Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Corona Sample Preparation
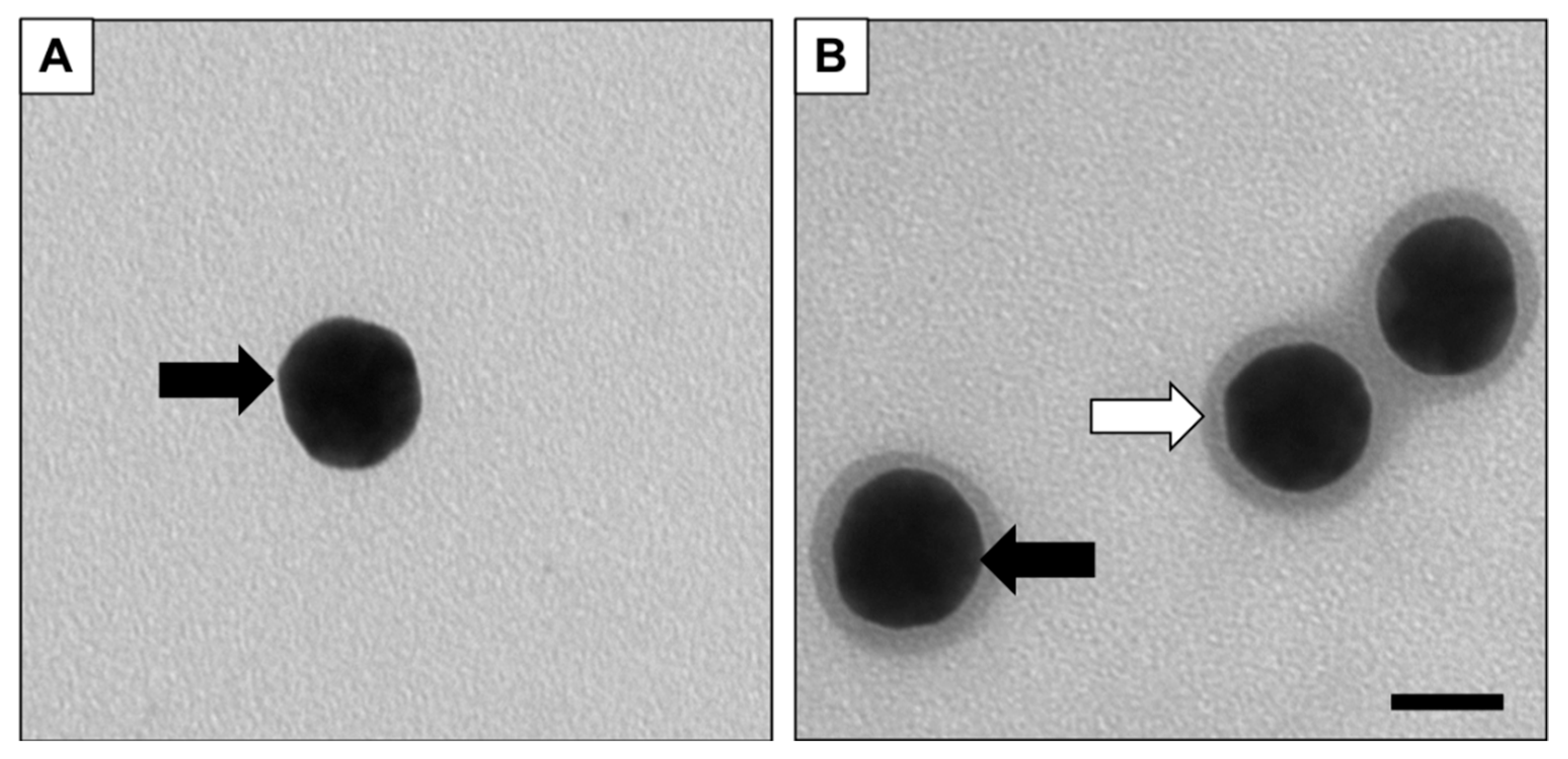
2.2. Characterization of the Nanoparticle System
2.3. Mass Spectrometry Data Collection
2.4. Data Flow
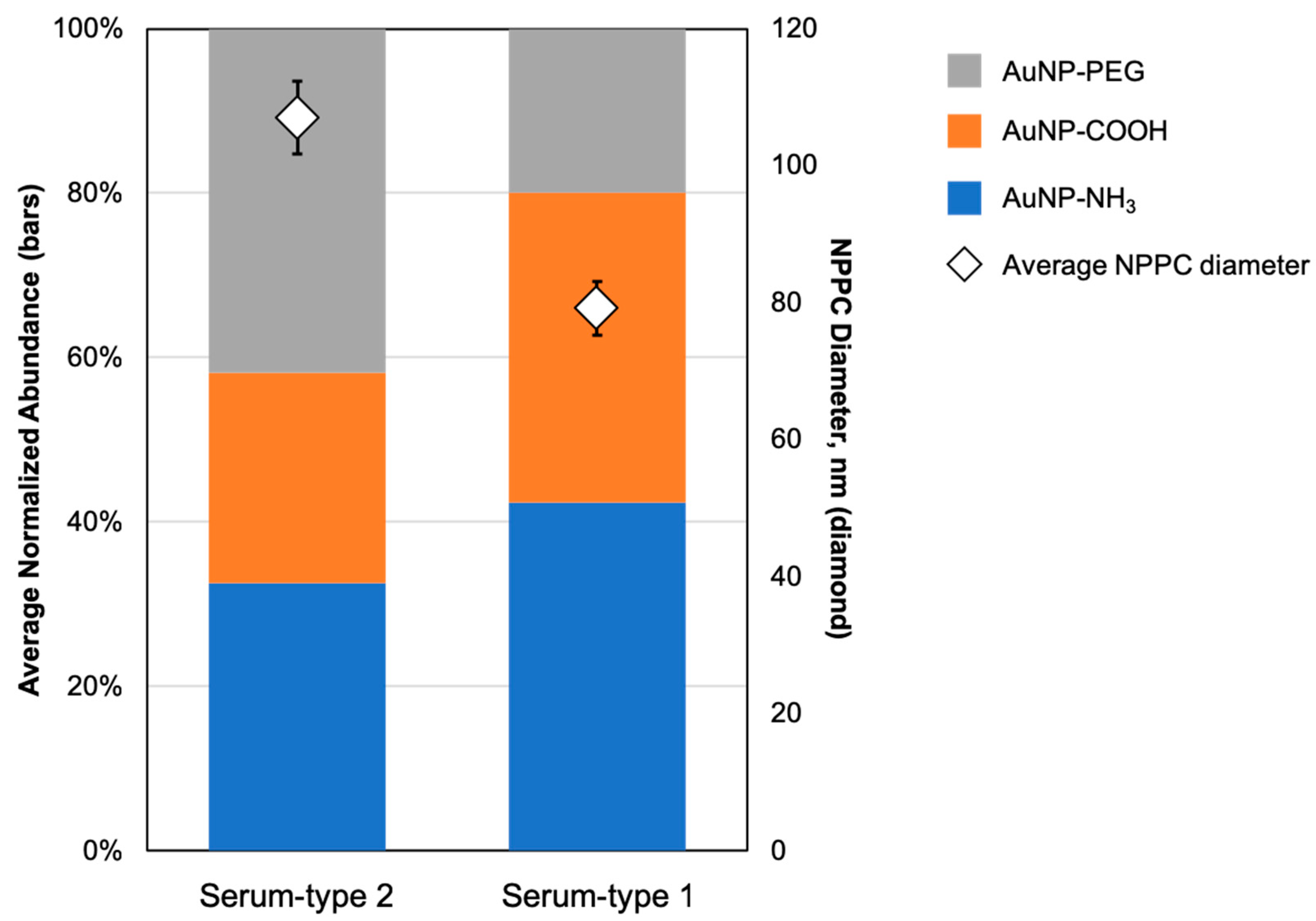
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cedervall, T.; Lynch, I.; Lindman, S.; Berggård, T.; Thulin, E.; Nilsson, H.; Dawson, K.A.; Linse, S. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 2050–2055. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Stigler, J.; Elia, G.; Lynch, I.; Cedervall, T.; Dawson, K.A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. USA 2008, 105, 14265–14270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical–chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Rose, J.; Plantevin, S.; Auffan, M.; Bottero, J.Y.; Vidaud, C. Protein corona formation for nanomaterials and proteins of a similar size: Hard or soft corona? Nanoscale 2013, 5, 1658–1668. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Silveira, C.P.; Durán, M.; Martinez, D.S.T. Silver nanoparticle protein corona and toxicity: A mini-review. J. Nanobiotechnol. 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Fenaroli, F.; Monopoli, M.P.; Åberg, C.; Dawson, K.A.; Salvati, A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano 2012, 6, 5845–5857. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, N.P.; Hurst, G.B.; Wang, W.; Foster, C.M.; Nallathamby, P.D.; Retterer, S.T. Dynamic development of the protein corona on silica nanoparticles: Composition and role in toxicity. Nanoscale 2013, 5, 6372–6380. [Google Scholar] [CrossRef] [PubMed]
- Shannahan, J.H.; Podila, R.; Aldossari, A.A.; Emerson, H.; Powell, B.A.; Ke, P.C.; Rao, A.M.; Brown, J.M. Formation of a protein corona on silver nanoparticles mediates cellular toxicity via scavenger receptors. Toxicol. Sci. 2014, 143, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Barrán-Berdón, A.L.; Pozzi, D.; Caracciolo, G.; Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Riccioli, A.; Palchetti, S.; Laganà, A. Time evolution of nanoparticle–protein corona in human plasma: Relevance for targeted drug delivery. Langmuir 2013, 29, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Corbo, C.; Molinaro, R.; Parodi, A.; Toledano Furman, N.E.; Salvatore, F.; Tasciotti, E. The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery. Nanomedicine 2016, 11, 81–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.; Ghosh, P.; Rotello, V.M. Functionalized gold nanoparticles for drug delivery. Nanomedicine 2007, 2, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Zhang, S.; Yang, Q.; Zhang, T.; Wei, X.Q.; Jiang, L.; Zhang, C.L.; Chen, Q.M.; Zhang, Z.R.; Lin, Y.F. Preformed albumin corona: A protective coating for nanoparticles based drug delivery system. Biomaterials 2013, 34, 8521–8530. [Google Scholar] [CrossRef] [PubMed]
- Salvati, A.; Pitek, A.S.; Monopoli, M.P.; Prapainop, K.; Bombelli, F.B.; Hristov, D.R.; Kelly, P.M.; Åberg, C.; Mahon, E.; Dawson, K.A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braydich-Stolle, L.K.; Schaeublin, N.M.; Murdock, R.C.; Jiang, J.; Biswas, P.; Schlager, J.J.; Hussain, S.M. Crystal structure mediates mode of cell death in TiO2 nanotoxicity. J. Nanopart. Res. 2009, 11, 1361–1374. [Google Scholar] [CrossRef]
- Kobayashi, N.; Naya, M.; Endoh, S.; Maru, J.; Yamamoto, K.; Nakanishi, J. Comparative pulmonary toxicity study of nano-TiO2 particles of different sizes and agglomerations in rats: Different short-and long-term post-instillation results. Toxicology 2009, 264, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Warheit, D.B.; Webb, T.R.; Reed, K.L.; Frerichs, S.; Sayes, C.M. Pulmonary toxicity study in rats with three forms of ultrafine-TiO2 particles: Differential responses related to surface properties. Toxicology 2007, 230, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Sayes, C.M.; Wahi, R.; Kurian, P.A.; Liu, Y.; West, J.L.; Ausman, K.D.; Warheit, D.B.; Colvin, V.L. Correlating nanoscale titania structure with toxicity: A cytotoxicity and inflammatory response study with human dermal fibroblasts and human lung epithelial cells. Toxicol. Sci. 2006, 92, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Powers, K.W.; Palazuelos, M.; Moudgil, B.M.; Roberts, S.M. Characterization of the size, shape, and state of dispersion of nanoparticles for toxicological studies. Nanotoxicology 2007, 1, 42–51. [Google Scholar] [CrossRef]
- Fujiwara, K.; Suematsu, H.; Kiyomiya, E.; Aoki, M.; Sato, M.; Moritoki, N. Size-dependent toxicity of silica nano-particles to Chlorella kessleri. J. Environ. Sci. Healthpart A 2008, 43, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Warheit, D.B.; Webb, T.R.; Sayes, C.M.; Colvin, V.L.; Reed, K.L. Pulmonary instillation studies with nanoscale TiO2 rods and dots in rats: Toxicity is not dependent upon particle size and surface area. Toxicol. Sci. 2006, 91, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Ghazani, A.A.; Chan, W.C.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006, 6, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Romoser, A.; Banerjee, N.; Zebda, R.; Sayes, C.M. The relationship between pH and zeta potential of ∼30 nm metal oxide nanoparticle suspensions relevant to in vitro toxicological evaluations. Nanotoxicology 2009, 3, 276–283. [Google Scholar] [CrossRef]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Warheit, D.B.; Webb, T.R.; Colvin, V.L.; Reed, K.L.; Sayes, C.M. Pulmonary bioassay studies with nanoscale and fine-quartz particles in rats: Toxicity is not dependent upon particle size but on surface characteristics. Toxicol. Sci. 2006, 95, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Huk, A.; Izak-Nau, E.; Reidy, B.; Boyles, M.; Duschl, A.; Lynch, I.; Dušinska, M. Is the toxic potential of nanosilver dependent on its size? Part. Fibre Toxicol. 2014, 11, 65. [Google Scholar] [CrossRef] [PubMed]
- Christensen, F.M.; Johnston, H.J.; Stone, V.; Aitken, R.J.; Hankin, S.; Peters, S.; Aschberger, K. Nano-silver: Feasibility and challenges for human health risk assessment based on open literature. Nanotoxicology 2010, 4, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.C.; Chan, A.W. Nanotoxicity: The growing need for in vivo study. Curr. Opin. Biotechnol. 2007, 18, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Docherty, S.L.; Vorderstrasse, A.; Brandon, D.; Johnson, C. Visualization of multidimensional data in nursing science. West. J. Nurs. Res. 2017, 39, 112–126. [Google Scholar] [CrossRef] [PubMed]
- Grossman, J.H.; McNeil, A.S.E. Nanotechnology in cancer medicine. Phys. Today 2012, 65, 38. [Google Scholar] [CrossRef]
- Tyner, K.M.; Zou, P.; Yang, X.; Zhang, H.; Cruz, C.N.; Lee, S.L. Product quality for nanomaterials: Current US experience and perspective. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Pissuwan, D.; Valenzuela, S.M.; Cortie, M.B. Therapeutic possibilities of plasmonically heated gold nanoparticles. Trends Biotechnol. 2006, 24, 62–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.E.; Chen, Z.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Han, G.; De, M.; Kim, C.K.; Rotello, V.M. Gold nanoparticles in delivery applications. Adv. Drug Deliv. Rev. 2008, 60, 1307–1315. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Ray, A.P. Multifunctional plasmonic shell-magnetic core nanoparticles for targeted diagnostics, isolations and photothermal treatment of tumor cells. Abstr. Pap. Am. Chem. Soc. 2012, 6, 1065–1073. [Google Scholar]
- Song, J.B.; Zhou, J.J.; Duan, H.W. Self-assembled plasmonic vesicles of SERS-encoded amphiphilic gold nanoparticles for cancer cell targeting and traceable intracellular drug delivery. J. Am. Chem. Soc. 2012, 134, 13458–13469. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.C.; Dou, S.; Xiong, M.H.; Sun, T.M.; Wang, J. Doxorubicin-tethered responsive gold nanoparticles facilitate intracellular drug delivery for overcoming multidrug resistance in cancer cells. ACS Nano 2011, 5, 3679–3692. [Google Scholar] [CrossRef] [PubMed]
- Aebersold, R.; Mann, M. Mass spectrometry-based proteomics. Nature 2003, 422, 198. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, E.P. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool opportunities and potential limitations. Mol. Cell. Proteom. 2004, 3, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Mann, M. Mass spectrometry–based proteomics turns quantitative. Nat. Chem. Biol. 2005, 1, 252. [Google Scholar] [CrossRef] [PubMed]
- Sayes, C.M.; Warheit, D.B. Characterization of nanomaterials for toxicity assessment. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2009, 1, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Yngard, R.A.; Lin, Y. Silver nanoparticles: Green synthesis and their antimicrobial activities. Adv. Colloid Interf. Sci. 2009, 145, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Docter, D.; Distler, U.; Storck, W.; Kuharev, J.; Wünsch, D.; Hahlbrock, A.; Knauer, S.K.; Tenzer, S.; Stauber, R.H. Quantitative profiling of the protein coronas that form around nanoparticles. Nat. Protoc. 2014, 9, 2030. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Murovic, J.A.; Tiel, R.L.; Kline, D.G. Management and outcomes in 318 operative common peroneal nerve lesions at the Louisiana State University Health Sciences Center. Neurosurgery 2004, 54, 1421–1428; discussion 1428–1429. [Google Scholar] [CrossRef] [PubMed]
- Whyte, A.M.; McNamara, D.; Rosenberg, I.; Whyte, A.W. Magnetic resonance imaging in the evaluation of temporomandibular joint disc displacement: A review of 144 cases. Int. J. Oral Maxillofac. Surg. 2006, 35, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, L.A.; Knilans, T.K. Syncope in children: Diagnostic tests have a high cost and low yield. J. Pediatrics 2005, 146, 355–358. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, R.V.; Nanda, A. Implanted ventricular shunts in the United States: The billion-dollar-a-year cost of hydrocephalus treatment. Neurosurgery 2005, 56, 139–144; discussion 144–145. [Google Scholar] [CrossRef] [PubMed]
- Welthagen, W.; Schnelle-Kreis, J.; Zimmermann, R. Search criteria and rules for comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry analysis of airborne particulate matter. J. Chromatogr. A 2003, 1019, 233–249. [Google Scholar] [CrossRef]
- Ewing, R.M.; Chu, P.; Elisma, F.; Li, H.; Taylor, P.; Climie, S.; McBroom-Cerajewski, L.; Robinson, M.D.; O’Connor, L.; Li, M.; et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007, 3, 89. [Google Scholar] [CrossRef] [PubMed]
- Friendly, M. Visualizing Categorical Data; SAS Institute: Cary, NC, USA, 2000. [Google Scholar]
- Howell, D.C. Statistical Methods for Psychology; Cengage Learning: Independence, KY, USA, 2009. [Google Scholar]
- Cox, D.R. Analysis of Binary Data; Taylor & Francis Group, Routledge: New York, NY, USA, 2018. [Google Scholar]
- Larose, D.T.; Larose, C.D. Discovering Knowledge in Data: An Introduction to Data Mining; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Keim, D.A.; Mansmann, F.; Thomas, J. Visual analytics: How much visualization and how much analytics? ACM Sigkdd Explor. Newsl. 2010, 11, 5–8. [Google Scholar] [CrossRef]
- Russom, P. Big data analytics. Tdwi Best Pract. Rep. 2011, 19, 1–34. [Google Scholar]
- Hegarty, M. The cognitive science of visual-spatial displays: Implications for design. Top. Cogn. Sci. 2011, 3, 446–474. [Google Scholar] [CrossRef] [PubMed]
- Moere, A.V.; Purchase, H. On the role of design in information visualization. Inf. Vis. 2011, 10, 356–371. [Google Scholar] [CrossRef]
- Aparicio, M.; Costa, C.J. Data visualization. Commun. Des. Q. Rev. 2015, 3, 7–11. [Google Scholar] [CrossRef]
- Ankley, G.T.; Daston, G.P.; Degitz, S.J.; Denslow, N.D.; Hoke, R.A.; Kennedy, S.W.; Miracle, A.L.; Perkins, E.J.; Snape, J.; Tillitt, D.E.; et al. Toxicogenomics in regulatory ecotoxicology. Environ. Sci. Technol. 2006, 40, 4055–4065. [Google Scholar] [CrossRef] [PubMed]
- Boverhof, D.R.; Zacharewski, T.R. Toxicogenomics in risk assessment: Applications and needs. Toxicol. Sci. 2005, 89, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Dowling, V.A.; Sheehan, D. Proteomics as a route to identification of toxicity targets in environmental toxicology. Proteomics 2006, 6, 5597–5604. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Spanke, S.; Rettenmeier, A.W. Protein expression profiling in chemical carcinogenesis: A proteomic-based approach. Proteomics 2011, 11, 644–656. [Google Scholar] [CrossRef] [PubMed]
- DeHaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Merrick, B.A.; Witzmann, F.A. The role of toxicoproteomics in assessing organ specific toxicity. Exp. Suppl. 2009, 99, 367–400. [Google Scholar] [Green Version]
- Rabilloud, T.; Lescuyer, P. Proteomics in mechanistic toxicology: History, concepts, achievements, caveats, and potential. Proteomics 2015, 15, 1051–1074. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Vanaerschot, M.; Jankevics, A.; Cuypers, B.; Breitling, R.; Dujardin, J.C. LC-MS metabolomics from study design to data-analysis: Using a versatile pathogen as a test case. Comput. Struct. Biotechnol. J. 2013, 4, e201301002. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, E.; Kaiser, D.L.; Fujita, D.; Van de Voorde, M. Metrology and Standardization for Nanotechnology: Protocols and Industrial Innovations; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Waters, M.D.; Fostel, J.M. Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 2004, 5, 936. [Google Scholar] [CrossRef] [PubMed]
- Hey, T.; Tansley, S.; Tolle, K.M. The Fourth Paradigm: Data-Intensive Scientific Discovery; Microsoft Research: Redmond, WA, USA, 2009; Volume 1. [Google Scholar]
- Oveland, E.; Muth, T.; Rapp, E.; Martens, L.; Berven, F.S.; Barsnes, H. Viewing the proteome: How to visualize proteomics data? Proteomics 2015, 15, 1341–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeanquartier, F.; Jean-Quartier, C.; Holzinger, A. Integrated web visualizations for protein-protein interaction databases. BMC Bioinform. 2015, 16, 195. [Google Scholar] [CrossRef] [PubMed]




| Instrument/Software | Online Database | ||||||
|---|---|---|---|---|---|---|---|
| Accession Number | Description | Average Charge (z) | Number of Peptides | Mass (Da) | Protein Abbreviation | Molecular Function | Biological Function |
| P30009 | Myristoylated alanine-rich C-kinase substrate | 5.71 | 2 | 29,795 | MARCS_RAT | Binds calmodulin, actin, and synapsin; filamentous (F) actin cross-linking protein | Actin filament organization; activation of phospholipase D activity |
| Q08368 | Acetyl-CoA decarbonylase/synthase complex subunit alpha 1 | 3.00 | 1 | 88,538 | ACDA1_METTE | Catalyzes acetyl-CoA cleavage; functions as carbon monoxide dehydrogenase | Involved in methanogenisis |
| Q02575 | Helix-loop-helix protein 1 | 5.33 | 1 | 14,618 | HEN1_HUMAN | DNA-binding protein; determines cell-type | Involved in cell differentiation/transcription |
| Q23679 | Mediator of RNA polymerase II transcription subunit 22 | 1.33 | 1 | 18,163 | MED22_CAEEL | Involved in regulated transcription of polymerase-dependent genes | Involved in transcription and transcription regulation |
| P55903 | Beta-insect depressant toxin BotIT4 | 2.00 | 2 | 6845 | SIX4_BUTOC | Affects sodium channel activation; active only in insects | Defense response |
| P81038 | Thrombin-like enzyme cerastotin | 1.20 | 3 | 11,168 | VSPA_CERCE | Cleaves fibrinogen; induces platelet aggregation | Activates detoxification |
| Q15528 | Mediator of RNA polymerase II transcription subunit 22 | 10 | 2 | 22,221 | MED22_HUMAN | Involved in regulated transcription of polymerase-dependent genes | Involved in transcription and transcription regulation |
| P0CG47 | Polyubiquitin-B | 1.75 | 4 | 25,762 | UBB_HUMAN | Activates protein kinases | Involved in DNA repair and cell-cycle regulation |
| P14111 | Kil protein | 5.80 | 1 | 6950 | VKIL_BPP22 | Essential for lytic growth | Expression causes filamentation and cell death |
| Q02155 | Hexokinase | 1.50 | 1 | 55,346 | HXK_PLAFA | Metabolizes carbohydrate | Participates in glycolysis |
| O42395 | Cellular nucleic acid-binding protein | 3.40 | 2 | 19,043 | CNBP_CHICK | DNA-binding protein; Represses sterol | Involved in transcription and transcription regulation |
| P49258 | Calmodulin-related protein 97A | 2.40 | 2 | 17,015 | CALL_DROME | Calcium-mediated signal transduction | Participates in actin filament-based movement |
| P15545 | Cytochrome c oxidase subunit 2 | 4.50 | 2 | 26,111 | COX2_STRPU | Catalyzes reduction of oxygen to water | Involved in electron transport and respiratory chain |
| P16527 | Myristoylated alanine-rich C-kinase substrate | 2.33 | 4 | 27,728 | MARCS_CHICK | Binds calmodulin, actin, and synapsin | Filamentous (F) actin cross-linking protein |
| Abbreviation | NH3-ST2 | NH3-ST1 | PEG-ST2 | PEG-ST1 | COOH-ST2 | COOH-ST1 |  |
| UBB_HUMAN | 4124 | 3575 | 12,800 | 4543 | 3729 | 3349 | |
| MARCS_RAT | 32,900 | 2622 | 18,700 | 11,700 | 896 | 5976 | |
| CALL_DROME | 28,900 | 48,500 | 18,500 | 18,600 | 61,300 | 13,900 | |
| VKIL_BPP22 | 391 | 311 | 629 | 135 | 2596 | 1315 | |
| COX2_STRPU | 737 | 167 | 930 | 469 | 22 | 98 | |
| CNBP_CHICK | 22,800 | 8270 | 9565 | 14,700 | 26,500 | 28,800 | |
| MARCS_CHICK | 11,000 | 1266 | 5820 | 3351 | 2889 | 914 | |
| MED22_CAEEL | 28,600 | 43,600 | 23,200 | 29,200 | 15,900 | 58,100 | |
| VSPA_CERCE | 10,400 | 1273 | 5528 | 5847 | 5618 | 11,900 | |
| SIX4_BUTOC | 2979 | 810 | 1697 | 2853 | 4300 | 1602 | |
| HEN1_HUMAN | 10,200 | 278 | 11,700 | 2197 | 268 | 3025 | |
| MED22_HUMAN | 23,000 | 7404 | 13,200 | 15,800 | 8656 | 32,700 | |
| ACDA1_METTE | 376 | 238 | 168 | 79 | 0 | 5 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stewart, M.; Mulenos, M.R.; Steele, L.R.; Sayes, C.M. Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics. Appl. Sci. 2018, 8, 2669. https://doi.org/10.3390/app8122669
Stewart M, Mulenos MR, Steele LR, Sayes CM. Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics. Applied Sciences. 2018; 8(12):2669. https://doi.org/10.3390/app8122669
Chicago/Turabian StyleStewart, Madison, Marina R. Mulenos, London R. Steele, and Christie M. Sayes. 2018. "Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics" Applied Sciences 8, no. 12: 2669. https://doi.org/10.3390/app8122669
APA StyleStewart, M., Mulenos, M. R., Steele, L. R., & Sayes, C. M. (2018). Differences among Unique Nanoparticle Protein Corona Constructs: A Case Study Using Data Analytics and Multi-Variant Visualization to Describe Physicochemical Characteristics. Applied Sciences, 8(12), 2669. https://doi.org/10.3390/app8122669






