Automatic Zebrafish Egg Phenotype Recognition from Bright-Field Microscopic Images Using Deep Convolutional Neural Network
Abstract
Featured Application
Abstract
1. Introduction
- Imbalanced training dataset. In biological research, it is difficult to collect a balanced number of fertilized and unfertilized egg samples as the training dataset. The imbalanced training set will result in insufficient classification ability for the category with fewer training samples, leading to unsuccessful network training.
- Small training dataset. The training of deep neural network requires no less than thousands of training samples. However, it is difficult to collect enough training data for a specific biological image analysis task. Small training sample set will lead to overfitting of the training data, hampering the generalization ability of the network.
- Subtle inter-class differences. In bright-field microscopic images, fertilized and unfertilized zebrafish eggs usually demonstrate subtle inter-class differences. This challenging problem becomes a technical bottleneck for automated zebrafish egg image analysis.
2. Materials and Methods
2.1. Data Collection
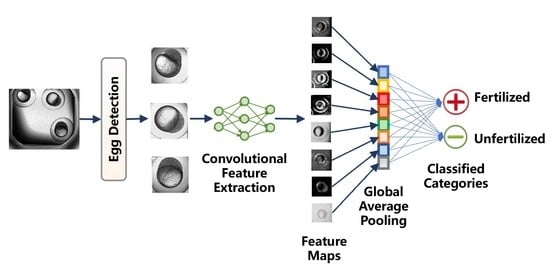
2.2. Method Workflow
2.3. Egg Detection
2.4. Convolutional Feature Extraction
2.5. Global Average Pooling Classifier
3. Results
3.1. Zebrafish Egg Classification Accuracy
3.2. Comparison between Regular Fully Connected Layers and Global Average Pooling
3.3. Comparison between Augmented Dataset and Original Dataset
3.4. Comparison with Other Zebrafish Embryo Microscopic Image Analysis Studies
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blaser, R.E.; Vira, D.G. Experiments on learning in zebrafish (Danio rerio): A promising model of neurocognitive function. Neurosci. Biobehav. Rev. 2014, 42, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; LaBarbera, D.V. 3D High-Content Screening of Organoids for Drug Discovery. In Chemistry, Molecular Sciences and Chemical Engineering; Elsevier Inc.: Amsterdam, The Netherlands, 2017; pp. 388–415. [Google Scholar] [CrossRef]
- Unser, M.; Sage, D.; Delgado-Gonzalo, R. Advanced image processing for biology, and the Open Bio Image Alliance (OBIA). In Proceedings of the 21st European Signal Processing Conference, Marrakech, Morocco, 9–13 September 2013; pp. 1–5. [Google Scholar]
- Liu, R.; Lin, S.; Rallo, R.; Zhao, Y.; Damoiseaux, R.; Xia, T.; Lin, S.; Nel, A.; Cohen, Y. Automated phenotype recognition for zebrafish embryo based in vivo high throughput toxicity screening of engineered nano-materials. PLoS ONE 2012, 7, e35014. [Google Scholar] [CrossRef] [PubMed]
- Jeanray, N.; Marée, R.; Pruvot, B.; Stern, O.; Geurts, P.; Wehenkel, L.; Muller, M. Phenotype classification of zebrafish embryos by supervised learning. PLoS ONE 2015, 10, e0116989. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, S.; Giles, C.L.; Tsoi, A.C.; Back, A.D. Face recognition: A convolutional neural-network approach. IEEE Trans. Neural Netw. 1997, 8, 98–113. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, X.; Tang, X. Deep convolutional network cascade for facial point detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, 23–28 June 2013; pp. 3476–3483. [Google Scholar]
- Hu, B.; Lu, Z.; Li, H.; Chen, Q. Convolutional neural network architectures for matching natural language sentences. Adv. Neural Info. Process. Syst. 2014, 3, 2042–2050. [Google Scholar]
- Kalchbrenner, N.; Grefenstette, E.; Blunsom, P. A convolutional neural network for modelling sentences. In Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics, Baltimore, ML, USA, 23–25 June 2014; pp. 655–665. [Google Scholar]
- Ji, S.; Xu, W.; Yang, M.; Yu, K. 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Tompson, J.; Jain, A.; LeCun, Y.; Bregler, C. Joint training of a convolutional network and a graphical model for human pose estimation. In Proceedings of the Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 1799–1807. [Google Scholar]
- Nielsen, M.A. Neural Networks and Deep Learning. 2015. Available online: http://neuralnetworksanddeeplearning.com/about.html (accessed on 15 August 2019).
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Info. Process. Syst. 2012, 25, 2012. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–July 2016; pp. 770–778. [Google Scholar]
- Angermueller, C.; Pärnamaa, T.; Parts, L.; Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 2016, 12, 878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, R.; Zeng, T.; Sun, Q.; Kumar, S.; Ye, J.; Ji, S. Deep model based transfer and multi-task learning for biological image analysis. IEEE Trans. Big Data 2016, 1475–1484. [Google Scholar] [CrossRef]
- Lin, S.; Zhao, Y.; Xia, T.; Meng, H.; Ji, Z.; Liu, R.; George, S.; Xiong, S.; Wang, X.; Zhang, H.; et al. High content screening in zebrafish speeds up hazard ranking of transition metal oxide nanoparticles. ACS Nano 2011, 5, 7284–7295. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Xception: Deep Learning with Separable Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Wager, S.; Wang, S.; Liang, P. Dropout training as adaptive regularization. In Proceedings of the 26th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013; pp. 351–359. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Min, L.; Qiang, C.; Yan, S. Network in Network. In Proceedings of the International Conference on Learning Representations, Banff, AL, Canada, 14–16 April 2014; pp. 1–8. [Google Scholar]
- Ravishankar, H.; Sudhakar, P.; Venkataramani, R.; Thiruvenkadam, S.; Annangi, P.; Babu, N.; Vaidya, V. Understanding the Mechanisms of Deep Transfer Learning for Medical Images. In Deep Learning and Data Labeling for Medical Applications, Proceedings of the International Workshop on Deep Learning in Medical Image Analysis, Athens, Greece, 21 October 2016; Carneiro, G., Mateus, D., Eds.; Springer International Publishing AG: Cham, Switzerland, 2016. [Google Scholar]
- Zhou, Z.; Shin, J.; Lei, Z.; Gurudu, S.; Gotway, M.; Liang, J. Fine-tuning Convolutional Neural Networks for Biomedical Image Analysis: Actively and Incrementally. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 22–25 July 2017; pp. 4761–4772. [Google Scholar]
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.W.; Chen, J.; Liu, X.; Pietikainen, M. Deep Learning for Generic Object Detection: A Survey. arXiv 2018, arXiv:1809.02165. [Google Scholar]







| Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Mean ± Std. |
|---|---|---|---|---|---|
| 93.3% | 93.2% | 98.8% | 93.7% | 95.9% | 95.0 ± 2.2% |
| Method | Sensitivity | Specificity | Precision | Accuracy |
|---|---|---|---|---|
| with Data Augmentation | 97.3% | 99.2% | 99.2% | 98.8% |
| without Data Augmentation | 68.0% | 99.6& | 99.4% | 83.8% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, S.; Long, L.; Lin, S.; Cong, F. Automatic Zebrafish Egg Phenotype Recognition from Bright-Field Microscopic Images Using Deep Convolutional Neural Network. Appl. Sci. 2019, 9, 3362. https://doi.org/10.3390/app9163362
Shang S, Long L, Lin S, Cong F. Automatic Zebrafish Egg Phenotype Recognition from Bright-Field Microscopic Images Using Deep Convolutional Neural Network. Applied Sciences. 2019; 9(16):3362. https://doi.org/10.3390/app9163362
Chicago/Turabian StyleShang, Shang, Ling Long, Sijie Lin, and Fengyu Cong. 2019. "Automatic Zebrafish Egg Phenotype Recognition from Bright-Field Microscopic Images Using Deep Convolutional Neural Network" Applied Sciences 9, no. 16: 3362. https://doi.org/10.3390/app9163362
APA StyleShang, S., Long, L., Lin, S., & Cong, F. (2019). Automatic Zebrafish Egg Phenotype Recognition from Bright-Field Microscopic Images Using Deep Convolutional Neural Network. Applied Sciences, 9(16), 3362. https://doi.org/10.3390/app9163362