Lactobacillus rhamnosus GG as Biosensor for Oral and Systemic Health Conditions: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth
2.2. Saliva Collection
2.3. Sample Preparation and Processing for Proteomic Analysis
2.3.1. Centrifugation and Sample Processing
2.3.2. RNA Extraction
2.3.3. Solution Digestion
2.3.4. Purification Protocol
2.3.5. High-Resolution Mass Spectrometry Analysis (HRMS)
2.3.6. Data Analysis
2.4. Statistical Analysis
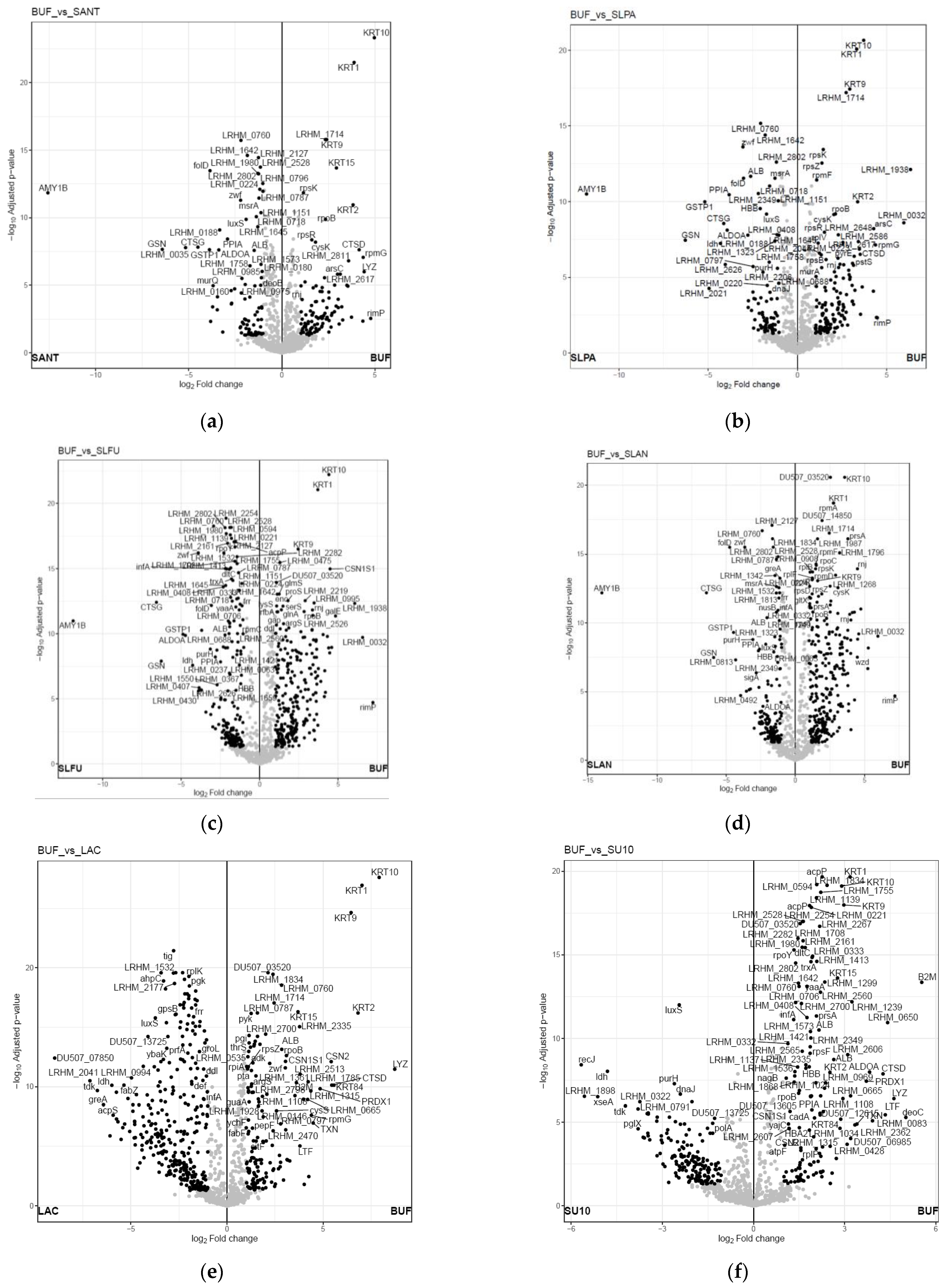
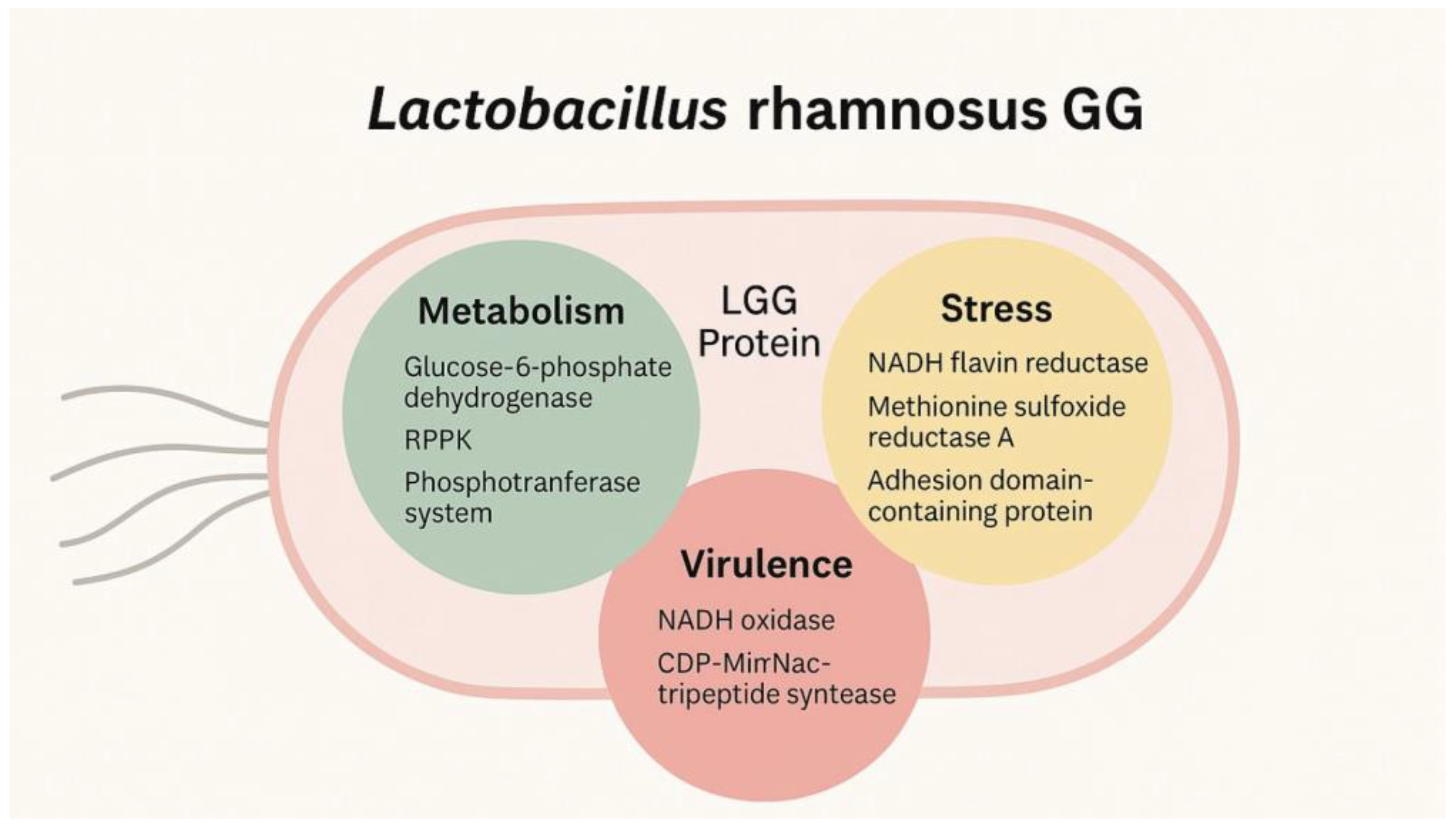
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LGG | Lactobacillus rhamnosus GG |
| EcN | Escherichia coli Nissle |
| MMP | matrix metalloproteinase |
| IL | interleukin |
| G6PD | Glucose-6-phosphate dehydrogenase |
| RPPK | Ribose-phosphate pyrophosphokinase |
| PRPP | 5-phosphate to phosphoribosyl pyrophosphate |
| MreB | Cell Shape-Determining Protein |
| Gro3P | Glycerol 3-phosphate |
| FMN | Flavin mononucleotide |
References
- Wang, J.; Schipper, H.M.; Velly, A.M.; Mohit, S.; Gornitsky, M. Salivary biomarkers of oxidative stress: A critical review. Free Radic. Biol. Med. 2015, 85, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldón, T. The human oral microbiome in health and disease: From sequences to ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Zaura, E.; Keijser, B.J.F.; Huse, S.M.; Crielaard, W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Belstrøm, D.; Jersie-Christensen, R.R.; Lyon, D.; Damgaard, C.; Jensen, L.J.; Holmstrup, P.; Olsen, J.V. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. PeerJ 2016, 4, e2433. [Google Scholar] [CrossRef]
- Börner, R.A.; Kandasamy, V.; Axelsen, A.M.; Nielsen, A.T.; Bosma, E.F. Genome editing of lactic acid bacteria: Opportunities for food, feed, pharma, and biotech. FEMS Microbiol. Lett. 2018, 366, fny291. [Google Scholar] [CrossRef]
- Sonnenborn, U. Escherichia coli strain Nissle 1917—From bench to bedside and back: History of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett. 2016, 363, fnw212. [Google Scholar] [CrossRef]
- Schultz, M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1012–1018. [Google Scholar] [CrossRef]
- Gleerup, H.S.; Hasselbalch, S.G.; Simonsen, A.H. Biomarkers for Alzheimer’s disease in saliva: A systematic review. Dis. Markers 2019, 2019, 4761054. [Google Scholar] [CrossRef]
- Sancesario, G.; Bernardini, S. AD biomarker discovery in CSF and alternative matrices. Clin. Biochem. 2019, 72, 52–57. [Google Scholar] [CrossRef]
- Schepici, G.; Silvestro, S.; Trubiani, O.; Bramanti, P.; Mazzon, E. Salivary biomarkers: Future approaches for early diagnosis of neurodegenerative diseases. Brain Sci. 2020, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Malon, R.S.; Sadir, S.; Balakrishnan, M.; Córcoles, E.P. Saliva-based biosensors: Noninvasive monitoring tool for clinical diagnostics. Biomed. Res. Int. 2014, 2014, 962903. [Google Scholar] [CrossRef] [PubMed]
- Huynh-Ba, G.; Kuonen, P.; Hofer, D.; Schmid, J.; Lang, N.P.; Salvi, G.E. The effect of periodontal therapy on the survival rate and incidence of complications of multirooted teeth with furcation involvement after an observation period of at least 5 years: A systematic review. J. Clin. Perio 2009, 36, 164–176. [Google Scholar] [CrossRef]
- Liotta, L.; Petricoin, E. Molecular profiling of human cancer. Nat. Rev. Genet. 2000, 1, 48–56. [Google Scholar] [CrossRef]
- Ideker, T.; Thorsson, V.; Ranish, J.A.; Christmas, R.; Buhler, J.; Eng, J.K.; Bumgarner, R.; Goodlett, D.R.; Aebersold, R.; Hood, L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 2001, 292, 929–934. [Google Scholar] [CrossRef]
- Adamu, J.Y.; Wawegama, N.K.; Browning, G.F.; Markham, P.F. Membrane proteins of Mycoplasma bovis and their role in pathogenesis. Res. Vet. Sci. 2013, 95, 321–325. [Google Scholar] [CrossRef]
- Mora-Montes, H.M. A perspective on the role of proteins and peptides in the virulence and pathogenesis. Curr. Protein Pept. Sci. 2019, 20, 960–961. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, Q.; Lin, Q.; Duan, Y. Emerging salivary biomarkers by mass spectrometry. Clin. Chim. Acta 2015, 438, 214–221. [Google Scholar] [CrossRef]
- Tuomola, E.M.; Ouwehand, A.C.; Salminen, S.J. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol. Med. Microbiol. 1999, 26, 137–142. [Google Scholar] [CrossRef]
- Lebeer, S. Molecular Study of Adaptation and Probiotic Factors in Lactobacillus Rhamnosus GG. Ph.D. Thesis, Faculty of Bio-engineering Sciences, KU Leuven, Leuven, Belgium, 2008. [Google Scholar]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef]
- Ionescu, A.; Brambilla, E.; Hahnel, S. Does recharging dental restorative materials with fluoride influence biofilm formation? Dent. Mater. 2019, 35, 1450–1463. [Google Scholar] [CrossRef] [PubMed]
- Corda, D.; Mosca, M.G.; Ohshima, N.; Grauso, L.; Yanaka, N.; Mariggiò, S. The emerging physiological roles of the glycerophosphodiesterase family. FEBS J. 2014, 281, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Hiraishi, M.; Haneoka, M.; Fujinaka, H.; Yano, Y. Protease inhibitor concentrations in the saliva of individuals experiencing oral dryness. BMC Oral Health 2021, 21, 661. [Google Scholar] [CrossRef]
- Akula, S.; Welinder, C.; Fu, Z.; Olsson, A.K.; Hellman, L. Identification of the Major Protein Components of Human and Cow Saliva. Int. J. Mol. Sci. 2023, 24, 16838. [Google Scholar] [CrossRef]
- Harry, E.; Monahan, L.; Thompson, L. Bacterial cell division: The mechanism and its precision. Int. Rev. Cytol. 2005, 253, 27–94. [Google Scholar] [CrossRef]
- Ren, J.; Sang, Y.; Lu, J.; Yao, Y. Protein acetylation and its role in bacterial virulence. Trends Microbiol 2017, 25, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Proctor, G.B.; Carpenter, G. The function of salivary proteins and the regulation of their secretion by salivary glands. Biomed. Rev. 1998, 9, 3–15. [Google Scholar] [CrossRef]
- Proctor, G.B.; Carpenter, G.H. Regulation of salivary gland function by autonomic nerves. Auton. Neurosci. 2007, 133, 3–18. [Google Scholar] [CrossRef]
- Ronner, P. DNA Repair and Therapy of Cancer. In Netter’s Essential Biochemistry; Elsevier: Philadelphia, PA, USA, 2018; Volume 1, pp. 10–12. ISBN 978-1-929007-63-9. [Google Scholar]
- Schwerdt, G.; Schulz, M.C.; Kopf, M.; Mildenberger, S.; Reime, S.; Gekle, M. Physiological regulation of oral saliva ion composition and flow rate are not coupled in healthy humans-Partial revision of our current knowledge required. Pflug. Arch. 2025, 477, 55–65. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tanaka, K.J.; Song, S.; Mason, K.; Pinkett, H.W. Selective substrate uptake: The role of ATP-binding cassette (ABC) importers in pathogenesis. Biochim. Biophys. Acta Biomembr. 2018, 1860, 868–877. [Google Scholar] [CrossRef]
- Browning, D.F.; Grainger, D.C.; Busby, S.J. Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression. Curr. Opin. Microbiol. 2010, 13, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Hołówka, J.; Zakrzewska-Czerwińska, J. Nucleoid-associated proteins: The small organizers that help to cope with stress. Front. Microbiol. 2020, 11, 590. [Google Scholar] [CrossRef] [PubMed]
- Kimata, K.; Takahashi, H.; Inada, T.; Postma, P.; Aiba, H. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 12914–12919. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, B.; De Mey, M. Biological Switches: Past and Future Milestones of Transcription Factor-Based Biosensors. ACS Synth. Biol. 2025, 14, 72–86. [Google Scholar] [CrossRef]
- Williams, P.; Bainton, N.J.; Swift, S.; Chhabra, S.R.; Winson, M.K.; Stewart, G.S.; Salmond, G.P.; Bycroft, B.W. Small molecule-mediated density-dependent control of gene expression in prokaryotes: Bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 1992, 100, 161–167. [Google Scholar] [CrossRef]
- Kimber, M.S.; Martin, F.; Lu, Y.; Houston, S.; Vedadi, M.; Dharamsi, A.; Fiebig, K.M.; Schmid, M.; Rock, C.O. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 52593–52602. [Google Scholar] [CrossRef]
- Foster, J.M.; Davis, P.J.; Raverdy, S.; Sibley, M.H.; Raleigh, E.A.; Kumar, S.; Carlow, C.K. Evolution of bacterial phosphoglycerate mutases: Non-homologous isofunctional enzymes undergoing gene losses, gains and lateral transfers. PLoS ONE 2010, 5, e13576. [Google Scholar] [CrossRef]
- Zhou, W.; Tsai, A.; Dattmore, D.A.; Stives, D.P.; Chitrakar, I.; D’alessandro, A.M.; Patil, S.; Hicks, K.A.; French, J.B. Crystal structure of E. coli PRPP synthetase. BMC Struct. Biol. 2019, 19, 1–7. [Google Scholar] [CrossRef]
- Schmid, J.; Heider, D.; Wendel, N.J.; Sperl, N.; Sieber, V. Bacterial glycosyltransferases: Challenges and opportunities of a highly diverse enzyme class toward tailoring natural products. Front. Microbiol. 2016, 18, 182. [Google Scholar] [CrossRef]
- Wang, D.; Kompaniiets, D.; Hu, Y.; Liu, B. Transcription and its regulation in bacteria. Front. Microbiol. 2023, 14, 1200443. [Google Scholar] [CrossRef]
- Khan, S.R.; Banerjee-Bhatnagar, N. Loss of catabolite repression function of HPr, the phosphocarrier protein of the bacterial phosphotransferase system, affects expression of the cry4A toxin gene in Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 2002, 184, 5410–5417. [Google Scholar] [CrossRef]
- Zhang, Y.; Bao, C.; Shen, L.; Tian, C.; Zang, X.; Chen, G.; Zhang, S. Microbial cold shock proteins: Overview of their function and mechanism of action. Protein Pept. Lett. 2022, 29, 133–142. [Google Scholar] [CrossRef]
- Smetana, J.H.C. Introduction to biomolecular structure and biophysics. In Principles of Protein Structure and Function; Misra, G., Ed.; Springer: Singapore, 2017; pp. 1–33. [Google Scholar]
- Surette, M.G.; Bassler, B.L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1998, 95, 7046–7050. [Google Scholar] [CrossRef] [PubMed]
- Hertzberger, R.; Arents, J.; Dekker, H.L.; Pridmore, R.D.; Gysler, C.; Kleerebezem, M.; de Mattos, M.J.T. H2O2 production in species of the Lactobacillus acidophilus group: A central role for a novel NADH-dependent flavin reductase. Appl. Environ. Microbiol. 2014, 80, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, B.M.; Stanton, C.; Fitzgerald, G.F.; Ross, R.P. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol. 2005, 71, 3060–3067. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, J.K.; Zhao, H. Identification and characterization of the flavin: NADH reductase (PrnF) involved in a novel two-component arylamine oxygenase. J. Bacteriol. 2007, 189, 8556–8563. [Google Scholar] [CrossRef]
- Oien, D.B.; Moskovitz, J. Genetic regulation of longevity and age-associated diseases through the methionine sulfoxide reductase system. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1756–1762. [Google Scholar] [CrossRef]
- Jalal, N.; Lee, S.F. The MsrAB reducing pathway of Streptococcus gordonii is needed for oxidative stress tolerance, biofilm formation, and oral colonization in mice. PLoS ONE 2020, 15, e0229375. [Google Scholar] [CrossRef]
- Weissbach, H.; Etienne, F.; Hoshi, T.; Heinemann, S.H.; Lowther, W.T.; Matthews, B.; John, G.S.; Nathan, C.; Brot, N. Peptide methionine sulfoxide reductase: Structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 2002, 397, 172–178. [Google Scholar] [CrossRef]
- Kok, M.; Bron, G.; Erni, B.; Mukhija, S. Effect of enzyme I of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology 2003, 149 Pt 9, 2645–2652. [Google Scholar] [CrossRef]
- Vadeboncoeur, C.; Pelletier, M. The phosphoenolpyruvate: Sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol. Rev. 1997, 19, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Fellers, R.T.; Greer, J.B.; LeDuc, R.D.; Thomas, P.M.; Kelleher, N.L. Proteoform-predictor: Increasing the Phylogenetic Reach of Top-Down Proteomics. J. Proteome Res. 2025, 24, 1861–1870. [Google Scholar] [CrossRef] [PubMed]
- Carballido-López, R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 2006, 70, 888–909. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.S.F. Utilizing Genetically Encoded Biosensors for Monitoring Microbial Stress at Different Scales. Ph.D. Thesis, Technical University of Denmark, Department of Biotechnology and Biomedicine, Lyngby, Denmark, 2022. [Google Scholar]
- Cumming, A.J.; Khananisho, D.; Balka, M.; Liljestrand, N.; Daley, D.O. Biosensor that detects stress caused by periplasmic proteins. ACS Synth. Biol. 2024, 13, 1477–1491. [Google Scholar] [CrossRef]
- Allen, R.G.; Tresini, M. Oxidative stress and gene regulation. Free Radic. Biol. Med. 2000, 28, 463–499. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mali Rai, P.; Ionescu, A.C.; Soggiu, A.; Panio, A.; Panda, S.; Savadori, P.; Tartaglia, G.M.; Del Fabbro, M.; Goker, F. Lactobacillus rhamnosus GG as Biosensor for Oral and Systemic Health Conditions: A Pilot Study. Appl. Sci. 2025, 15, 4809. https://doi.org/10.3390/app15094809
Mali Rai P, Ionescu AC, Soggiu A, Panio A, Panda S, Savadori P, Tartaglia GM, Del Fabbro M, Goker F. Lactobacillus rhamnosus GG as Biosensor for Oral and Systemic Health Conditions: A Pilot Study. Applied Sciences. 2025; 15(9):4809. https://doi.org/10.3390/app15094809
Chicago/Turabian StyleMali Rai, Pooja, Andrei Cristian Ionescu, Alessio Soggiu, Antonella Panio, Sourav Panda, Paolo Savadori, Gianluca Martino Tartaglia, Massimo Del Fabbro, and Funda Goker. 2025. "Lactobacillus rhamnosus GG as Biosensor for Oral and Systemic Health Conditions: A Pilot Study" Applied Sciences 15, no. 9: 4809. https://doi.org/10.3390/app15094809
APA StyleMali Rai, P., Ionescu, A. C., Soggiu, A., Panio, A., Panda, S., Savadori, P., Tartaglia, G. M., Del Fabbro, M., & Goker, F. (2025). Lactobacillus rhamnosus GG as Biosensor for Oral and Systemic Health Conditions: A Pilot Study. Applied Sciences, 15(9), 4809. https://doi.org/10.3390/app15094809








