Extracting Retinal Anatomy and Pathological Structure Using Multiscale Segmentation
Abstract
1. Introduction
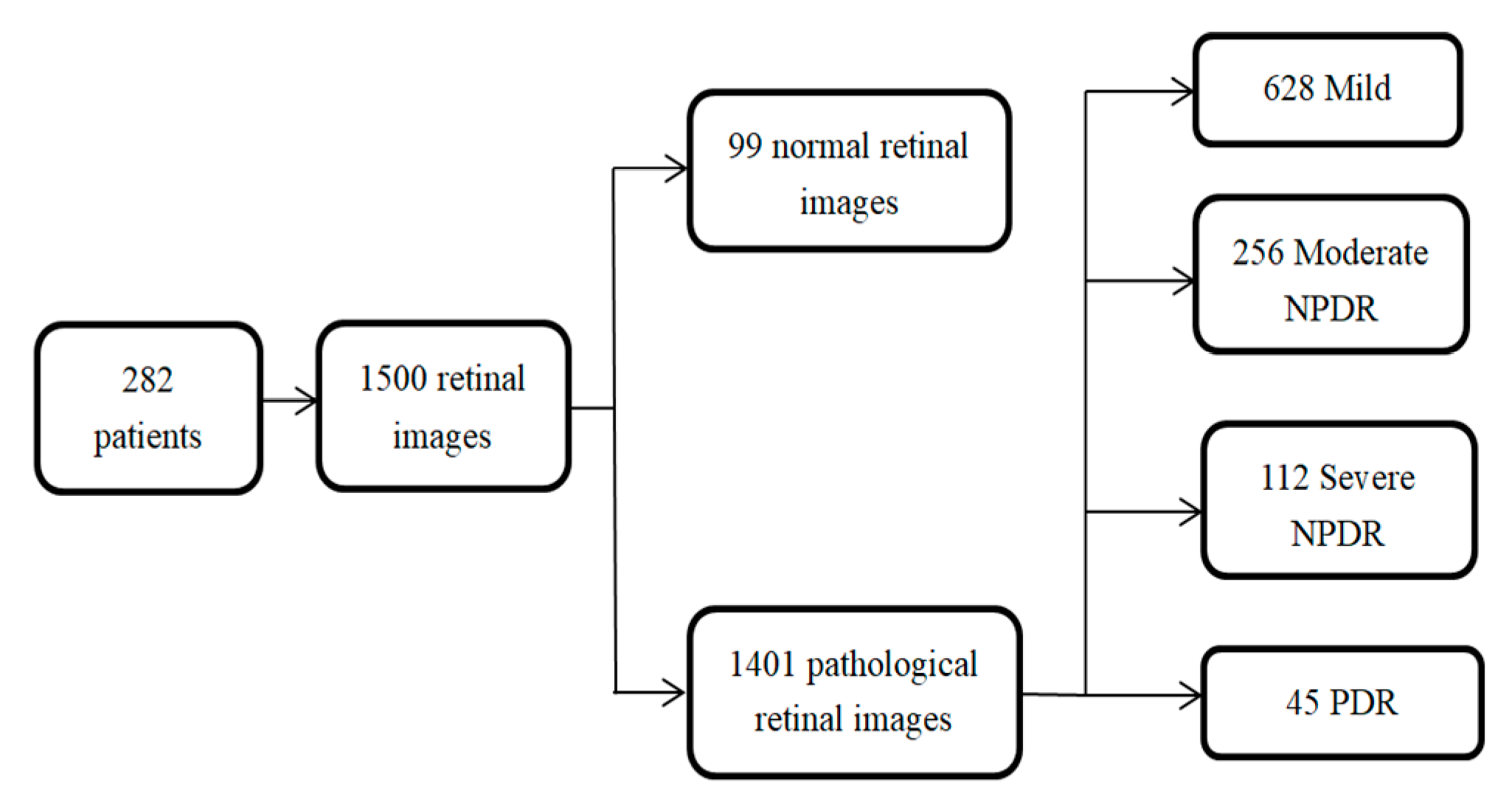
- A task-specific network that is designed to compute the retinal anatomy and pathological structure of a fundus. We collected a large-scale retinal dataset composed of 1500 images from 282 proliferative diabetic retinopathy (PDR) and non-proliferative diabetic retinopathy (NPDR) patients. PDR and NPDR are indicators for judging diabetic retinopathy grades. The difference between PDR and NPDR lies in blood vessel formation. We also know that PDR develops after NPDR. Therefore, we design an adaptive thresholding method to segment the FOV boundary from the background of the retinal fundus image. The design of the network can often support the boundary FOV segmentation and the automated segmentation training system, which is consistent with clinical requirements.
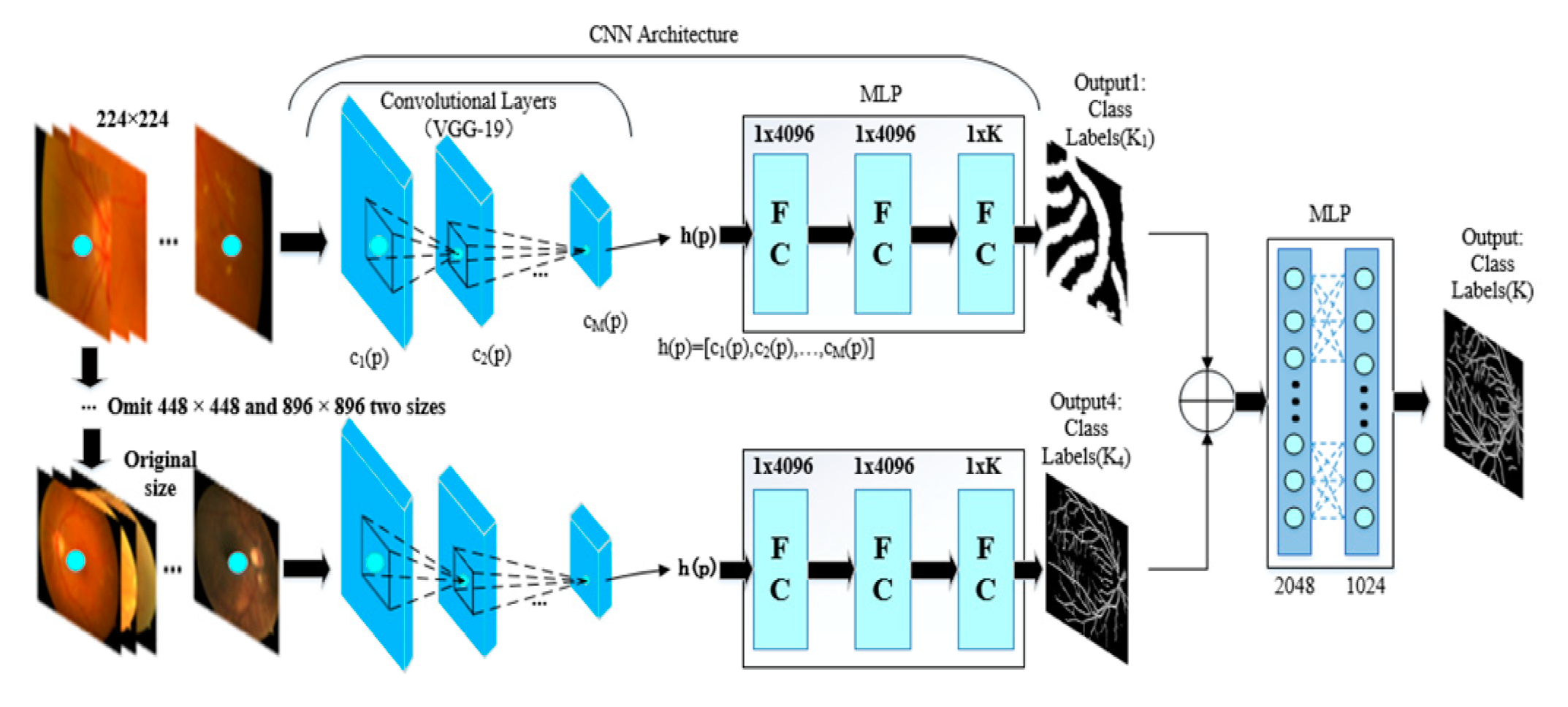
- A multiscale network is proposed to achieve the final segmentation step, e.g., a four-input branch that enhances the integrity of segmentation and the ability to generalize the training model. The purpose does not neglect the integrity of the structure while training. The network is also error-tolerable, which means that the diagnostic accuracy can be guaranteed even when one of the branches is not accurate enough.
2. Methodology
2.1. PixelNet
2.2. Multiscale Segmentation Network
3. Experiments
3.1. Data Preparation
3.1.1. Public Database
3.1.2. Optimized Database
3.2. Segmentation Criteria
3.3. Training
3.4. Comparison to other Segmentation Networks
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Abràmoff, M.D.; Folk, J.C.; Han, D.P.; Walker, J.D.; Williams, D.F.; Russell, S.R.; Lamard, M. Automated analysis of retinal images for detection of referable diabetic retinopathy. JAMA Ophthalmol. 2013, 131, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L. Research on Vascular Segmentation Technology in Fundus Images; Nanjing Aerospace University: Nanjing, China, 2011. [Google Scholar]
- Yao, C. Research and Application of Fundus Image Segmentation Method; Beijing Jiao Tong University: Beijing, China, 2009. [Google Scholar]
- Ganjee, R.; Azmi, R.; Gholizadeh, B. An Improved Retinal Vessel Segmentation Method Based on High Level Features for Pathological Images. J. Med. Syst. 2014, 38, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Nayebifar, B.; Abrishami, M.H. A novel method for retinal vessel tracking using particle filters. In Computers in Biology & Medicine; McGraw-Hill: New York, NY, USA, 1965; Volume 43, pp. 541–548. [Google Scholar]
- Ricci, E.; Perfetti, R. Retinal Blood Vessel Segmentation Using Line Operators and Support Vector Classification. IEEE Trans. Med. Imaging 2017, 26, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Liu, L.; Zhang, Z. Superpixel-Based Segmentation for 3D Prostate MR Images. IEEE Trans. Med. Imaging 2016, 35, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.C.T.; Benameur, S.; Mignotte, M.; Lavoie, F. Superpixel and multi-atlas based fusion entropic model for the segmentation of X-ray images. Med. Image Anal. 2018, 48, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, D. Watershed Segmentation for Breast Tumor in 2-D Sonography. Ultrasound Med. Biol. 2004, 30, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Alireza, B.; Mohammad, A.P.; Alireza, R. Automatic liver segmentation in MRI images using an iterative watershed algorithm and artificial neural network. Biomed. Signal Process. Control 2012, 7, 429–437. [Google Scholar]
- Ciecholewski, M. Malignant and Benign Mass Segmentation in Mammograms Using Active Contour Methods. Symmetry 2017, 9, 277. [Google Scholar] [CrossRef]
- Ciecholewski, M.; Jan Henryk, S. Semi–Automatic Corpus Callosum Segmentation and 3D Visualization Using Active Contour Methods. Symmetry 2018, 10, 589. [Google Scholar] [CrossRef]
- Zhao, Y.; Rada, L.; Chen, K.; Harding, S.P.; Zheng, Y. Automated Vessel Segmentation Using Infinite Perimeter Active Contour Model with Hybrid Region Information with Application to Retinal Images. IEEE Trans. Med. Imaging 2015, 34, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, A.F.; Yassine, I.A.; Fahmy, A.S. Convolutional neural networks for deep feature learning in retinal vessel segmentation. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 385–388. [Google Scholar]
- Staal, J.; Abràmoff, M.D.; Niemeijer, M. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Xu, Y.; Wong, D.W.K.; Liu, J. Retinal vessel segmentation via deep learning network and fully-connected conditional random fields. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Praha, Czech Republic, 9–16 April 2016; pp. 698–701. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation, IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 142–158. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Proceedings of the International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Kirbas, C.; Quek, F. A review of vessel extraction techniques and algorithms. ACM Comput. Surv. 2004, 36, 81–121. [Google Scholar] [CrossRef]
- Aayush, B.; Chen, X.; Bryan, R.; Abhinav, G.; Ramanan, D. Marr Revisited: 2D-3D Model Alignment via Surface Normal Prediction. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2281. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Bottou, L. Large-scale machine learning with stochastic gradient descent. In Proceedings of the COMPSTAT’2010; Physica-Verlag HD: Heidelberg, Germany, 2010; pp. 177–186. [Google Scholar]
- Dean, J.; Corrado, G.; Monga, R.; Chen, K.; Devin, M.; Mao, M.; Senior, A.; Tucker, P.; Yang, K.; Le, Q.V.; et al. Large scale distributed deep networks. In Advances in Neural Information Processing Systems; Mit Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Bordes, A.; Bottou, L.; Gallinari, P. Careful quasi-newton stochastic gradient descent. J. Mach. Learn. Res. 2009, 10, 1737–1754. [Google Scholar]
- Staal, J.J.; Abramoff, M.D.; Niemeijer, M.; Viergever, M.A.; Ginneken, B. DRIVE: Digital Retinal Images for Vessel Extraction. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Kauppi, T.; Kalesnykiene, V.; Kamarainen, J.-K.; Lensu, L.; Sorri, I.; Raninen, A.; Voutilainen, R.; Pietilä, J.; Kälviäinen, H.; Uusitalo, H. DIARETDB1: Standard Diabetic Retinopathy Database. Available online: http://www.it.lut.fi/project/imageret/diaretdb1/ (accessed on 9 September 2007).
- Messidor, Methods to Evaluate Segmentation and Indexing Techniques in the Field of Retinal Ophthalmology. Available online: http://www.adcis.net/en/Download-Third-Party/Messidor.html (accessed on 9 October 2014).
- Decencière, E.; Cazuguel, G.; Zhang, X.; Thibault, G.; Klein, J.C.; Meyer, F.; Marcotegui, B.; Quellec, G.; Lamard, M.; Danno, R.; et al. TeleOphta: Machine learning and image processing methods for teleophthalmology. Irbm 2013, 34, 196–203. [Google Scholar] [CrossRef]
- Retina (STARE). Available online: http://www.ces.clemson.edu/~ahoover/stare/ (accessed on 4 April 2000).
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Gao, W.; Zhou, Z.H. Dropout Rademacher Complexity of Deep Neural Networks. Sci. Chin. Inf. Sci. 2016, 59, 1–12. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Y. George Papandreou, Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the European conference on computer vision (ECCV), Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar]








| K1 | K2 | K3 | K4 | Binary | |
|---|---|---|---|---|---|
| Accuracy | 0.9551 | 0.9433 | 0.9356 | 0.9233 | 0.9271 |
| Sensitivity | 0.9432 | 0.9315 | 0.9235 | 0.9126 | 0.9182 |
| Specificity | 0.9491 | 0.9345 | 0.9239 | 0.9138 | 0.9158 |
| Index | Se | Sp | Acc |
|---|---|---|---|
| Database labeling | 0.9386 | 0.9978 | 0.9982 |
| Multi-seg-CNN | 0.9130 | 0.9940 | 0.9973 |
| PixelNet | 0.9134 | 0.9934 | 0.9974 |
| Unet | 0.8894 | 0.9870 | 0.9954 |
| Index | Se | Sp | Acc |
|---|---|---|---|
| Database labeling | 0.8936 | 0.9278 | 0.9282 |
| Multi-seg-CNN | 0.9130 | 0.9240 | 0.9273 |
| PixelNet | 0.7940 | 0.8344 | 0.8456 |
| Unet | 0.8394 | 0.8970 | 0.8933 |
| DeepLabv3+ | 0.7826 | 0.8332 | 0.8421 |
| Index | Se | Sp | Acc |
|---|---|---|---|
| Database labeling | 0.8986 | 0.9378 | 0.9352 |
| Multi-seg-CNN | 0.9030 | 0.9340 | 0.9386 |
| PixelNet | 0.8234 | 0.8434 | 0.8574 |
| Unet | 0.8354 | 0.8873 | 0.9004 |
| DeepLabv3+ | 0.8125 | 0.8336 | 0.8533 |
| Index | Se | Sp | Acc |
|---|---|---|---|
| Database labeling | 0.8736 | 0.9159 | 0.9188 |
| Multi-seg-CNN | 0.8835 | 0.9140 | 0.9043 |
| PixelNet | 0.7854 | 0.8334 | 0.8455 |
| Unet | 0.8594 | 0.8860 | 0.8953 |
| DeepLabv3+ | 0.7432 | 0.8128 | 0.8356 |
| Methods | Unet | PixelNet | DeepLabv3+ | Multi-Seg-CNN | ||||
|---|---|---|---|---|---|---|---|---|
| Severity Levels | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity |
| Cup optic disc | 0.892 | 0.991 | 0.913 | 0.992 | - | - | 0.915 | 0.994 |
| Macula | 0.896 | 0.992 | 0.917 | 0.994 | - | - | 0.918 | 0.997 |
| Vessel | 0.841 | 0.901 | 0.794 | 0.835 | 0.782 | 0.835 | 0.914 | 0.921 |
| Exudate | 0.843 | 0.892 | 0.828 | 0.842 | 0.813 | 0.831 | 0.907 | 0.932 |
| Roth spots | 0.866 | 0.897 | 0.793 | 0.837 | 0.741 | 0.816 | 0.882 | 0.917 |
| Total result | 0.864 | 0.932 | 0.844 | 0.896 | 0.777 | 0.823 | 0.902 | 0.948 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, L.; Che, H.; Xiao, Z.; Liu, Y. Extracting Retinal Anatomy and Pathological Structure Using Multiscale Segmentation. Appl. Sci. 2019, 9, 3669. https://doi.org/10.3390/app9183669
Geng L, Che H, Xiao Z, Liu Y. Extracting Retinal Anatomy and Pathological Structure Using Multiscale Segmentation. Applied Sciences. 2019; 9(18):3669. https://doi.org/10.3390/app9183669
Chicago/Turabian StyleGeng, Lei, Hengyi Che, Zhitao Xiao, and Yanbei Liu. 2019. "Extracting Retinal Anatomy and Pathological Structure Using Multiscale Segmentation" Applied Sciences 9, no. 18: 3669. https://doi.org/10.3390/app9183669
APA StyleGeng, L., Che, H., Xiao, Z., & Liu, Y. (2019). Extracting Retinal Anatomy and Pathological Structure Using Multiscale Segmentation. Applied Sciences, 9(18), 3669. https://doi.org/10.3390/app9183669




