Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Rhamnolipid-Producing Bacterial Strain Isolation and Screening
2.1.1. Soil Sampling and Strain Isolation
2.1.2. Screening for the Presence of the RhlAB operon
2.1.3. Screening of the Bacterial Strains for Biosurfactants Production Ability
2.1.4. Selected Bacterial Strain Identification
2.2. Rhamnolipid Production of the Isolate Under Laboratory Conditions
2.2.1. Optimization of Rhamnolipid Production
2.2.2. Extraction, Partial Purification, and Characterization of Rhamnolipids
2.3. Soil Bioremediation Study
2.3.1. Experimental Design
2.3.2. Estimation of Bacterial Abundance and Activity in Treated Soils
2.3.3. Estimation of the Total Petroleum Hydrocarbon Content in Soil
2.3.4. Germination Assay
2.4. Statistical Analysis
3. Results and Discussion
3.1. The Characteristics of Isolated Rhamnolipid-Producing Bacterial Strain
3.1.1. Genetic Characterization of the Strains
3.1.2. Production of Biosurfactant by the Isolate under Optimized Conditions
3.1.3. The Characteristics of Rhamnolipids Produced by the Isolate
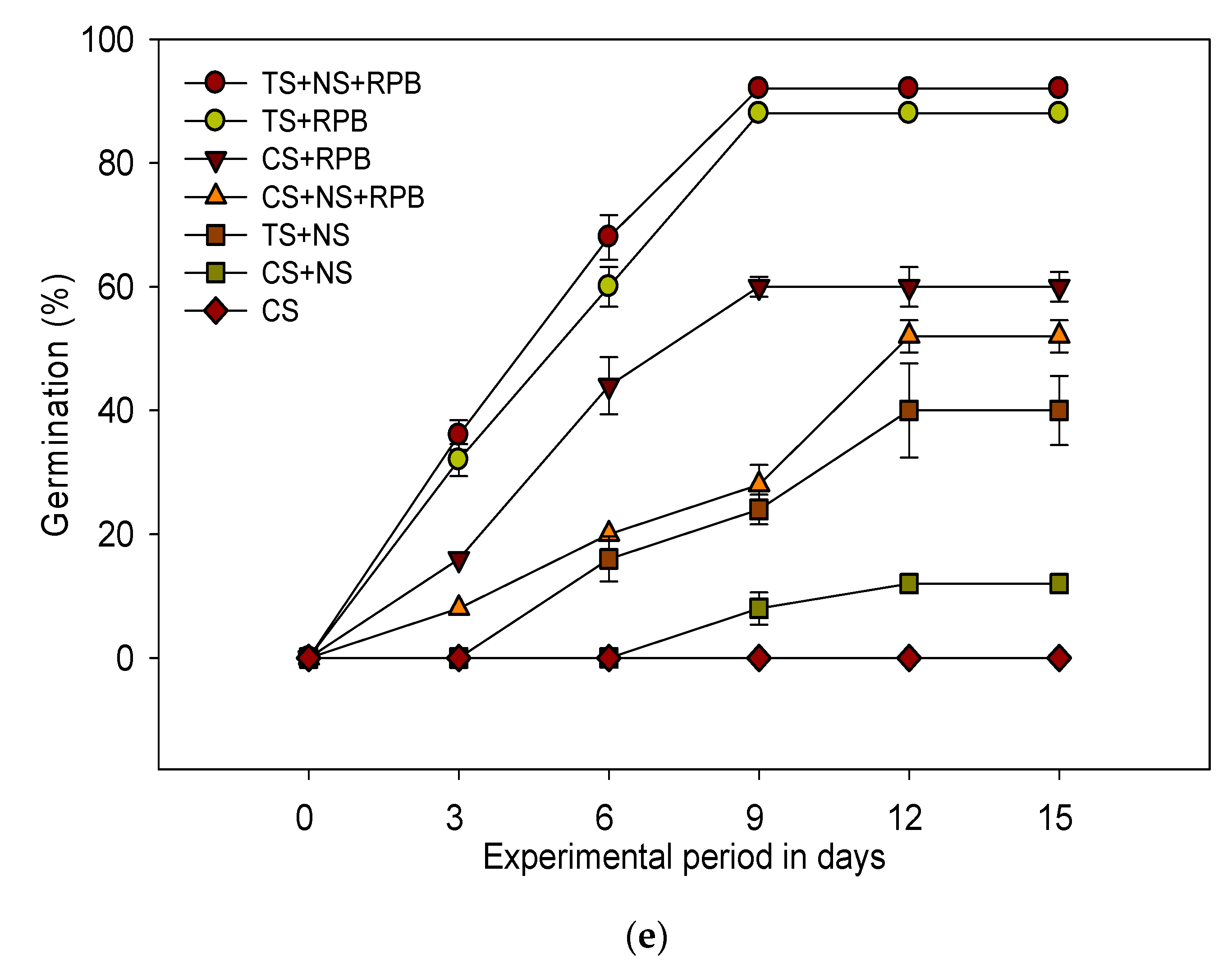
3.2. Treatment Efficiency of the Contaminated Soil Under Different Bioremediation Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abioye, O. Biological remediation of hydrocarbon and heavy metals contaminated soil. In Soil Contamination; Pascucci, Ed.; InTech: Vienna, Austria, 2011; pp. 127–142. [Google Scholar]
- Mori, Y.; Suetsugu, A.; Matsumoto, Y.; Fujihara, A.; Suyama, K. Enhancing bioremediation of oil-contaminated soils by controlling nutrient dispersion using dual characteristics of soil pore structure. Ecol. Eng. 2013, 51, 237–243. [Google Scholar] [CrossRef]
- Juhanson, J.; Truu, J.; Heinaru, E.; Heinaru, A. Survival and catabolic performance of introduced Pseudomonas strains during phytoremediation and bioaugmentation field experiment. FEMS Microbiol. Ecol. 2009, 70, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Truu, J.; Truu, M.; Espenberg, M.; Nõlvak, H.; Juhanson, J. Phytoremediation and plant-assisted bioremediation in soil and treatment wetlands: A review. Open Biotechnol. J. 2015, 9, 85–92. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, A.R.; Wick, L.Y.; Harms, H. Principles of microbial PAH-degradation in soil. Environ. Pollut. 2005, 133, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.M.; Kügler, J.H.; Henkel, M.; Gerlitzki, M.; Hörmann, B.; Pöhnlein, M.; Syldatk, C.; Hausmann, R. Rhamnolipids--next generation surfactants? J. Biotechnol. 2012, 162, 366–380. [Google Scholar] [CrossRef]
- Liu, H.; Xu, J.; Liang, R.; Liu, J. Characterization of the medium- and long-chain n-alkanes degrading Pseudomonas aeruginosa strain SJTD-1 and its alkane hydroxylase genes. PLoS ONE 2014, 9, e105506. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Fiechter, A.; Reiser, J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 1994, 269, 19787–19795. [Google Scholar]
- Wittgens, A.; Kovacic, F.; Müller, M.M.; Gerlitzki, M.; Santiago-Schübel, B.; Hofmann, D.; Tiso, T.; Blank, L.M.; Henkel, M.; Hausmann, R.; et al. Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 2016. [Google Scholar] [CrossRef]
- Benincasa, M.; Abalos, A.; Oliveira, I.; Manresa, A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Antonie Leeuwenhoek 2004, 85, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.S.M.; Thahira-Rahman, J.; Lakshmanaperumalsamy, P.; Banat, I.M. Towards efficient crude oil degradation by a mixed bacterial consortium. Bioresour. Technol. 2002, 85, 257–261. [Google Scholar] [CrossRef]
- Van Dyke, M.I.; Couture, P.; Brauer, M.; Lee, H.; Trevors, J.T. Pseudomonas aeruginosa UG2 rhamnolipid biosurfactants: Structural characterization and their use in removing hydrophobic compounds from soil. Can. J. Microbiol. 1993, 39, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Hošková, M.; Schreiberová, O.; Ježdík, R.; Chudoba, J.; Masák, J.; Sigler, K.; Rezanka, T. Characterization of rhamnolipids produced by non-pathogenic Acinetobacter and Enterobacter bacteria. Bioresour. Technol. 2013, 130, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Rooney, A.P.; Price, N.P.J.; Ray, K.J.; Kuo, T.M. Isolation and characterization of rhamnolipid-producing bacterial strains from a biodiesel facility. FEMS Microbiol. Lett. 2009, 295, 82–87. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nalini, S.; Parthasarathi, R. Production and characterization of rhamnolipids produced by Serratia rubidaea SNAU02 under solid-state fermentation and its application as biocontrol agent. Bioresour. Technol. 2014, 173, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Trögl, J.; Esuola, C.; Kříženecká, S.; Kuráň, P.; Seidlová, L.; Veronesi-Dáňová, P.; Popelka, J.; Babalola, O.; Hrabák, P.; Czinnerová, M.; et al. Biodegradation of High Concentrations of Aliphatic Hydrocarbons in Soil from a Petroleum Refinery: Implications for Applicability of New Actinobacterial Strains. Appl. Sci. 2018, 8, 1855. [Google Scholar] [CrossRef]
- Bodour, A.; Miller-Maier, R. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 1998, 32, 273–280. [Google Scholar] [CrossRef]
- Youssef, N.; Duncan, K.; Nagle, D. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 2004, 56, 339–347. [Google Scholar] [CrossRef]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure–function relationship of lipopeptide biosurfactants. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Cooper, D.; Goldenberg, B. Surface-active agents from two Bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar]
- Plaza, G.; Zjawiony, I.; Banat, I. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated bioremediated soils. J. Pet. Sci. Eng. 2006, 50, 71–77. [Google Scholar]
- Eden, P.A.; Schmidt, T.M.; Blakemore, R.P.; Pace, N.R. Phylogenetic Analysis of Aquaspirillum magnetotacticum Using Polymerase Chain Reaction-Amplified 16s rRNA-Specific DNA. Int. J. Syst. Bacteriol. 1991, 41, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, E.V.; Bemiller, J.N. Constituent analysis of glycosaminoglycans. In Methods in Carbohydrate Chemistry; Academic Press: New York, NY, USA, 1980; Volume 3, pp. 89–96. [Google Scholar]
- Rufino, R.D.; de Luna, J.M.; de Campos Takaki, G.M.; Sarubbo, L.A. Characterization and properties of the biosurfactant produced by Candida lipolytica UCP 0988. Electron. J. Biotechnol. 2014, 17, 34–38. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Amro, M.M.; Bock, M.; Boseker, K.; Fredrickson, H.L.; Kessel, D.G.; Timmis, K.N. The potential of Bacillus licheniformis strains for in situ enhanced oil recovery. J. Pet. Sci. Eng. 1997, 18, 147–160. [Google Scholar] [CrossRef]
- Smyth, T.J.; Perfumo, A.; Mcclean, S.; Banat, I.M. Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3687–3704. [Google Scholar]
- Sharma, D.; Saharan, B.S.; Chauhan, N.; Procha, S.; Lal, S. Isolation and functional characterization of novel biosurfactant produced by Enterococcus faecium. Springerplus 2015, 4, 1–14. [Google Scholar] [CrossRef]
- Yin, H.; Qiang, J.; Jia, Y.; Ye, J.; Peng, H.; Qin, H.; Zhang, N.; He, B. Characteristics of biosurfactant produced by Pseudomonas aeruginosa S6 isolated from oil-containing wastewater. Process Biochem. 2009, 44, 302–308. [Google Scholar] [CrossRef]
- Silva, Í.S.; dos Santos, E.D.C.; de Menezes, C.R.; de Faria, A.F.; Franciscon, E.; Grossman, M.; Durrant, L.R. Bioremediation of a polyaromatic hydrocarbon contaminated soil by native soil microbiota and bioaugmentation with isolated microbial consortia. Bioresour. Technol. 2009, 100, 4669–4675. [Google Scholar] [CrossRef]
- Benincasa, M. Rhamnolipid produced from agroindustrial wastes enhances hydrocarbon biodegradation in contaminated soil. Curr. Microbiol. 2007, 54, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Bekku, Y.; Koizumi, H.; Oikawa, T.; Iwaki, H. Examination of four methods for measuring soil respiration. Appl. Soil Ecol. 1997, 5, 247–254. [Google Scholar] [CrossRef]
- Pepper, I.L.; Gerba, C.P.; Brendecke, J. Environmental Microbiology: A Laboratory Manual; Academic Press Inc.: New York, NY, USA, 2000. [Google Scholar]
- Peng, S.; Zhou, Q.; Cai, Z.; Zhang, Z. Phytoremediation of petroleum contaminated soils by Mirabilis Jalapa L. in a greenhouse plot experiment. J. Hazard. Mater. 2009, 168, 1490–1496. [Google Scholar] [CrossRef]
- Marecik, R.; Biegańska-Marecik, R. Phytoremediation of industrial wastewater containing nitrates, nitroglycerin, and nitroglycol. Polish J. Environ. Stud. 2013, 22, 773–780. [Google Scholar]
- Gerdes, B.; Brinkmeyer, R.; Dieckmann, G.; Helmke, E. Influence of crude oil on changes of bacterial communities in Arctic sea-ice. FEMS Microbiol. Ecol. 2005, 53, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Rathour, R.; Gupta, J.; Tyagi, B.; Kumari, T.; Thakur, I.S. Biodegradation of pyrene in soil microcosm by Shewanella sp. ISTPL2, a psychrophilic, alkalophilic and halophilic bacterium. Bioresour. Technol. Rep. 2018, 4, 129–136. [Google Scholar] [CrossRef]
- Ram, G.; Joe, M.M.; Devraj, S.; Benson, A. Rhamnolipid production using Shewanella seohaensis BS18 and evaluation of its efficiency along with phytoremediation and bioaugmentation for bioremediation of hydrocarbon contaminated soils. Int. J. Phytoremediation 2019, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Martín-Gil, J.; Ramos-Sánchez, M.C.; Martín-Gil, F.J. Shewanella putrefaciens in a fuel-in-water emulsion from the Prestige oil spill. Antonie Van Leeuwenhoek 2004, 86, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Hassanshahian, M. Isolation and characterization of biosurfactant producing bacteria from Persian Gulf (Bushehr provenance). Mar. Pollut. Bull. 2014, 86, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Juárez, K.; Díaz, R.; Soberón-Chávez, G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 2003, 149, 3073–3081. [Google Scholar] [CrossRef]
- Soberón-Chávez, G.; Aguirre-Ramírez, M.; Sánchez, R. The Pseudomonas aeruginosa RhlA enzyme is involved in rhamnolipid and polyhydroxyalkanoate production. J. Ind. Microbiol. Biotechnol. 2005, 32, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Abalos, A.; Maximo, F.; Manresa, M.; Bastida, J. Utilization of response surface methodology to optimize the culture media for the production of rhamnolipids by Pseudomonas aeruginosa AT10. J. Chem. Technol. Biotechnol. 2002, 77, 777–784. [Google Scholar] [CrossRef]
- Zhao, F.; Zhou, J.; Han, S.; Ma, F.; Zhang, Y.; Zhang, J. Medium factors on anaerobic production of rhamnolipids by Pseudomonas aeruginosa SG and a simplifying medium for in situ microbial enhanced oil recovery applications. World J. Microbiol. Biotechnol. 2016, 32, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Box, G.; Hunter, W.; Hunter, J. Statistics for Experimenters; John Wiley & Sons: New York, NY, USA, 1978. [Google Scholar]
- Keeler, S.J.; Hendrickson, E.R.; Hnatow, L.L.; Jackson, S.C. Identification, Characterization, and Application of Shewanella Putrefaciens (LH4:18), Useful in Microbially Enhanced Oil Release. U.S. Patent 7,776,795, 17 August 2010. [Google Scholar]
- Heidelberg, J.F.; Paulsen, I.T.; Nelson, K.E.; Gaidos, E.J.; Nelson, W.C.; Read, T.D.; Eisen, J.A.; Seshadri, R.; Ward, N.; Methe, B.; et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 2002, 20, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Leitermann, F.; Syldatk, C.; Hausmann, R. Fast quantitative determination of microbial rhamnolipids from cultivation broths by ATR-FTIR Spectroscopy. J. Biol. Eng. 2008, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- García-Ruiz, R.; Ochoa, V.; Hinojosa, M.B.; Carreira, J.A. Suitability of enzyme activities for the monitoring of soil quality improvement in organic agricultural systems. Soil Biol. Biochem. 2008, 40, 2137–2145. [Google Scholar] [CrossRef]
- Gil-Sotres, F.; Trasar-Cepeda, C.; Leirós, M.C.; Seoane, S. Different approaches to evaluating soil quality using biochemical properties. Soil Biol. Biochem. 2005, 37, 877–887. [Google Scholar] [CrossRef]
- Truu, M.; Truu, J.; Ivask, M. Soil microbiological and biochemical properties for assessing the effect of agricultural management practices in Estonian cultivated soils. Eur. J. Soil Biol. 2008, 44, 231–237. [Google Scholar] [CrossRef]
- Dilly, O.; Blume, H.P.; Munch, J.C. Soil microbial activities in Luvisols and Anthrosols during 9 years of region-typical tillage and fertilisation practices in northern Germany. Biogeochemistry 2003, 65, 319–339. [Google Scholar] [CrossRef]
- Masciandaro, G.; Ceccanti, B.; Ronchi, V.; Bauer, C. Kinetic parameters of dehydrogenase in the assessment of the response of soil to vermicompost and inorganic fertilisers. Biol. Fertil. Soils 2000, 32, 479–483. [Google Scholar] [CrossRef]
- Ławniczak, Ł.; Marecik, R.; Chrzanowski, Ł. Contributions of biosurfactants to natural or induced bioremediation. Appl. Microbiol. Biotechnol. 2013, 97, 2327–2339. [Google Scholar] [CrossRef] [PubMed]
- Tahseen, R.; Afzal, M.; Iqbal, S.; Shabir, G.; Khan, Q.M.; Khalid, Z.M.; Banat, I.M. Rhamnolipids and nutrients boost remediation of crude oil-contaminated soil by enhancing bacterial colonization and metabolic activities. Int. Biodeterior. Biodegrad. 2016, 115, 192–198. [Google Scholar] [CrossRef]
- Szulc, A.; Ambrożewicz, D.; Sydow, M.; Ławniczak, Ł. The influence of bioaugmentation and biosurfactant addition on bioremediation efficiency of diesel-oil contaminated soil: Feasibility during field studies. J. Environ. Manag. 2014, 132, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yin, R.; Lin, X.; Liu, W.; Chen, R.; Li, X. Interactive effect of biosurfactant and microorganism to enhance phytoremediation for removal of aged polycyclic aromatic hydrocarbons from contaminated soils. J. Heal. Sci. 2010, 56, 257–266. [Google Scholar] [CrossRef]
- Fadhile Almansoory, A.; Abu Hasan, H.; Idris, M.; Sheikh Abdullah, S.R.; Anuar, N. Potential application of a biosurfactant in phytoremediation technology for treatment of gasoline-contaminated soil. Ecol. Eng. 2015, 84, 113–120. [Google Scholar] [CrossRef]





| Source | Sum of Squares | Degrees of Freedom | Mean Square | f Value | p-Value |
|---|---|---|---|---|---|
| Model | 9.73 | 14 | 0.69 | 30.73 | <0.0001 |
| A-Crude oil | 6.51 | 1 | 6.51 | 287.93 | <0.0001 |
| B-Inoculation load | 1.35 | 1 | 1.35 | 59.87 | <0.0001 |
| C-Temperature | 0.07 | 1 | 0.07 | 3.11 | 0.0980 |
| D-pH | 0.01 | 1 | 0.01 | 0.46 | 0.5076 |
| AB | 0.46 | 1 | 0.46 | 20.15 | 0.0004 |
| AC | 5.62 | 1 | 5.62 | 0.25 | 0.6252 |
| AD | 5.62 | 1 | 5.62 | 0.25 | 0.6252 |
| BC | 0.01 | 1 | 0.01 | 0.69 | 0.4189 |
| BD | 6.25 | 1 | 6.25 | 0.02 | 0.8702 |
| CD | 0.01 | 1 | 0.01 | 0.69 | 0.4189 |
| A^2 | 0.87 | 1 | 0.87 | 38.49 | <0.0001 |
| B^2 | 0.02 | 1 | 0.02 | 0.96 | 0.3428 |
| C^2 | 0.37 | 1 | 0.37 | 16.22 | 0.0011 |
| D^2 | 0.37 | 1 | 0.37 | 16.22 | 0.0011 |
| Residual | 0.34 | 15 | 0.02 | ||
| Lack of Fit | 0.29 | 10 | 0.02 | 3.01 | 0.1179 |
| Pure Error | 0.04 | 5 | 9.66 | ||
| Cor Total | 10.07 | 29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joe, M.M.; Gomathi, R.; Benson, A.; Shalini, D.; Rengasamy, P.; Henry, A.J.; Truu, J.; Truu, M.; Sa, T. Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil. Appl. Sci. 2019, 9, 3773. https://doi.org/10.3390/app9183773
Joe MM, Gomathi R, Benson A, Shalini D, Rengasamy P, Henry AJ, Truu J, Truu M, Sa T. Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil. Applied Sciences. 2019; 9(18):3773. https://doi.org/10.3390/app9183773
Chicago/Turabian StyleJoe, Manoharan Melvin, Ram Gomathi, Abitha Benson, Devaraj Shalini, Parthasarathi Rengasamy, Allen John Henry, Jaak Truu, Marika Truu, and Tongmin Sa. 2019. "Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil" Applied Sciences 9, no. 18: 3773. https://doi.org/10.3390/app9183773
APA StyleJoe, M. M., Gomathi, R., Benson, A., Shalini, D., Rengasamy, P., Henry, A. J., Truu, J., Truu, M., & Sa, T. (2019). Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil. Applied Sciences, 9(18), 3773. https://doi.org/10.3390/app9183773








