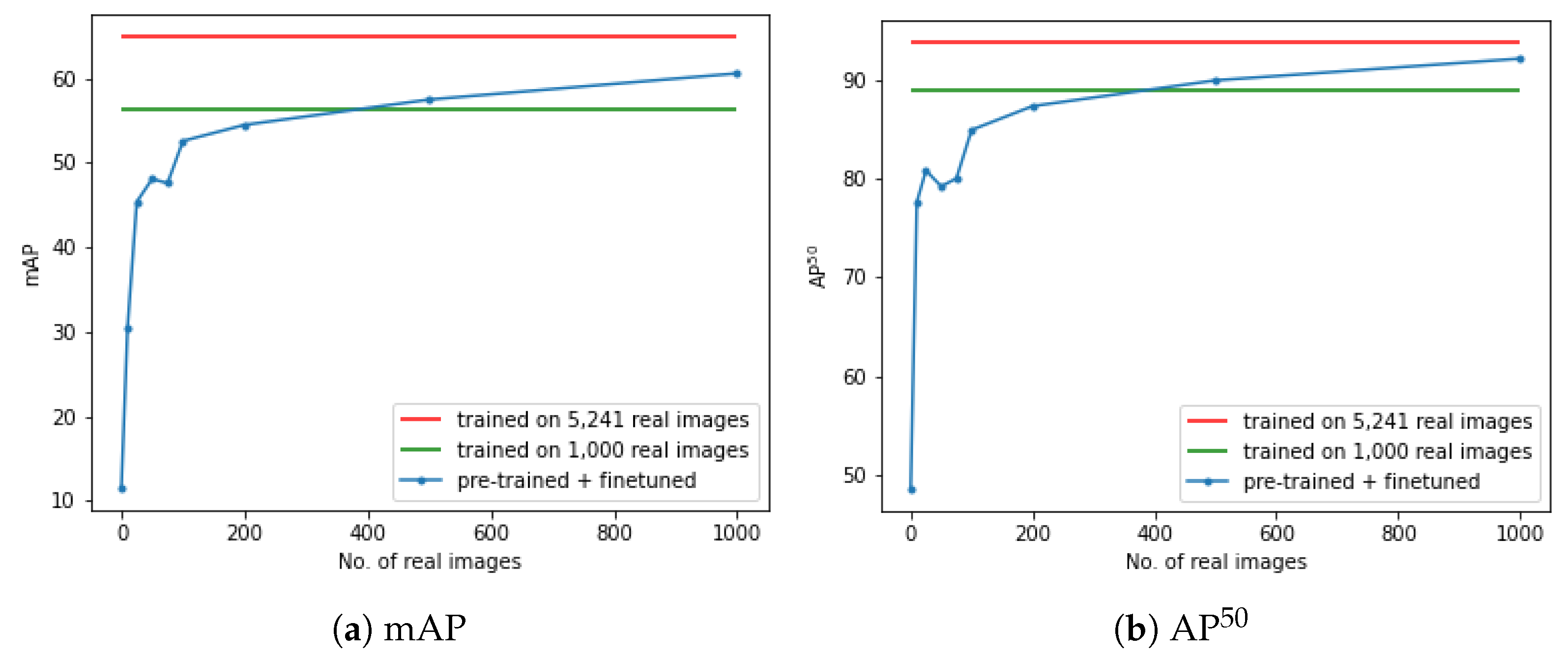
4.2.3. Evaluation of the Minimal Number of Real Images
4.3. State-of-the-Art Comparison
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geissmann, Q. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS ONE 2013, 8, e54072. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.M.; Torelli, A.; Wolf, I.; Gretz, N. AutoCellSeg: Robust automatic colony forming unit (CFU)/cell analysis using adaptive image segmentation and easy-to-use post-editing techniques. Sci. Rep. 2018, 8, 7302. [Google Scholar] [CrossRef] [PubMed]
- Pawłowski, J.; Majchrowska, S.; Golan, T. Generation of microbial colonies dataset with deep learning style transfer. Sci. Rep. 2022, 12, 5212. [Google Scholar] [CrossRef] [PubMed]
- Majchrowska, S.; Pawłowski, J.; Guła, G.; Bonus, T.; Hanas, A.; Loch, A.; Pawlak, A.; Roszkowiak, J.; Golan, T.; Drulis-Kawa, Z. AGAR a microbial colony dataset for deep learning detection. arXiv 2021, arXiv:2108.01234. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. (NeurIPS) 2017, 30, 6000. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations (ICLR), Virtual, 26 April–1 May 2020. [Google Scholar]
- Ebert, N.; Stricker, D.; Wasenmüller, O. PLG-ViT: Vision Transformer with Parallel Local and Global Self-Attention. Sensors 2023, 23, 3447. [Google Scholar] [CrossRef]
- Ebert, N.; Stricker, D.; Wasenmüller, O. Transformer-based Detection of Microorganisms on High-Resolution Petri Dish Images. In Proceedings of the International Conference on Computer Vision Workshops (ICCVW), Paris, France, 2–6 October 2023. [Google Scholar]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Naets, T.; Huijsmans, M.; Smyth, P.; Sorber, L.; de Lannoy, G. A Mask R-CNN approach to counting bacterial colony forming units in pharmaceutical development. arXiv 2021, arXiv:2103.05337. [Google Scholar]
- Goceri, E. Medical image data augmentation: Techniques, comparisons and interpretations. Artif. Intell. Rev. 2023, 56, 12561–12605. [Google Scholar] [CrossRef]
- Andreini, P.; Bonechi, S.; Bianchini, M.; Mecocci, A.; Scarselli, F. Image generation by GAN and style transfer for agar plate image segmentation. Comput. Methods Programs Biomed. 2020, 184, 105268. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P. MSG-GAN based synthesis of brain MRI with meningioma for data augmentation. In Proceedings of the International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 2–4 July 2020. [Google Scholar]
- Ghiasi, G.; Cui, Y.; Srinivas, A.; Qian, R.; Lin, T.Y.; Cubuk, E.D.; Le, Q.V.; Zoph, B. Simple copy-paste is a strong data augmentation method for instance segmentation. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the International Conference on Computer Vision (ICCV), Paris, France, 2–6 October 2023. [Google Scholar]
- Alexander, N.; Glick, D. Automatic counting of bacterial cultures—A new machine. IRE Trans. Med. Electron. 1958, PGME-12, 89–92. [Google Scholar] [CrossRef]
- Mansberg, H. Automatic particle and bacterial colony counter. Science 1957, 126, 3278. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Lombardi, S.; Signoroni, A. Bacterial colony counting with convolutional neural networks in digital microbiology imaging. Pattern Recognit. 2017, 61, 629–640. [Google Scholar] [CrossRef]
- Andreini, P.; Bonechi, S.; Bianchini, M.; Mecocci, A.; Scarselli, F. A deep learning approach to bacterial colony segmentation. In Proceedings of the International Conference on Artificial Neural Networks (ICANN), Rhodes, Greece, 4–7 October 2018. [Google Scholar]
- Ramesh, N.; Tasdizen, T. Cell segmentation using a similarity interface with a multi-task convolutional neural network. J. Biomed. Health Inform. 2018, 23, 1457–1468. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Liu, S.J.; Huang, P.C.; Liu, X.S.; Lin, J.J.; Zou, Z. A two-stage deep counting for bacterial colonies from multi-sources. Appl. Soft Comput. 2022, 130, 109706. [Google Scholar] [CrossRef]
- Cai, Z.; Vasconcelos, N. Cascade r-cnn: Delving into high quality object detection. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. Trans. Pattern Anal. Mach. Intell. 2017, 39.6, 1137. [Google Scholar] [CrossRef]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018. [Google Scholar]
- Feng, C.; Zhong, Y.; Gao, Y.; Scott, M.R.; Huang, W. Tood: Task-aligned one-stage object detection. In Proceedings of the International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- Hendrycks, D.; Mu, N.; Cubuk, E.D.; Zoph, B.; Gilmer, J.; Lakshminarayanan, B. AugMix: A Simple Data Processing Method to Improve Robustness and Uncertainty. In Proceedings of the International Conference on Learning Representations (ICLR), Virtual, 26 April–1 May 2020. [Google Scholar]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. Cutmix: Regularization strategy to train strong classifiers with localizable features. In Proceedings of the International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Oehri, S.; Ebert, N.; Abdullah, A.; Stricker, D.; Wasenmüller, O. GenFormer—Generated Images are All You Need to Improve Robustness of Transformers on Small Datasets. In Proceedings of the International Conference on Pattern Recognition (ICPR), Kolkata, India, 1–5 December 2024. [Google Scholar]
- Sarıyıldız, M.B.; Alahari, K.; Larlus, D.; Kalantidis, Y. Fake it till you make it: Learning transferable representations from synthetic ImageNet clones. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Vancouver, BC, Canada, 17–24 June 2023. [Google Scholar]
- Reichardt, L.; Uhr, L.; Wasenmüller, O. Text3DAug – Prompted Instance Augmentation for LiDAR Perception. In Proceedings of the International Conference on Intelligent Robots and Systems (IROS), Abu Dhabi, United Arab Emirates, 14–18 October 2024. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Alyafi, B.; Diaz, O.; Marti, R. DCGANs for realistic breast mass augmentation in x-ray mammography. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 15–20 February 2020. [Google Scholar]
- Desai, S.D.; Giraddi, S.; Verma, N.; Gupta, P.; Ramya, S. Breast cancer detection using gan for limited labeled dataset. In Proceedings of the International Conference on Computational Intelligence and Communication Networks (CICN), Bhimtal, India, 25–26 September 2020. [Google Scholar]
- Shen, T.; Hao, K.; Gou, C.; Wang, F.Y. Mass image synthesis in mammogram with contextual information based on GANs. Comput. Methods Programs Biomed. 2021, 202, 106019. [Google Scholar] [CrossRef]
- Li, Q.; Yu, Z.; Wang, Y.; Zheng, H. TumorGAN: A multi-modal data augmentation framework for brain tumor segmentation. Sensors 2020, 20, 4203. [Google Scholar] [CrossRef]
- Neelima, G.; Chigurukota, D.R.; Maram, B.; Girirajan, B. Optimal DeepMRSeg based tumor segmentation with GAN for brain tumor classification. Biomed. Signal Process. Control 2022, 74, 103537. [Google Scholar] [CrossRef]
- Bissoto, A.; Perez, F.; Valle, E.; Avila, S. Skin lesion synthesis with generative adversarial networks. In Proceedings of the OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis, Granada, Spain, 16–20 September 2018. [Google Scholar]
- Ghorbani, A.; Natarajan, V.; Coz, D.; Liu, Y. Dermgan: Synthetic generation of clinical skin images with pathology. In Proceedings of the Machine Learning for Health Workshop, Virtual, 11 December 2020. [Google Scholar]
- Dar, S.U.H.; Ghanaat, A.; Kahmann, J.; Ayx, I.; Papavassiliou, T.; Schoenberg, S.O.; Engelhardt, S. Investigating data memorization in 3d latent diffusion models for medical image synthesis. arXiv 2023, arXiv:2307.01148. [Google Scholar]
- Sagers, L.W.; Diao, J.A.; Melas-Kyriazi, L.; Groh, M.; Rajpurkar, P.; Adamson, A.S.; Rotemberg, V.; Daneshjou, R.; Manrai, A.K. Augmenting medical image classifiers with synthetic data from latent diffusion models. arXiv 2023, arXiv:2308.12453. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved training of wasserstein gans. Adv. Neural Inf. Process. Syst. (NeurIPS) 2017, 360, 5769. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Qiu, J. Ultralytics YOLOv8. 2023. Available online: https://github.com/ultralytics/ultralytics (accessed on 16 December 2024).
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision (ECCV), Zurich, Switzerland, 6–12 September 2014. [Google Scholar]

| Class | Full Set | FT-Set | ||
|---|---|---|---|---|
| AP50 | mAP | AP50 | mAP | |
| S. aureus | 94.9 | 62.0 | 89.2 | 54.6 |
| B. subtilis | 97.9 | 72.1 | 95.0 | 64.5 |
| P. aeruginosa | 96.2 | 67.0 | 93.1 | 59.7 |
| E. coli | 97.7 | 74.2 | 94.2 | 65.8 |
| C. albicans | 81.9 | 49.2 | 73.4 | 37.3 |
| Mean | 93.7 | 64.9 | 89.0 | 56.4 |
| S. aureus | B. subtilis | P. aeruginosa | E. coli | C. albicans | Mean | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP50 | mAP | AP50 | mAP | AP50 | mAP | AP50 | mAP | AP50 | mAP | AP50 | mAP | |
| Baseline | 89.2 | 54.6 | 95.0 | 64.5 | 93.1 | 59.7 | 94.2 | 65.8 | 73.4 | 37.3 | 89.0 | 56.4 |
| (I) Raw data | 92.3 | 56.9 | 96.7 | 66.5 | 94.4 | 61.8 | 95.4 | 68.2 | 77.1 | 41.0 | 91.2 | 58.9 |
| (II) No overlapping colonies | 91.6 | 55.7 | 95.5 | 66.1 | 94.5 | 61.7 | 95.2 | 67.9 | 76.3 | 40.8 | 90.6 | 58.4 |
| (III) Pairs of overlapping colonies | 93.1 | 58.9 | 96.7 | 67.2 | 94.7 | 62.8 | 95.9 | 69.6 | 78.9 | 43.9 | 91.9 | 60.5 |
| (IV) Color differentiation | 92.9 | 58.4 | 96.9 | 67.0 | 94.8 | 62.9 | 96.1 | 69.3 | 78.6 | 42.7 | 91.9 | 60.1 |
| (V) Two classes max | 93.3 | 59.0 | 97.1 | 68.2 | 94.9 | 63.1 | 96.2 | 69.3 | 79.2 | 43.3 | 92.1 | 60.6 |
| (VI) Big clusters of colonies | 93.6 | 59.2 | 97.1 | 67.7 | 94.8 | 63.6 | 96.0 | 70.1 | 78.9 | 42.6 | 92.1 | 60.6 |
| (VII) Dish color differentiation | 92.6 | 57.6 | 96.9 | 66.9 | 94.6 | 62.3 | 96.0 | 69.0 | 77.5 | 40.5 | 91.5 | 59.3 |
| (VIII) Opacity 90% | 93.2 | 57.8 | 97.2 | 67.3 | 95.0 | 62.9 | 96.1 | 69.8 | 78.2 | 41.3 | 91.9 | 59.8 |
| Class | Both | Dark | Bright | |||
|---|---|---|---|---|---|---|
| AP | mAP | AP | mAP | AP | mAP | |
| S. aureus | 93.6 | 59.2 | 93.7 | 59.7 | 91.2 | 56.1 |
| B. subtilis | 97.1 | 67.7 | 97.2 | 67.7 | 96.4 | 65.9 |
| P. aeruginosa | 94.8 | 63.6 | 95.0 | 63.3 | 94.6 | 61.1 |
| E. coli | 96.0 | 70.1 | 96.2 | 70.1 | 95.7 | 68.1 |
| C. albicans | 78.9 | 42.6 | 78.4 | 42.3 | 75.1 | 37.0 |
| Mean | 92.1 | 60.6 | 92.1 | 60.6 | 90.6 | 57.9 |
| Method | Params | Synth. Data | Metric | |
|---|---|---|---|---|
| AP50 | mAP | |||
| Full Dataset | ||||
| Faster-RCNN * [4] | 41.5 M | 76.7 | 49.3 | |
| Cascade-RCNN * [4] | 69.2 M | 79.2 | 51.6 | |
| Faster-RCNN * [8] | 41.5 M | 80.2 | 56.0 | |
| AttnPAFPN * [8] | 32.8 M | 96.3 | 68.2 | |
| YOLOv8 [48] | 3.2 M | 93.7 | 64.9 | |
| 500 real images | ||||
| Faster-RCNN * [8] | 41.5 M | 78.8 | 53.4 | |
| AttnPAFPN * [8] | 32.8 M | 92.3 | 62.9 | |
| YOLOv8 [48] | 3.2 M | 84.7 | 53.0 | |
| YOLOv8 + Ours | 3.2 M | ✓ | 89.9 | 57.5 |
| 100 real images | ||||
| Faster-RCNN * [3] | 41.5 M | ✓ | - | 40.1 |
| Cascade-RCNN * [3] | 69.2 M | ✓ | - | 41.6 |
| YOLOv8 [48] | 3.2 M | 73.4 | 44.6 | |
| YOLOv8 + Ours | 3.2 M | ✓ | 84.9 | 52.6 |
| 50 real images | ||||
| Faster-RCNN * [8] | 41.5 M | 70.6 | 41.2 | |
| AttnPAFPN * [8] | 32.8 M | 72.5 | 42.8 | |
| YOLOv8 [48] | 3.2 M | 60.8 | 36.0 | |
| YOLOv8 + Ours | 3.2 M | ✓ | 79.2 | 48.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mennemann, K.; Ebert, N.; Reichardt, L.; Wasenmüller, O. Enhancing Colony Detection of Microorganisms in Agar Dishes Using SAM-Based Synthetic Data Augmentation in Low-Data Scenarios. Appl. Sci. 2025, 15, 1260. https://doi.org/10.3390/app15031260
Mennemann K, Ebert N, Reichardt L, Wasenmüller O. Enhancing Colony Detection of Microorganisms in Agar Dishes Using SAM-Based Synthetic Data Augmentation in Low-Data Scenarios. Applied Sciences. 2025; 15(3):1260. https://doi.org/10.3390/app15031260
Chicago/Turabian StyleMennemann, Kim, Nikolas Ebert, Laurenz Reichardt, and Oliver Wasenmüller. 2025. "Enhancing Colony Detection of Microorganisms in Agar Dishes Using SAM-Based Synthetic Data Augmentation in Low-Data Scenarios" Applied Sciences 15, no. 3: 1260. https://doi.org/10.3390/app15031260
APA StyleMennemann, K., Ebert, N., Reichardt, L., & Wasenmüller, O. (2025). Enhancing Colony Detection of Microorganisms in Agar Dishes Using SAM-Based Synthetic Data Augmentation in Low-Data Scenarios. Applied Sciences, 15(3), 1260. https://doi.org/10.3390/app15031260







