Multi-Class Sleep Stage Analysis and Adaptive Pattern Recognition
Abstract
:1. Introduction
2. Methods
2.1. PSG Data Acquisition
- Wake stage (Wake): normal body functions,
- No Rapid Eye Movement 1 (NonREM1): the initial sleep stage with eyes closed,
- No Rapid Eye Movement 2 (NonREM2): the light sleep stage with slower heart rate and body temperature going down,
- No Rapid Eye Movement 3 (NonREM3): the deep sleep stage during which the body repairs and regrows tissues,
- Rapid Eye Movement (REM): the specific sleep period with faster brain activities, faster breathing and heart rate (period of dreams).
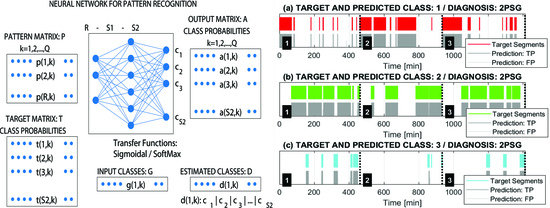
2.2. Pattern Matrix Construction
2.3. Machine Learning for Pattern Recognition
3. Results
- the EEG channels only, with patterns evaluated as mean energies in 5 selected frequency bands (1–4, 4–8, 8–12, 12–16, 16–20 Hz),
- additional features evaluated from the 4 frequency bands (0.05–0.2, 0.2–0.3, 0.3–0.4, 0.4–0.8 Hz) of the flow channel and 3 frequency bands (4–15, 15–30, 30–40 Hz) of the electro-oculogram channel.
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Looney, D.; Goverdovsky, V.; Rosenzweig, I.; Morrell, M.; Mandic, D. Wearable In-Ear Encephalography Sensor for Monitoring Sleep. Ann. Am. Thorac. Soc. 2016, 13, 2230–2233. [Google Scholar] [CrossRef] [PubMed]
- Procházka, A.; Schätz, M.; Centonze, F.; Kuchyňka, J.; Vyšata, O.; Vališ, M. Extraction of Breathing Features Using MS Kinect for Sleep Stage Detection. Signal Image Video Process. 2016, 10, 1278–1286. [Google Scholar] [CrossRef]
- Procházka, A.; Schätz, M.; Vyšata, O.; Vališ, M. Microsoft Kinect Visual and Depth Sensors for Breathing and Heart Rate Analysis. Sensors 2016, 16, 996. [Google Scholar] [CrossRef] [PubMed]
- Liaw, S.; Chen, J. Characterizing sleep stages by the fractal dimensions of electroencephalograms. Biostat. Biom. 2017, 2, 555584:1–555584:7. [Google Scholar]
- Kianzad, R.; Kordy, H. Automatic Sleep Stages Detection Based on EEG Signals Using Combination of Classifiers. J. Electr. Comput. Eng. Innov. 2013, 1, 99–105. [Google Scholar]
- Metsis, V.; Kosmopoulos, D.; Athitsos, V.; Makedon, F. Non-invasive analysis of sleep patterns via multimodal sensor input. Pers. Ubiquitous Comput. 2014, 18, 19–26. [Google Scholar] [CrossRef]
- Diykh, M.; Li, Y.; Wen, P. EEG Sleep Stages Classification Based on Time Domain Features and Structural Graph Similarity. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 24, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Fraiwan, L.; Lweesy, K.; Khasawneh, N.; Fraiwan, M.; Wenz, H.; Dickhaus, H. Classification of Sleep Stages Using Multi-wavelet Time Frequency Entropy and LDA. Methods Inf. Med. 2010, 49, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Lajnef, T.; Chaibi, S.; Ruby, P.; Aguera, P.; Eichenlaub, J.; Samet, M.; Kachouri, A.; Jerbi, K. Learning machines and sleeping brains: Automatic sleep stage classification using decision-tree multi-class support vector machines. J. Neurosci. Methods 2015, 250, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.; Long, X.; Radha, M.; Haakma, R.; Aarts, R.; Rolink, J. Sleep stage classification with ECG and respiratory effort. Physiol. Meas. 2015, 36, 2027–2040. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sotelo, J.; Osorio-Forero, A.; Jiménez-Rodríguez, A.; Cuesta-Frau, D.; Cirugeda-Roldán, E.; Peluffo, D. Automatic Sleep Stages Classification Using EEG Entropy Features and Unsupervised Pattern Analysis Techniques. Entropy 2014, 16, 6573–6589. [Google Scholar] [CrossRef]
- Sharma, R.; Pachori, R.; Upadhyay, A. Automatic sleep stages classification based on iterative filtering of electroencephalogram signals. Neural Comput. Appl. 2017, 28, 2959–2978. [Google Scholar] [CrossRef]
- Huang, C.S.; Lin, C.L.; Ko, L.W.; Liu, S.Y.; Sua, T.P.; Lin, C.T. A hierarchical classification system for sleep stage scoring via forehead EEG signals. In Proceedings of the IEEE Symposium on Computational Intelligence, Cognitive Algorithms, Mind, and Brain (CCMB), Singapore, 16–19 April 2013; pp. 1–5. [Google Scholar]
- Zhu, G.; Li, Y.; Wen, P.P. Analysis and Classification of Sleep Stages Based on Difference Visibility Graphs From a Single-Channel EEG Signal. IEEE J. Biomed. Health Inform. 2014, 18, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.-T.; Kuo, C.-E.; Zeng, J.-H.; Liang, S.-F. A transition-constrained discrete hidden Markov model for automatic sleep staging. Biomed. Eng. Online 2012, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Liu, K.; Zhang, X.; Chen, L.; Xian, X. An Obstructive Sleep Apnea Detection Approach Using a Discriminative Hidden Markov Model From ECG Signals. IEEE Trans. Biomed. Eng. 2016, 63, 1532–1542. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Pan, W.; Li, Y.; Jiang, Q.; Liu, G. A method to detect sleep apnea based on deep neural network and hidden Markov model using single-lead ECG signal. Neurocomputing 2018, 294, 94–101. [Google Scholar] [CrossRef]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. Control 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A Deep Learning Architecture for Temporal Sleep Stage Classification Using Multivariate and Multimodal Time Series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Boostani, R.; Karimzadeh, F.; Nami, M. A comparative review on sleep stage classification methods in patients and healthy individuals. Comput. Methods Programs Biomed. 2017, 140, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Aboalayon, K.; Faezipour, M.; Almuhammadi, W.; Saeid Moslehpour, S. Sleep Stage Classification Using EEG Signal Analysis: A Comprehensive Survey and New Investigation. Entropy 2016, 18, 272. [Google Scholar] [CrossRef]
- Peker, M. An efficient sleep scoring system based on EEG signal using complex-valued machine learning algorithms. Neurocomputing 2016, 207, 165–177. [Google Scholar] [CrossRef]
- Krefting, D.; Jansen, C.; Penzel, T.; Han, F.; Kantelhardt, J.W. Age and gender dependency of physiological networks in sleep. Physiol. Meas. 2017, 38, 959–975. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoo, S. Electroencephalography Analysis Using Neural Network and Support Vector Machine during Sleep. Engineering 2013, 5, 88–92. [Google Scholar] [CrossRef]
- Ozsen, S. Classification of sleep stages using class-dependent sequential feature selection and artificial neural network. Neural Comput. Appl. 2013, 23, 1239–1250. [Google Scholar] [CrossRef]
- Langkvist, A.; Karlsson, A.; Loutfi, A. Sleep Stage Classification Using Unsupervised Feature Learning. Adv. Artif. Neural Syst. 2012, 12, 1–9. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Yang, Y.T.; Wang, J.S.; Hsu, C.Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Hashizaki, M.; Nakajima, H.; Kume, K. Monitoring of Weekly Sleep Pattern Variations at Home with a Contactless Biomotion Sensor. Sensors 2014, 15, 18950–18964. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Peker, M.; Cavusoglu, A.; Celebi, F. A Comparative Study on Classification of Sleep Stage Based on EEG Signals Using Feature Selection and Classification Algorithms. J. Med. Syst. 2014, 38, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Bušková, J.; Ibarburu, V.; Šonka, K.; Ružička, E. Screening for REM sleep behavior disorder in the general population. Sleep Med. 2016, 24, 147. [Google Scholar] [CrossRef] [PubMed]
- Tagluk, E.; Sezgin, N.; Akin, M. Estimation of Sleep Stages by an Artificial Neural Network Employing EEG, EMG and EOG. J. Med. Syst. 2010, 34, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Kouchaki, K.S.; Eftaxias, K.; Sanei, S. An Adaptive Filtering Approach Using Supervised SSA for Identification of Sleep Stages From EEG. Front. Biomed. Technol. 2014, 1, 233–239. [Google Scholar]
- Venkatesh, K.; Poonguzhali, S.; Mohanavelu, K.; Adalarasu, K. Sleep Stages Classification Using Neural Network with Single Channel EEG. Int. J. Recent Adv. Eng. Technol. 2014, 2, 1–8. [Google Scholar]
- Ahuja, Y.; Yadav, S. Multiclass classification and support vector machine. Glob. J. Comput. Sci. Technol. 2012, 12, 15–19. [Google Scholar]
- Ou, G.; Murphey, Y. Multi-class pattern classification using neural networks. Pattern Recognit. 2007, 40, 4–8. [Google Scholar] [CrossRef]
- Procházka, A.; Vyšata, O.; Ťupa, O.; Mareš, J.; Vališ, M. Discrimination of Axonal Neuropathy Using Sensitivity and Specificity Statistical Measures. Neural Comput. Appl. 2014, 25, 1349–1358. [Google Scholar] [CrossRef]
- Procházka, A.; Kuchyňka, J.; Suarez Araujo, C.P.; Vyšata, O. Adaptive Segmentation of Multimodal Polysomnography Data for Sleep Stages Detection. In Proceedings of the 22nd International Conference on Digital Signal Processing, London, UK, 23–25 August 2017; pp. 1–4. [Google Scholar]
- Dursun, M.; Ozsen, S.; Yucelbas, C.; Yucelbas, S.; Tezel, G.; Kuccukturk, S.; Yosunkaya, S. A new approach to eliminating EOG artifacts from the sleep EEG signals for the automatic sleep stage classification. Neural Comput. Appl. 2017, 28, 3095–3112. [Google Scholar] [CrossRef]
- Saeid, S. Adaptive Processing of Brain Signals; John Wiley & Sons Ltd.: Chichester, UK, 2013. [Google Scholar]
- Saeid, S.; Chambers, J. EEG Signal Processing; John Wiley & Sons Ltd.: Chichester, UK, 2007. [Google Scholar]
- Chaparro-Vargas, R.; Cvetkovic, D. A single-trial toolbox for advanced sleep polysomnographic preprocessing. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan, 3–7 July 2013; pp. 5829–5832. [Google Scholar]
- Mares, J.; Vysata, O.; Prochazka, A.; Valis, M. Age-Dependent Complex Noise Fluctuation in the Brain. Physiol. Meas. 2013, 34, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Procházka, A.; Vyšata, O.; Vališ, M.; Ťupa, O.; Schatz, M.; Mařík, V. Bayesian classification and analysis of gait disorders using image and depth sensors of Microsoft Kinect. Digit. Signal Process. 2015, 47, 169–177. [Google Scholar] [CrossRef]
- Everson, R.; Fieldsend, J. Multi-class ROC analysis from a multi-objective optimisation perspective. Pattern Recognit. Lett. 2006, 27, 918–927. [Google Scholar] [CrossRef] [Green Version]
- Carlos, J.; Caballero, F.; Martinez, F.; Hervas, C.; Gutierrez, P. Sensitivity versus Accuracy in Multiclass Problems Using Memetic Pareto Evolutionary Neural Network. IEEE Trans. Neural Netw. 2010, 21, 750–770. [Google Scholar]
- Landgrebe, T.; Duin, R. Approximating the multiclass ROS by pairwise analysis. Pattern Recognit. Lett. 2007, 28, 1747–1758. [Google Scholar] [CrossRef]
- Espiritu, J. Aging-Related Sleep Changes. Clin. Geriatr. Med. 2008, 24, 1–14. [Google Scholar] [CrossRef] [PubMed]






| Diagnosis | M-Male | |||
| Number | Mean Age (Year) | STD | ||
| 1 | Healthy individuals | 36 | 40.0 | 15.9 |
| 2 | Sleep Apnea (SA) | 48 | 50.3 | 14.2 |
| 3 | Restless Leg (RL) Syndrome | 8 | 59.9 | 9.1 |
| 4 | SA and RL Syndrome | 19 | 58.3 | 9.0 |
| Diagnosis | F: Females | |||
| Number | Mean Age (Year) | STD | ||
| 1 | Healthy individuals | 27 | 40.8 | 14.7 |
| 2 | Sleep Apnea (SA) | 29 | 54.7 | 14.4 |
| 3 | RL Syndrome | 8 | 48.9 | 12.6 |
| 4 | RL Syndrome and SA | 9 | 52.7 | 20.4 |
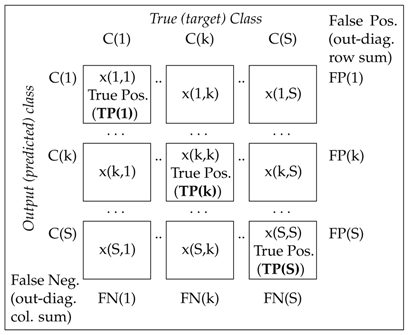
| Characteristics | Definition | Comment |
|---|---|---|
Precision of class k (pos. predict. value) | Probability of correct classification of class k related to the number of instants classified to class k | |
Sensitivity of class k (True positive rate, recall) | Probability of correct classification of class k related to the number of instants belonging to class k | |
Specificity of class k (True negative rate) | , | Probability of incorrect classification of class k related to the number of instants not classified to class k |
False positive rate | Probability of positive classification for the negative set (1-specificity) | |
Accuracy | Probability of global correct classification |
| Method | Parameters | Accuracy (%) | Cross-Valid. |
|---|---|---|---|
| 79.58 | 0.13 | ||
| Neural Net | 87.08 | 0.10 | |
| 88.33 | 0.12 | ||
| k-Nearest | 82.9 | 0.35 | |
| Neigbour | 77.9 | 0.29 | |
| Decision Tree | 85.7 | 0.14 |
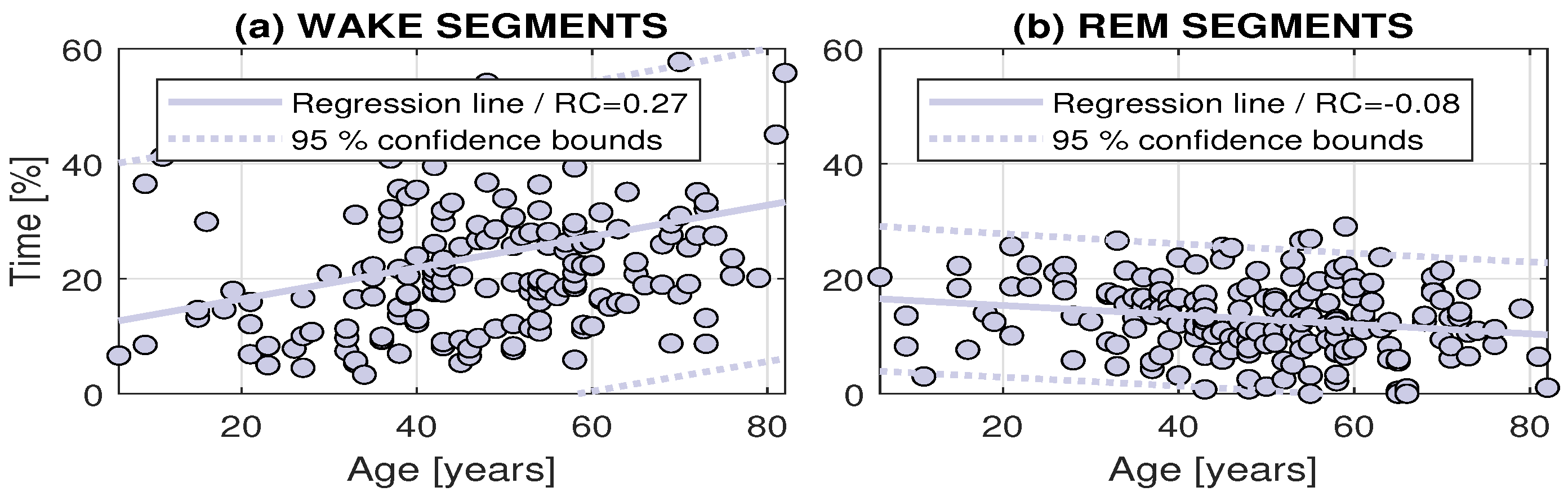
| Diagnosis | Wake Segments | REM Segments | |||
|---|---|---|---|---|---|
| RC | Confidence Bounds | RC | Confidence Bounds | ||
| All | 0.272 | (0.145, 0.399) | −0.082 | (−0.141, −0.023) | |
| 1 | 0.437 | (0.227, 0.648) | −0.094 | (−0.198, 0.010) | |
| 2 | 0.248 | (0.055, 0.442) | −0.119 | (−0.216, −0.022) | |
| 3 | −0.064 | (−0.687, 0.559) | 0.009 | (−0.334, 0.353) | |
| 4 | 0.178 | (−0.377, 0.734) | 0.057 | (−0.133, 0.246) | |
| Diagnosis (Male) | Precission (%) | Recall (%) | Acc. | Perf. | 10-Fold | ||||||
| C1 | C2 | C3 | C1 | C2 | C3 | Cross-Valid. | |||||
| 1 | Healthy Ind. | - F5 | 89.4 | 90.1 | 81.8 | 89.9 | 92.6 | 71.7 | 88.9 | 0.0977 | 0.15 |
| - F12 | 89.8 | 90.7 | 79.5 | 91.6 | 92.2 | 70.3 | 89.1 | 0.0941 | 0.12 | ||
| 2 | Sleep Apnea | - F5 | 92.9 | 94.8 | 79.7 | 94.8 | 95.4 | 72.5 | 92.7 | 0.0632 | 0.11 |
| - F12 | 93.8 | 95.4 | 81.4 | 95.3 | 95.6 | 76.5 | 93.5 | 0.0598 | 0.07 | ||
| 3 | RL Syndrome | - F5 | 84.5 | 94.7 | 81.7 | 88.8 | 92.7 | 82.5 | 90.1 | 0.0845 | 0.13 |
| - F12 | 86.0 | 95.0 | 84.3 | 89.3 | 93.5 | 85.1 | 91.1 | 0.0758 | 0.10 | ||
| 4 | SA and RL Syn. | - F5 | 91.5 | 88.1 | 76.7 | 86.8 | 94.0 | 62.5 | 87.8 | 0.0996 | 0.19 |
| - F12 | 90.9 | 89.3 | 83.2 | 88.3 | 94.2 | 67.6 | 89.1 | 0.0973 | 0.15 | ||
| Diagnosis (Female) | Precission (%) | Recall (%) | Acc. | Perf. | 10-Fold | ||||||
| C1 | C2 | C3 | C1 | C2 | C3 | Cross-Valid. | |||||
| 1 | Healthy Ind. | - F5 | 82.9 | 90.6 | 72.5 | 77.5 | 92.1 | 73.8 | 86.0 | 0.1186 | 0.16 |
| - F12 | 83.0 | 91.9 | 76.9 | 81.8 | 92.5 | 76.2 | 87.6 | 0.1055 | 0.14 | ||
| 2 | Sleep Apnea | - F5 | 86.0 | 91.7 | 73.5 | 90.6 | 93.3 | 62.4 | 88.0 | 0.1061 | 0.17 |
| - F12 | 87.5 | 92.2 | 74.7 | 92.4 | 93.4 | 64.1 | 88.7 | 0.1015 | 0.15 | ||
| 3 | RL Syndrome | - F5 | 85.9 | 92.0 | 81.3 | 86.3 | 93.1 | 75.9 | 89.3 | 0.1006 | 0.15 |
| - F12 | 87.5 | 92.2 | 80.0 | 86.3 | 93.5 | 76.7 | 89.6 | 0.0953 | 0.12 | ||
| 4 | SA and RL Syn. | - F5 | 83.0 | 89.7 | 81.9 | 84.0 | 91.4 | 72.6 | 86.9 | 0.1063 | 0.21 |
| - F12 | 84.0 | 90.9 | 83.9 | 86.1 | 91.9 | 75.3 | 88.1 | 0.1049 | 0.14 | ||
| Training Set | Target Class | |||||
| k | 1 | 2 | 3 | |||
| Output Class | 1 | 4644 | 601 | 236 | 837 | 84.7% |
| 2 | 368 | 10,971 | 534 | 902 | 92.4% | |
| 3 | 152 | 405 | 1958 | 557 | 77.9% | |
| 520 | 1006 | 770 | ACC: 88.4% | |||
| 89.9% | 91.6% | 71.8% | Error: 11.6% | |||
| Validation Set | Target Class | |||||
| k | 1 | 2 | 3 | |||
| Output Class | 1 | 1007 | 144 | 38 | 182 | 84.7% |
| 2 | 85 | 2304 | 114 | 199 | 92.0% | |
| 3 | 44 | 95 | 421 | 139 | 75.2% | |
| 129 | 239 | 152 | ACC: 87.8% | |||
| 88.6% | 90.6% | 73.5% | Error: 12.2% | |||
| Test Set | Target Class | |||||
| k | 1 | 2 | 3 | |||
| Output Class | 1 | 1007 | 155 | 49 | 204 | 83.2% |
| 2 | 76 | 2337 | 106 | 182 | 92.8% | |
| 3 | 34 | 79 | 416 | 113 | 78.6% | |
| 110 | 234 | 155 | ACC: 88.3% | |||
| 90.2% | 90.9% | 72.9% | Error: 11.7% | |||
| Sex | Accuracy (%) | 10-Fold Cross-Valid. | Class | Precission (%) | ||||
|---|---|---|---|---|---|---|---|---|
| F5 | F12 | F5 | F12 | F5 | F12 | |||
| Male | 89.9 | 90.7 | 0.15 | 0.11 | Class 1: Wake | 87.0 | 87.8 | |
| Female | 87.6 | 88.5 | 0.17 | 0.14 | Class 2: NonREM | 91.5 | 92.2 | |
| Mean | 88.7 | 89.6 | 0.16 | 0.12 | Class 3: REM | 78.6 | 80.5 | |
| Reference | Sleep Stages | Signals and Features | Model | Dataset | Accuracy |
|---|---|---|---|---|---|
| Fraiwan et al. [8] 2010 | Wake, REM NonREM1-4 | EEG (21 time-freq. features) | wavelet entropy and discriminant analysis | 32 subjects | 84.0 % |
| Pan et al. [15] 2012 | Wake, REM, SWS NonREM1-2 | EEG, EOG, EMG (13 energy features) | discrete hidden Markov model | 20 subjects | 85.3 % |
| Hsu et al. [27] 2013 | Wake, REM NonREM1-2 | EEG (6 energy features) | Elman recurrent neural classifier | 8 subjects | 87.2 % |
| Peker [22] 2016 | Wake, REM NonREM1-4 | EEG (8 complex features) | complex-valued neural networks | 8 subjects | 91.6 % |
| Sors et al. [18] 2018 | Wake, REM NonREM1-4 | raw EEG (no features selected) | deep convolutional neural network | 5728 segments | 87.0 % |
| Current Study | Wake, REM NonREM | EEG, Flow, EOG (12 energy features) | Bayesian neural network classifier | 184 subjects (own data) | 89.6 % |
| Current Study | Wake, REM NonREM | EEG (5 energy features) | Bayesian neural network classifier | 184 subjects (own data) | 88.7 % |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Procházka, A.; Kuchyňka, J.; Vyšata, O.; Cejnar, P.; Vališ, M.; Mařík, V. Multi-Class Sleep Stage Analysis and Adaptive Pattern Recognition. Appl. Sci. 2018, 8, 697. https://doi.org/10.3390/app8050697
Procházka A, Kuchyňka J, Vyšata O, Cejnar P, Vališ M, Mařík V. Multi-Class Sleep Stage Analysis and Adaptive Pattern Recognition. Applied Sciences. 2018; 8(5):697. https://doi.org/10.3390/app8050697
Chicago/Turabian StyleProcházka, Aleš, Jiří Kuchyňka, Oldřich Vyšata, Pavel Cejnar, Martin Vališ, and Vladimír Mařík. 2018. "Multi-Class Sleep Stage Analysis and Adaptive Pattern Recognition" Applied Sciences 8, no. 5: 697. https://doi.org/10.3390/app8050697
APA StyleProcházka, A., Kuchyňka, J., Vyšata, O., Cejnar, P., Vališ, M., & Mařík, V. (2018). Multi-Class Sleep Stage Analysis and Adaptive Pattern Recognition. Applied Sciences, 8(5), 697. https://doi.org/10.3390/app8050697