Microbial Profiling of Smear-Ripened Cheeses: Identification of Starter Cultures and Environmental Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Processing and DNA Extraction
2.2. Microbiota Composition Determined by 16S rRNA Gene Sequencing and Sequencing of Internal Transcribed Spacer Regions
2.3. Data Analysis and Statistics
2.4. Total Bacteria Count Determination and Microbiota Cultivation
2.5. Sanger Sequencing of Full-Length 16S rRNA PCR Products
3. Results
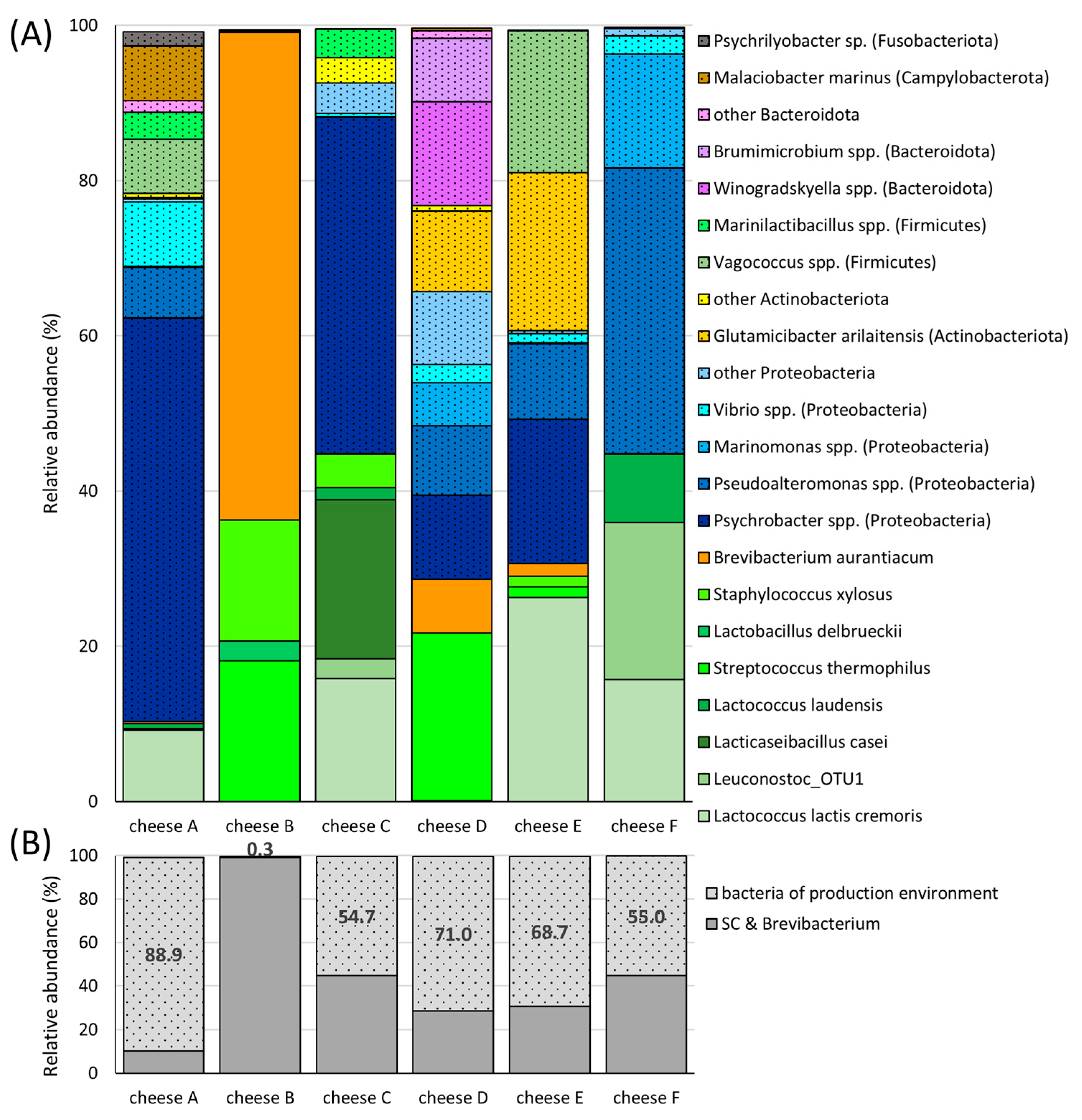
3.1. Microbiota Composition of Smear-Ripened Cheeses Determined by 16S rRNA Gene Sequencing
3.2. Bacterial Starter and Adjunct Cultures in Smear-Ripened Cheeses
3.3. Bacteria of the Production Environment in Smear-Ripened Cheeses
3.4. Microbiota Composition Determined by Sequencing of Internal Transcribed Spacer Regions
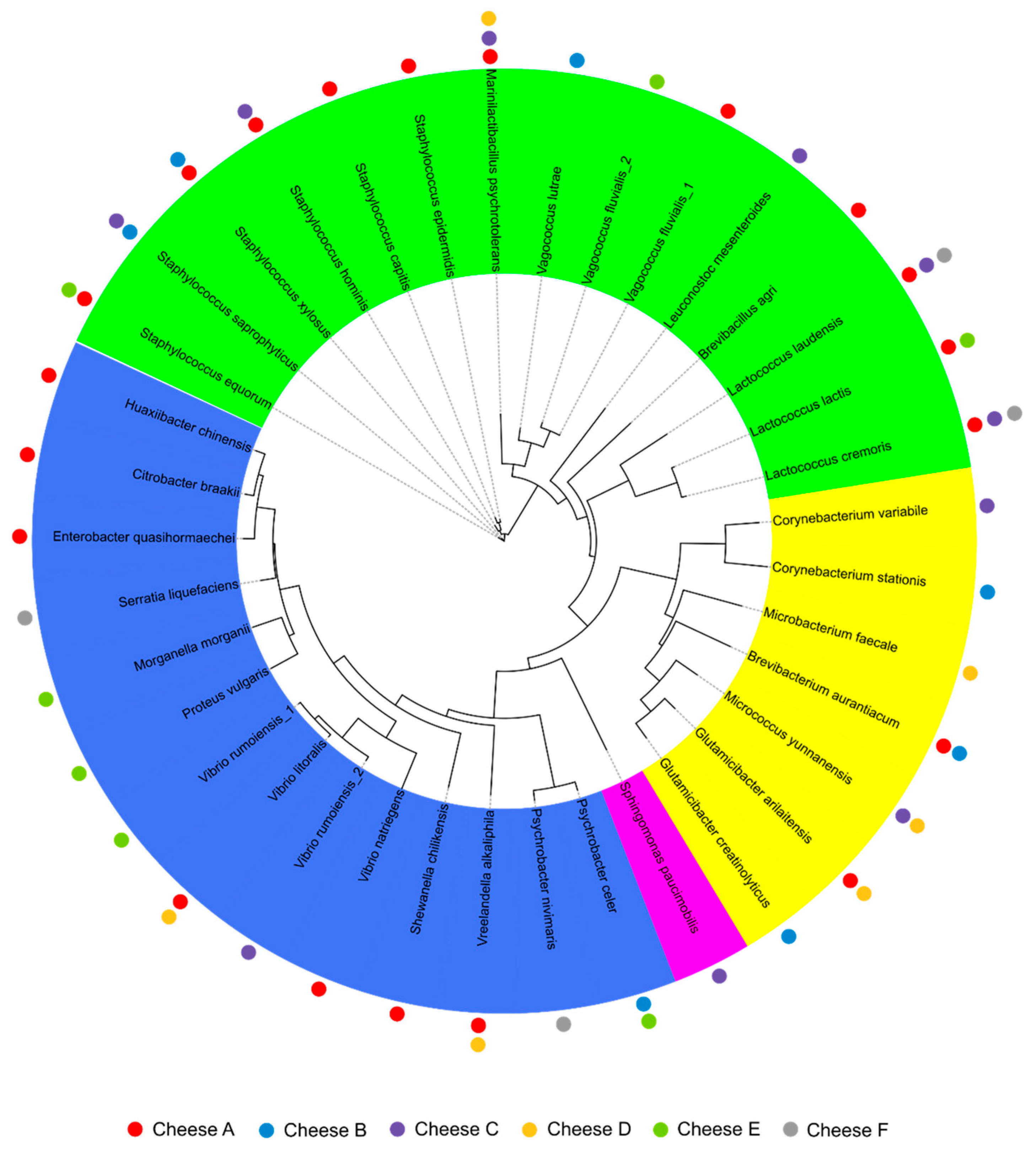
3.5. Microbiota Composition Determined by Cultivation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BHI | Brain heart infusion |
| ITS | Internal transcribed spacer |
| LAB | Lactic acid bacteria |
| OTU | Operational taxonomic unit |
| PCA | Plate count agar |
| PCoA | Principal component analysis |
| SCs | Starter cultures |
References
- Fox, P.F.; McSweeney, P.L. Cheese: An Overview. Cheese 2017, 5–21. [Google Scholar] [CrossRef]
- Brennan, N.M.; Cogan, T.M.; Loessner, M.; Scherer, S. Bacterial Surface-Ripened Cheeses. In Cheese: Chemistry, Physics and Microbiology; Fox, P.F., McSweeney, P.L.H., Cogan, T.M., Guinee, T.P., Eds.; Academic Press: Cambridge, MA, USA, 2004; Volume 2, pp. 199–225. [Google Scholar] [CrossRef]
- Bockelmann, W. Secondary Cheese Starter Cultures. In Technology of Cheesemaking; Wiley-Blackwell: Hoboken, NJ, USA, 2010; pp. 193–230. [Google Scholar]
- Bockelmann, W.; Willems, K.P.; Neve, H.; Heller, K.H. Cultures for the Ripening of Smear Cheeses. Int. Dairy J. 2005, 15, 719–732. [Google Scholar] [CrossRef]
- Corsetti, A.; Rossi, J.; Gobbetti, M. Interactions between yeasts and bacteria in the smear surface-ripened cheeses. Int. J. Food Microbiol. 2001, 69, 1–10. [Google Scholar]
- Mounier, J.; Gelsomino, R.; Goerges, S.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Scherer, S.; Swings, J.; Fitzgerald, G.F.; Cogan, T.M. Surface Microflora of Four Smear-Ripened Cheeses. Appl. Environ. Microbiol. 2005, 71, 6489–6500. [Google Scholar] [CrossRef]
- Korena, K.; Krzyzankova, M.; Florianova, M.; Karasova, D.; Babak, V.; Strakova, N.; Juricova, H. Microbial Succession in the Cheese Ripening Process-Competition of the Starter Cultures and the Microbiota of the Cheese Plant Environment. Microorganisms 2023, 11, 1735. [Google Scholar] [CrossRef] [PubMed]
- Ritschard, J.; Schuppler, M. The Microbial Diversity on the Surface of Smear-Ripened Cheeses and Its Impact on Cheese Quality and Safety. Foods 2024, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Mills, D.A. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 2013, 79, 5214–5223. [Google Scholar]
- Mariani, C.; Briandet, R.; Chamba, J.F.; Notz, E.; Carnet-Pantiez, A.; Eyoug, R.; Oulahal, N. Biofilm ecology of wooden shelves used in ripening the French raw milk smear cheese Reblochon de Savoie. J. Dairy Sci. 2007, 90, 1653–1661. [Google Scholar]
- Irlinger, F.; Mounier, J. Microbial Interactions in Cheese: Implications for Cheese Quality and Safety. Curr. Opin. Biotechnol. 2009, 20, 142–148. [Google Scholar]
- Karasova, D.; Crhanova, M.; Babak, V.; Jerabek, M.; Brzobohaty, L.; Matesova, Z.; Rychlik, I. Development of Piglet Gut Microbiota at the Time of Weaning Influences Development of Postweaning Diarrhea—A Field Study. Res. Vet. Sci. 2021, 135, 59–65. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.; Potter, S.; Finn, R.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) V6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Feurer, C.; Vallaeys, T.; Corrieu, G.; Irlinger, F. Does Smearing Inoculum Reflect the Bacterial Composition of the Smear at the End of the Ripening of a French Soft, Red-Smear Cheese? J. Dairy Sci. 2004, 87, 3189–3197. [Google Scholar] [CrossRef] [PubMed]
- Irlinger, F.; Monnet, C. Temporal Differences in Microbial Composition of Époisses Cheese Rinds during Ripening and Storage. J. Dairy Sci. 2021, 104, 7500–7508. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhao, G.; Gu, L.; Solem, C. A Novel Approach for Accelerating Smear Development on Bacterial Smear-Ripened Cheeses Reduces Ripening Time and Inhibits the Growth of Listeria and Other Unwanted Microorganisms on the Rind. LWT 2022, 170, 114109. [Google Scholar] [CrossRef]
- Bockelmann, W. Smear-ripened cheeses. In Encyclopaedia of Dairy Sciences; Roginski, H., Fuquay, J.W., Fox, P.F., Eds.; Academic Press: London, UK, 2002; pp. 391–401. [Google Scholar]
- Monnet, C.; Dugat-Bony, E.; Swennen, D.; Beckerich, J.M.; Irlinger, F.; Fraud, S.; Bonnarme, P. Investigation of the Activity of the Microorganisms in a Reblochon-Style Cheese by Metatranscriptomic Analysis. Front. Microbiol. 2016, 7, 536. [Google Scholar] [CrossRef]
- Bertuzzi, A.S.; Walsh, A.M.; Sheehan, J.J.; Cotter, P.D.; Crispie, F.; McSweeney, P.L.H.; Kilcawley, K.N.; Rea, M.C. Omics-Based Insights into Flavor Development and Microbial Succession within Surface-Ripened Cheese. MSystems 2018, 3, e00211-17. [Google Scholar] [CrossRef]
- Winkler, H.; Bobst, C.; Amrein, R. Kasepflege ohne Farbe. ALP Forum 2004, 5, 1–7. [Google Scholar]
- Irlinger, F.; Yung, S.A.Y.I.; Sarthou, A.-S.; Delbès-Paus, C.; Montel, M.-C.; Coton, E.; Coton, M.; Helinck, S. Ecological and Aromatic Impact of Two Gram-Negative Bacteria (Psychrobacter Celer and Hafnia Alvei) Inoculated as Part of the Whole Microbial Community of an Experimental Smear Soft Cheese. Int. J. Food Microbiol. 2012, 153, 332–338. [Google Scholar] [CrossRef]
- Unno, R.; Suzuki, T.; Matsutani, M.; Ishikawa, M. Evaluation of the Relationships Between Microbiota and Metabolites in Soft-Type Ripened Cheese Using an Integrated Omics Approach. Front. Microbiol. 2021, 12, 681185. [Google Scholar] [CrossRef]
- Salazar, J.K.; Carstens, C.K.; Ramachandran, P.; Shazer, A.G.; Narula, S.S.; Reed, E.; Ottesen, A.; Schill, K.M. Metagenomics of Pasteurized and Unpasteurized Gouda Cheese Using Targeted 16S RDNA Sequencing. BMC Microbiol. 2018, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for Detection of Subpopulations of Bacteria Not Previously Associated with Artisanal Cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [PubMed]
- Almeida, M.; Hébert, A.; Abraham, A.-L.; Rasmussen, S.; Monnet, C.; Pons, N.; Delbès, C.; Loux, V.; Batto, J.-M.; Leonard, P. Construction of a Dairy Microbial Genome Catalog Opens New Perspectives for the Metagenomic Analysis of Dairy Fermented Products. BMC Genom. 2014, 15, 1101. [Google Scholar]
- Feurer, C.; Irlinger, F.; Spinnler, H.; Glaser, P.; Vallaeys, T. Assessment of the Rind Microbial Diversity in a Farmhouse-Produced VS a Pasteurized Industrially Produced Soft Red-Smear Cheese Using Both Cultivation and RDNA-Based Methods. J. Appl. Microbiol. 2004, 97, 546–556. [Google Scholar] [CrossRef] [PubMed]
- von Neubeck, M.; Baur, C.; Krewinkel, M.; Stoeckel, M.; Kranz, B.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015, 211, 57–65. [Google Scholar]
- Kothe, C.I.; Mohellibi, N.; Renault, P. Revealing the Microbial Heritage of Traditional Brazilian Cheeses through Metagenomics. Food Res. Int. 2022, 157, 111265. [Google Scholar] [CrossRef]
- Marchand, S.; De Block, J.; De Jonghe, V.; Coorevits, A.; Heyndrickx, M.; Herman, L. Biofilm Formation in Milk Production and Processing Environments; Influence on Milk Quality and Safety. Compr. Rev. Food Sci. Food Saf. 2012, 11, 133–147. [Google Scholar] [CrossRef]
- Mounier, J.; Coton, M.; Irlinger, F.; Landaud, S.; Bonnarme, P. Smear-Ripened Cheeses. In Cheese; Elsevier: Amsterdam, The Netherlands, 2017; pp. 955–996. [Google Scholar]
- Unno, R.; Matsutani, M.; Suzuki, T.; Kodama, K.; Matsushita, H.; Yamasato, K.; Koizumi, Y.; Ishikawa, M. Lactic Acid Bacterial Diversity in Brie Cheese Focusing on Salt Concentration and PH of Isolation Medium and Characterisation of Halophilic and Alkaliphilic Lactic Acid Bacterial Isolates. Int. Dairy J. 2020, 109, 104757. [Google Scholar] [CrossRef]
- Suzuki, T.; Matsutani, M.; Matsuyama, M.; Unno, R.; Matsushita, H.; Sugiyama, M.; Yamasato, K.; Koizumi, Y.; Ishikawa, M. Growth and Metabolic Properties of Halophilic and Alkaliphilic Lactic Acid Bacterial Strains of Marinilactibacillus Psychrotolerans Isolated from Surface-Ripened Soft Cheese. Int. Dairy J. 2021, 112, 104840. [Google Scholar] [CrossRef]
- Florianova, M.; Korena, K.; Juricova, H. Whole-Genome Analysis of Methicillin-Resistant and Methicillin-Sensitive Staphylococcus Aureus in Dry-Fermented Salami. LWT Food Sci. Technol. 2022, 170, 114042. [Google Scholar] [CrossRef]
- Irlinger, F. Safety assessment of dairy microorganisms: Coagulase-negative staphylococci. Int. J. Food Microbiol. 2008, 126, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, C.W. Cheese and Microbes; Wiley Online Library: Hoboken, NJ, USA, 2014; ISBN 1-55581-859-5. [Google Scholar]
- Rea, M.; Görges, S.; Gelsomino, R.; Brennan, N.; Mounier, J.; Vancanneyt, M.; Scherer, S.; Swings, J.; Cogan, T.M. Stability of the Biodiversity of the Surface Consortia of Gubbeen, a Red-Smear Cheese. J. Dairy Sci. 2007, 90, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Ruaro, A.; Andrighetto, C.; Torriani, S.; Lombardi, A. Biodiversity and Characterization of Indigenous Coagulase-Negative Staphylococci Isolated from Raw Milk and Cheese of North Italy. Food Microbiol. 2013, 34, 106–111. [Google Scholar]
- Eliskases-Lechner, F.; Ginzinger, W. The Bacterial Flora of Surface-Ripened Cheeses with Special Regard to Coryneforms. Le Lait 1995, 75, 571–584. [Google Scholar]
- Bockelmann, W.; Hoppe-Seyler, T.; Krusch, U.; Hoffmann, W.; Heller, K. The Microflora of Tilsit Cheese. Part 2. Development of a Surface Smear Starter Culture. Food Nahr. 1997, 41, 213–218. [Google Scholar]
- Monnet, C.; Loux, V.; Gibrat, J.-F.; Spinnler, E.; Barbe, V.; Vacherie, B.; Gavory, F.; Gourbeyre, E.; Siguier, P.; Chandler, M. The Arthrobacter arilaitensis Re117 genome sequence reveals its genetic adaptation to the surface of cheese. PLoS ONE 2010, 5, e15489. [Google Scholar]
- Yesilmen, S.; Vural, A.; Erkan, M.E.; Yildirim, I.H. Prevalence and Antimicrobial Susceptibility of Arcobacter Species in Cow Milk, Water Buffalo Milk and Fresh Village Cheese. Int. J. Food Microbiol. 2014, 188, 11–14. [Google Scholar]
- Scarano, C.; Giacometti, F.; Manfreda, G.; Lucchi, A.; Pes, E.; Spanu, C.; De Santis, E.P.L.; Serraino, A. Arcobacter Butzleri in Sheep Ricotta Cheese at Retail and Related Sources of Contamination in an Industrial Dairy Plant. Appl. Environ. Microbiol. 2014, 80, 7036–7041. [Google Scholar]
- Serraino, A.; Giacometti, F. Occurrence of Arcobacter Species in Industrial Dairy Plants. J. Dairy Sci. 2014, 97, 2061–2065. [Google Scholar]
- Liu, M.; Wei, G.; Lai, Q.; Huang, Z.; Li, M.; Shao, Z. The First Host-Associated Anaerobic Isolate of Psychrilyobacter Provides Insights into Its Potential Roles in the Abalone Gut. bioRxiv 2022. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, S.; Lv, J.; Sun, Z.; Xu, W.; Ji, C.; Liang, H.; Li, S.; Yu, C.; Lin, X. Microbial Succession and the Changes of Flavor and Aroma in Chouguiyu, A Traditional Chinese Fermented Fish. Food Biosci. 2020, 37, 100725. [Google Scholar]
- Schön, K.; Schornsteiner, E.; Dzieciol, M.; Wagner, M.; Müller, M.; Schmitz-Esser, S. Microbial Communities in Dairy Processing Environment Floor-Drains Are Dominated by Product-Associated Bacteria and Yeasts. Food Control 2016, 70, 210–215. [Google Scholar] [CrossRef]
- Justé, A.; Thomma, B.; Lievens, B. Recent Advances in Molecular Techniques to Study Microbial Communities in Food-Associated Matrices and Processes. Food Microbiol. 2008, 25, 745–761. [Google Scholar] [PubMed]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011, 150, 81–94. [Google Scholar] [CrossRef]
- Mayo, B.; Rodríguez, J.; Vázquez, L.; Flórez, A.B. Microbial Interactions Within the Cheese Ecosystem and Their Application to Improve Quality and Safety. Foods 2021, 10, 602. [Google Scholar] [CrossRef]


| Cheese | Country of Origin | Cheese Type | No of Samples | Proteins (g/100 g) | Carbohydrates (g/100 g) | Fat (g/100 g) | Salt (g/100 g) |
|---|---|---|---|---|---|---|---|
| A | CZ | Romadur | 3 | 22 | 1.5 | 20 | 2 |
| B | CZ | Quargel | 3 | 28 | 2.6 | 0.5 | 4.5 |
| C | CZ | Beer cheese | 4 | 22.7 | 1.4 | 22 | 2.8 |
| D | DE | Limburger | 3 | 23 | <0.5 | 19 | 2.3 |
| E | FR | Munster | 2 | 20 | 1 | 27 | 1.9 |
| F | FR | Petit Tourtain | 1 | 17 | <0.5 | 33 | 1.2 |
| Cheese | No of OTUs | No of Isolates |
|---|---|---|
| A | 32 | 20 |
| B | 11 | 7 |
| C | 23 | 10 |
| D | 40 | 6 |
| E | 23 | 7 |
| F | 19 | 4 |
| Cheese | CFU/g |
|---|---|
| A | 1.3–2.3 × 108 |
| B | 1.2–2.8 × 109 |
| C | 2.9–4.0 × 108 |
| D | 1.7–3.9 × 107 |
| E | 6.9–8.4 × 108 |
| F | 2.7 × 108 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korena, K.; Klimesova, A.; Florianova, M.; Krzyzankova, M.; Karasova, D.; Babak, V.; Juricova, H. Microbial Profiling of Smear-Ripened Cheeses: Identification of Starter Cultures and Environmental Microbiota. Appl. Sci. 2025, 15, 3787. https://doi.org/10.3390/app15073787
Korena K, Klimesova A, Florianova M, Krzyzankova M, Karasova D, Babak V, Juricova H. Microbial Profiling of Smear-Ripened Cheeses: Identification of Starter Cultures and Environmental Microbiota. Applied Sciences. 2025; 15(7):3787. https://doi.org/10.3390/app15073787
Chicago/Turabian StyleKorena, Kristyna, Anna Klimesova, Martina Florianova, Miroslava Krzyzankova, Daniela Karasova, Vladimir Babak, and Helena Juricova. 2025. "Microbial Profiling of Smear-Ripened Cheeses: Identification of Starter Cultures and Environmental Microbiota" Applied Sciences 15, no. 7: 3787. https://doi.org/10.3390/app15073787
APA StyleKorena, K., Klimesova, A., Florianova, M., Krzyzankova, M., Karasova, D., Babak, V., & Juricova, H. (2025). Microbial Profiling of Smear-Ripened Cheeses: Identification of Starter Cultures and Environmental Microbiota. Applied Sciences, 15(7), 3787. https://doi.org/10.3390/app15073787






