Abstract
Developing new vaccine candidates is considered the best strategy for protecting poultry against artificial haemagglutinin (A/H5N1) strains. The transient expression system in plants has been a very efficient method for rapidly producing haemagglutinin-based recombinant vaccines. In this study, two novel artificial trimeric haemagglutinin constructs representing A/H5N1 strains that were detected in poultry from 2005 to 2015 in Vietnam, H5.c1 (representing all of the subclades 1.1, 1.1.1, and 1.1.2) and H5.c2 (representing all of the subclades 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c), were designed for transient expression in Nicotiana benthamiana via agroinfiltration. However, only the H5.c1 protein, which showed the best expression and biofunction via the haemagglutination test, was selected for purification by immobilized metal ion affinity chromatography (IMAC). The trimeric structure of the IMAC-purified H5.c1 protein was well characterized by cross-linking reaction and size exclusion chromatography. An indirect ELISA and Western blot analysis of vaccinated mouse sera demonstrated that the H5.c1 protein strongly induced HA-specific Immunoglobulin G (IgG) immune responses. Notably, the H5.c1 protein induced strongly neutralizing antibodies against homologous H5.c1 protein and that of three heterologous native strains of clade, 1, 1.1, and 2.3.2.1c, in haemagglutination inhibition assays. Therefore, the plant-based artificial H5.c1 protein can be a promising vaccine candidate for conferring poultry resistance against A/H5N1 viruses in Vietnam.
1. Introduction
Asian highly pathogenic avian influenza (HPAI) A (H5N1) virus is potentially able to cause a global pandemic due to its relatively large spread via avian hosts and its potential to directly infect humans [1]. In Vietnam, over 2750 outbreaks of H5N1 occurring in poultry were reported to the World Organisation for Animal Health (OIE) from the end of 2003 to 03 July 2019, among which one outbreak killed and disposed of 1120 birds in 2019 [2]. Influenza haemagglutinin (HA), a homotrimeric type I transmembrane protein, is one of the major surface glycoproteins of H5N [3]. The viral HA protein plays a critical role in the infectivity and pathogenicity of influenza virus by binding specifically to sialic acid on glycoconjugates of host membrane proteins and enabling virus entry into host cells [3]. Many studies have shown that protection provided by influenza vaccines is mediated by mainly an anti-HA neutralizing antibody [4]. Therefore, HA has been reported as a major target antigen for influenza vaccine development [5].
However, H5N1 HA antigenic variation occurs frequently. A/H5N1 evolved into 10 distinct clades (0–9) that are divided into many subclades [6]. In Vietnam, six clades (clades 1, 2, 3, 5, 7, and 8) exist, among which clade 1 (subclades 1.1, 1.1.1, and 1.1.2) and clade 2 (subclades 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c) are the most common [6]. In particular, one or more strains could circulate at the same time, in the same area, or both. Current strain-specific vaccines (for example, NIBRG–14 or RE–6) have limited effectiveness in the protection of poultry. The timely update of seasonal vaccines, which is necessary to ensure protection, requires significant time and effort. Recent studies of a universal influenza vaccine showed its potential efficacy through enhanced breadth and durability compared to those of alternate seasonal influenza vaccines [7,8,9,10,11,12]. In 2008, Crevar and Ross demonstrated that a polyvalent influenza virus-like particle (VLP) vaccine in which haemagglutinin represented different clades of A/H5N1 was a potential approach to elicit an immunity response against isolates from emerging clades and subclades of A/H5N1 [7]. Following studies reported that a vaccine containing haemagglutinin designed by a methodology called computationally optimized broadly reactive antigen (COBRA) elicited a broad immunity response against multiple H5N1 isolates from different clades [8,9,10,11]. The COBRA HA VLP vaccines elicited higher antibody titres than both a polyvalent HA VLP vaccine and a homologous vaccine [8,9,10]. In 2018, Pardi and co-workers demonstrated that the highly conserved influenza virus haemagglutinin stalk domain is one of the promising targets for a universal vaccine. They generated a formulated modified mRNA encoding full-length HA in lipid nanoparticles (LNPs) and subsequently vaccinated rabbits, mice, and ferrets. Immunization with HA mRNA–LNPs induced HA stalk-specific antibody responses against the HA stalk domain of homologous, heterologous, and heterosubtypic influenza viruses in these animals [13]. Therefore, universal influenza vaccines protecting poultry against many H5N1 variants will become a new potential trend in vaccine development.
Based on this trend, we designed two novel artificial HA proteins (including H5.c1 representing all of the A/H5N1 subclades 1.1, 1.1.1, and 1.1.2 and H5.c2 representing all of the A/H5N1 subclades 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c). Each amino acid site of the artificial HA was determined from a comparison of all of the HA sequences of A/H5N1 recorded in poultry from 2005 to 2015 in Vietnam, selecting for either the most common or most recent epitope. The aim of our research was to generate artificial HA proteins that will elicit broadly antigenic responses to protect poultry against many A/H5N1 strains of clades 1.1, 1.1.1, and 1.1.2 or 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c that have recently circulated. Transient expression systems in plants using agroinfiltration have been developed as a very safe and efficient methods to rapidly produce plant-based subunit vaccines because of their low production and infrastructure cost, ease of scale up, high stability and long shelf life [14,15,16,17]. Several studies demonstrated that trimeric HA had the ability to efficiently elicit neutralizing antibodies in vaccinated animals, whereas monomer haemagglutinins did not [18,19,20]. To produce a stable homotrimeric HA, an artificial trimeric motif (GCN4–pII) was used [21] and fused to the HA proteins at the C–terminal end. The artificial HA proteins were transiently expressed in N. benthamiana via agroinfiltration. In the current paper, we present initial results of expression, structural, and functional characterization and of specific antibody responses in mice for the trimerized artificial HA proteins.
2. Materials and Methods
2.1. Artificial Haemagglutinin
The complete HA sequences of 90, 38, 34, 19, 143, 28, and 165 of A/H5N1 subclades 1.1, 1.1.1, 1.1.2, 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c, respectively, that were detected in poultry from 2005 to 2015 in Vietnam were collected from GenBank, National Center for Biotechnology Information (NCBI) and Global Initiative on Sharing All Influenza Data (GISAID). The artificial HA was designed in two steps (as shown in Figure 1). In the first step, all of the HA sequences of each subclade were aligned by MegAlign/Lasergene software version 7.0.0. A primary artificial HA was designed by selection for the most conserved, common, and recent amino acids from the occurring strains. In the second step, using the same rules, the final artificial HA sequences (including H5.c1 representing the subclade group of 1.1, 1.1.1, and 1.1.2 strains and H5.c2 representing the subclade group of 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c strains) were designed by comparing all of the primary HA sequences of each subclade group together. The highest sequence identity of artificial H5.c1 and H5.c2 sequences when compared with all the HA sequences in the BLAST/NCBI search at the nucleotide and amino acid levels was 99.41% and 99.65%, respectively. For expression in N. benthamiana, the H5.c1 and H5.c2 amino acid sequences were codon optimized and synthesized commercially by GENECUST EUROPE (in Luxembourg, Germany).
Figure 1.
Schematic of designing artificial haemagglutinin (HA) sequences of artificial haemagglutinin (A/H5N1) from poultry in Vietnam.
2.2. Construction of Plant Expression Vectors
The complete artificial HA sequences were commercially provided in pUC57 vectors. To express and multimerize HA proteins, HA proteins were inserted into pRTRA vectors that contained trimerization (GCN4-pII) domains [22]. The resulting vectors, pRTRA_35S_LeB4SP_His_HA_cmyc_KDEL, were used for the expression of trimer HA proteins under the control of the CaMV 35S Pro promoter and CaMV 35S Term terminator (as shown in Figure 2). In this vector, the HA proteins were fused to a c–myc tag for detection by Western blot, to a His tag for purification by IMAC and to the legumin B4 signal peptide and the KDEL motif for promoting these proteins retention in the endoplasmic reticulum. The expression cassettes of HA proteins in the pRTRA vectors were sub-cloned into pCB301 vectors [23] at HindIII restriction sites. Finally, the pCB301_35S_LeB4SP_His_HA_cmyc_KDEL vectors were transferred into the Agrobacterium tumefaciens strain CV58 pGV2260.
Figure 2.
Expression cassettes of haemagglutinin (HA) proteins in plants.
2.3. Agrobacterium Infiltration
The expression of recombinant protein by agrobacterium infiltration was based on the protocol of Phan and colleagues [23,24]. Agrobacteria containing the shuttle vectors for the expression of recombinant HcPro [25,26] and HA proteins were precultivated independently in 50 ml of LB medium containing 50 μg/mL kanamycin, 50 μg/mL carbenicillin, and 50 μg/mL rifampicin at 28 °C overnight at a speed of 140 rpm. Then, 200 ml of new LB medium including the appropriate antibiotics was added into the preculture. The bacteria were then collected by centrifugation (4000 g, 30 min, 4 °C) and resuspended in infiltration buffer containing 10 mM 2–(N–morpholino) ethanesulfonic acid (MES), 10 mM MgSO4, pH 5.6 after 24 h of cultivation. These agrobacteria were mixed and diluted in infiltration buffer to a final OD600 of 0.8–1.0. The Agrobacterium suspension solution was used for infiltration into N. benthamiana plants (5–6 weeks old) by vacuum for 2 min. Subsequently, the plants were introduced into the greenhouse at 21 °C, with 16 h light per day. Leaf samples were harvested post infiltration for five days and stored at −80 °C until use.
2.4. Purification of Protein by Immobilized Metal Ion Affinity Chromatography (IMAC)
IMAC was based on the method described by Phan and colleagues [24], with some modifications. Eighty grams of tobacco leaf samples were ground in liquid nitrogen and blended in 240 mL of 50 mM Tris buffer (pH 8.0). The plant extracts were clarified by centrifugation (23000 rpm, 30 min, 4 °C). The supernatant solution was transferred into a new centrifuge tube and underwent repeated centrifugation under the same conditions. Subsequently, the extracts were combined with 20 mL of Ni–NTA agarose resin. After mixing for 30 min at 4 °C, the mixture was loaded on a chromatography column. For H5.c1 protein purification, the different concentration of imidazole (30 mM, 25 mM, 20 mM, or 5 mM imidazole) in 2L of washing buffer (50 mM NaH2PO4, 300 mM NaCl) was tested Finally, H5.c1 protein was released from the column by adding 20 ml of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 50 mM imidazole with artificial H5.c1, pH 8.0), concentrated in PEG 6000 and dialyzed against PBS. The native H5TG protein was purified successfully with 30 mM and 125 mM imidazole at the washing step and elution step, respectively [24]. Protein contents of purification steps were determined by Bradford assay [27]. The concentrated HA proteins were added 50% glycerol (w/v). The IMAC-purified HA proteins were stored at −20 °C and were then used for mouse immunization.
2.5. Cross-Linking Reaction Using Bis[sulfosuccinimidyl] Suberate (BS3)
A cross-linking reaction was carried out to determine the multimeric form of the IMAC-purified HA proteins based on the protocol of Weldon and colleagues [19]. In brief, 1 μg of IMAC-purified HA proteins was mixed with BS3 to a final concentration of 5 mM, and incubated at room temperature for 30 min. To stop the reaction, 1 M Tris–HCl pH 8.0 was added until a final concentration of 50 mM and incubated at room temperature for 15 min. Then, proteins were loaded on a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) under reducing conditions, blotted, and detected by Western blot.
2.6. Purification of Protein by Size Exclusion Chromatography (SEC)
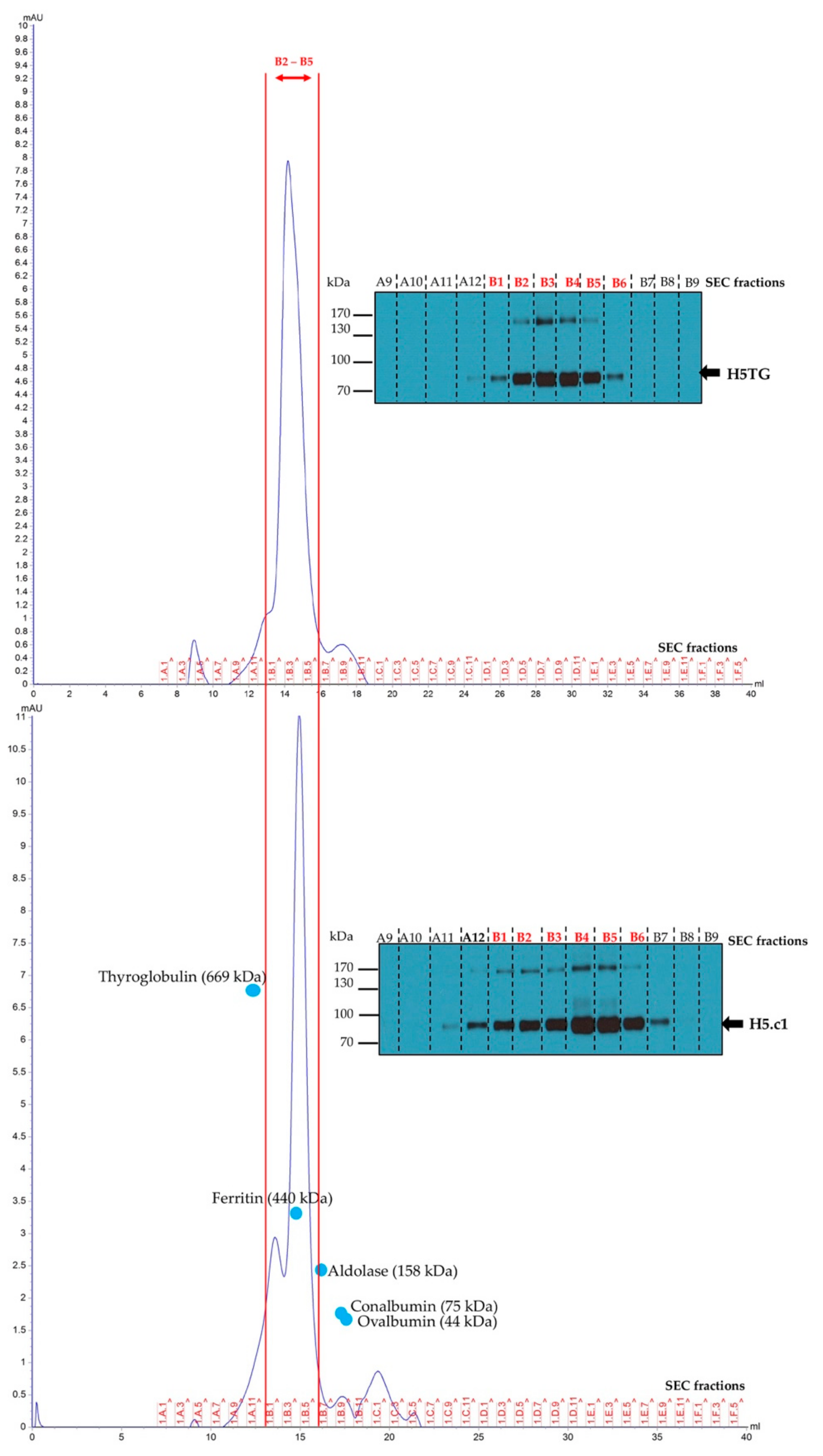
The SEC was based on the method described by Phan and colleagues [24]. Equal amounts of purified artificial H5.c1 and native H5TG were introduced into a Superose™ 6 10/300GL column (GE Healthcare) which was pre-equilibrated with phosphate-buffered saline (PBS). To estimate the molecular weight of the HA proteins, a high molecular weight kit containing standard proteins with molecular weights (44–2000 kDa) was loaded onto the column. The fractions were collected for Western blot analysis, collecting the oligomeric H5 fractions (B1 to B6) for concentrating. The concentrated HA proteins were then used for ELISA and Western blotting to detect HA-specific IgG mouse antibody production.
2.7. SDS–PAGE and Western Blotting
Proteins in crude extracts, purified artificial H5.c1 and native H5TG proteins were loaded on reducing SDS–PAGE (10% polyacrylamide) gels and then transferred to nitrocellulose membranes. The Western blotting procedure was performed using a monoclonal anti–c–myc antibody as a primary antibody according to the protocol described by Gahrtz and Conrad in 2009 [28]. Next, a secondary antibody (sheep anti-mouse IgG, horseradish peroxidase-linked whole antibody, GE Healthcare UK limited, Little Chalfont Buckinghamshire, UK) was added, and then the signals were detected by enhanced chemiluminescence (ECL)-based detection.
To detect the HA-specific IgG mouse antibody, 500 ng of each of the SEC-purified artificial H5.c1 and native H5TG proteins were separated on 4 lanes of a reducing SDS–PAGE gel (10% polyacrylamide) and transferred to a nitrocellulose membrane. The membrane was blocked with a 5% (w/v) fat-free milk powder dissolved in a PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) for 2 h. Then, each lane of the membrane was isolated by cutting and incubated at room temperature for 90 min with a 1:200 dilution of a mixture of sera from five mice of each group (artificial H5.c1, native H5TG and PBS groups) after second and third immunizations. Next, the membranes were incubated with a 1:2000 dilution of an HRP-conjugated goat anti-mouse IgG secondary antibody for 1 h at room temperature. Specific signals were detected by incubating membranes with 3,3–diaminobenzidine (DAB, Thermo Scientific Pierce) dissolved in 0.05 M Tris–HCl and 0.04% hydrogen peroxide for 10 min in the dark.
2.8. Mouse Immunization
The mouse immunization was based on the procedure described by Pham and colleagues [29]. Six- to eight-week-old female BALB/c mice (five per group) were given a subcutaneous injection on a schedule of 1, 14, and 28 days. Each mouse was injected with 2.5 μg of each IMAC-purified artificial H5.c1 and native H5TG protein. In the negative control group, mice were vaccinated with PBS. The antigens were mixed with complete Freund’s adjuvant and incomplete Freund’s adjuvant in the first immunization and the booster immunizations (the second and third), respectively. The mice were bled via the retro-orbital sinus after the second and the third immunization for seven days. Mouse sera were collected separately for haemagglutinin inhibition, indirect ELISA tests and Western blot. All procedures were in accordance with the “3Rs”, relevant national legislation and rules of IBT, Vietnam, on the use of animals for research.
2.9. Haemagglutination and Haemagglutinin Inhibition Assay
The haemagglutination and haemagglutinin inhibition assays followed a standard procedure of the Office International des Epizooties (OIE) [30]. The dilution that induced complete haemagglutination was defined as one haemagglutination unit (HAU) [30]. The homologous purified HA.c1 and H5.c2 proteins and three heterologous inactivated A/H5N1 strains produced by reverse genetics (with HA of A/Vietnam/1194/2004(H5N1), clade 1; A/duck/Vietnam/ST0970/2009(H5N1), clade 1.1; and A/duck/Vietnam/HT2/2014(H5N1), clade 2.3.2.1c) were used for the HI assay. A 25 μL aliquot of serum from a single mouse was added to the first well of a microtitre plate containing 25 μL of PBS. Two-fold serial dilutions were then made across the row of 8 wells. A 25 μL volume containing 4 HAUs of the purified HA proteins or heterologous inactivated strains was introduced into each well and incubated for 30 min at 25 °C. Finally, 25 μL of 1% chicken red blood cells was placed in each well, and the plates were incubated for 30 min at 25 °C. The HI titre is defined as the reciprocal of the highest dilution of serum that could completely inhibit haemagglutination [30].
2.10. Indirect ELISA
The indirect ELISA was based on the method described by Pham and colleagues [29]. First, 100 ng of SEC-purified artificial H5.c1 and native H5TG in phage PBS (100 mM NaCl, 32 mM Na2HPO4, 17 mM Na2HPO4, pH 7.2) was added into microtitre plates (ImmunoPlateMaxisorp, Nalgen Nunc International, Roskilde, Denmark) that were incubated at room temperature overnight. The plates were then blocked with 3% (w/v) bovine serum albumin (BSA) and 0.05% (v/v) Tween20 in PBS (PBST) for 2 h. Next, 100 µL of the mouse serum at a dilution of 16 x 10−3 was applied and incubated for 1 h at 25 °C. Each serum dilution was repeated four times. The plates were washed 5 times with PBST and then incubated with a horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG dilution (2000 times in 1% (w/v) BSA in PBST for 1 h at 25 °C. Subsequently, 1-Step Substrate Solution (Thermo Fisher Scientific, Lithuania) was introduced as the enzymatic substrate. Finally, the reaction was stopped via 1 M HCl after 20 min of incubation. All mouse sera of all the plates were measured under the same ELISA conditions. The absorbance signal was determined at 450 nm. Control values for BSA at the solid phase were subtracted. The ELISA values of all mouse sera were normalized by use of an internal control serum of mouse number 2 of group 1 after the 3 immunizations with 16 × 10−3 dilution. The normalized ELISA values of all mouse sera at the 16 × 10−3 dilution were used for statistical analysis in this paper.
2.11. Statistical Analysis
Statistical analyses for ELISA were carried out using a Welch’s t-test, and HI assays were analyzed with the Mann–Whitney rank–sum test in Sigma Plot software. The difference between the mean of sample data was compared and was defined as the sample’s mean (X) ± the standard deviation (SD). P values that were less than 0.05 are presented as significant differences.
3. Results
3.1. Expression and Functional Characterization of Haemagglutinin (HA) Proteins in Plant Crude Extract
The protein three-dimensional structure regulates the functions of that protein. Therefore, the more similar the structure of protein molecules are, the more biologically similar functions are available. The structure of the protein provides much more insight into its biological function than its sequence. In this study, four H5 sequences were submitted to http://raptorx.uchicago.edu/ to predict the structure of these proteins, including two sequences of the clade 1 group (artificial H5.c1 and native H5TG sequences, with identity at the amino acid level of 98.4%) and two sequences of the clade 2 group (artificial H5.c2 and native H5Dk sequences, with identity at the amino acid level of 99.4%). The results showed that the three-dimensional structures of two HA proteins in each of the clade 1 and clade 2 groups are similar (as shown in Appendix A, Figure A1), although in the amino acid sequence of HA, each group has two different importance amino acid sites (341 and 156 of clade 1 group; and 99 and 140 of the clade 2 group). The differences in cleavage sites and antigenic sites between artificial and native H5 sequences are described in yellow and green color, respectively, and all of the H5 sequences removed at the cleavage site 342–344/345 are shown in Table 1.

Table 1.
The amino acid sequence at sites of protease cleavage, receptor binding, epitopes of antigenic sites, and variable glycosylation of haemagglutinin (HA) proteins.
Native HA proteins have been expressed successfully in plants [21,24]. In this study, artificial HA proteins were also expressed and detected in the crude extracts of tobacco leaves (as shown in Figure 3a). However, the expression of artificial H5.c2 was lower than that of the remaining proteins. The apparent molecular weight shown in Figure 3a is higher than the expected sizes (67.15 kDa) predicted from the polypeptide sequence of H5 by Lasergene software. This can be explained by the fact that glycosylation influences the running behaviour during the electrophoretic separation [31]. Therefore, in the haemagglutination test, the HA titre of artificial H5.c2 was also lower than that of the remaining proteins (as shown in Figure 3b). The artificial H5.c1 protein, which had a similar expression and HI titre to native H5TG and H5Dk proteins, was selected for future experiments.
Figure 3.
Expression of haemagglutinin (HA) proteins in the crude extracts of tobacco leaves measured using Western blot (a); the functional characterization of HA proteins in the crude extracts of tobacco leaves performed using a haemagglutination assay (b).
3.2. Functional and Structural Characterization of the Purified Haemagglutinin (HA) Proteins
To precisely detect the function and structure of the recombinant artificial H5.c1 and native H5TG proteins, which were transiently expressed in N. benthamiana, their purification is essential. These HA proteins were purified by IMAC. IMAC is a commonly employed method to purify recombinant proteins containing polyhistidine amino acid residues based on the strong specific interaction between these amino acids and immobilized metal ions (Co2+, Ni2+, Cu2+, and Zn2+) [32]. To obtain HA proteins from transiently transformed N. benthamiana leaves, these proteins were expressed as fusion proteins containing a N-terminal 6×His-tag. The plant extract containing HA proteins was then incubated with Ni–NTA agarose. The HA proteins containing sequences of consecutive histidine residues were efficiently retained on IMAC column matrices. For washing and elution steps in the IMAC column matrices, 5 and 50 mM imidazole were tested for the artificial H5.c1 protein, while 30 and 125 mM imidazole were used for the native H5TG, respectively [24]. Optimization of imidazole concentration for purification of H5.c1 by IMAC was shown in Figure S1. The IMAC-purified artificial H5.c1 protein was compared to the IMAC-purified native H5TG protein for functional and structural characterization, as shown in Figure 4. The results showed that both of these HA proteins had a similar haemagglutination function, with only 0.23 μg of the purified HA proteins stopping the agglutination of rooster blood cells (as shown in Figure 4a).
Figure 4.
Functional characterization of the immobilized metal ion affinity chromatography (IMAC)-purified haemagglutinin (HA) proteins (a); structural characterization of the IMAC-purified HA proteins by a cross-linking reaction using bis[sulfosuccinimidyl suberate (BS3) (b).
The trimeric HA exhibits epitopes that could induce neutralizing antibodies; therefore, it is important to retain the stable trimeric HA state [20]. In this study, the trimerization domain (GCN4–pII) was used to produce stable homotrimeric HA proteins. The multimeric states of IMAC-purified HA proteins were determined by cross-linking using BS3, a homobifunctional and water-soluble crosslinker [33,34]. In the previous study, the oligomeric status of H5 protein fused GCN4–pII was demonstrated by the cross-linking reaction with BS3, and the gel mobility of (H5pII)3 was approximately consistent with the expected size of a trimer [21]. Similarly, analysis by denaturing SDS–PAGE and Western blot indicated that the recombinant artificial H5.c1 and native H5TG proteins were observed as a mixture of multimeric protein without BS3 (as shown in Figure 4b, lanes 1, 3) and only trimeric proteins with BS3 (as shown in Figure 4, lanes 2, 4). These results were also similar to the study of Weldon and colleagues. This result indicated that the modification of the HA proteins at the C-terminus with the GCN4–pII motif stabilized the trimeric structure of the secreted recombinant HA protein [19].
The results were obtained when characterizing the structure of IMAC-purified artificial H5.c1 and native H5TG proteins by SEC, as shown in Figure 5. The HA proteins presented from A11 to B7 fractions, in which most of the HA proteins eluted in the B1 to B6 fractions, which were fractions containing standard proteins with high molecular weights (160–500 kDa). This result indicated that most of the purified H5 proteins were oligomeric. The purified oligomeric H5 proteins from the B1 to B6 fractions were collected and then used for ELISA and Western blotting to detect HA-specific IgG mouse antibody production. The purity of collected fractions were confirmed by SDS–PAGE and Coomassie stain (Figure S2).
Figure 5.
Structural characterization of the purified haemagglutinin (HA) proteins by size exclusion chromatography (SEC) and the A9–B9 SEC fractions analyzed by SDS–PAGE 10% under reducing conditions and Western blot using an anti–c–myc antibody.
3.3. The Artificial H5.c1 Protein Strongly Elicited Haemagglutinin (HA)-Specific IgG and Neutralizing Antibodies
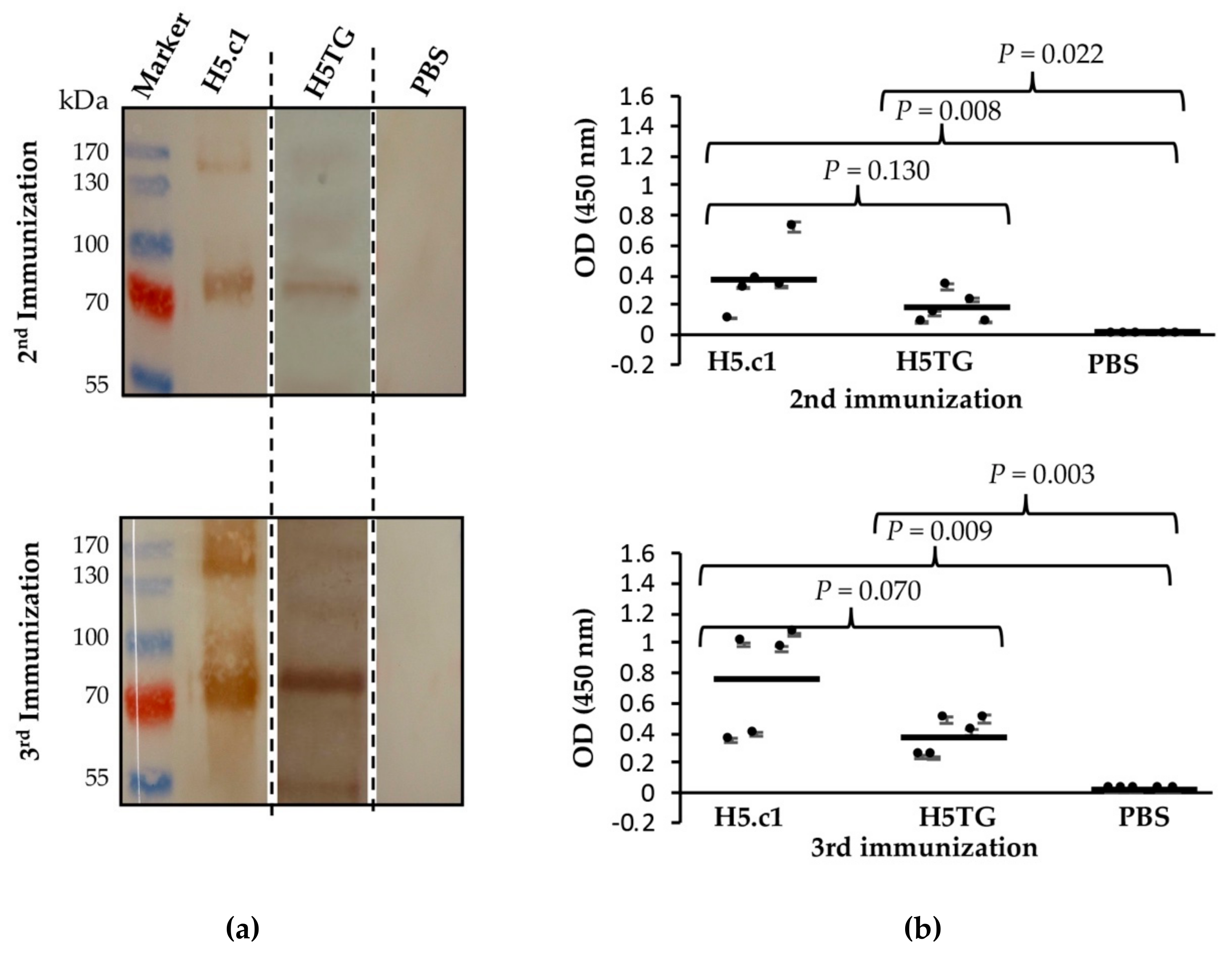
Immunogenicity of the artificial H5.c1 protein and native H5TG was tested by immunization of mice. Five BALB/c mice from each group were immunized with IMAC-purified artificial H5.c1 protein (group 1), IMAC-purified native H5TG protein (group 2), or PBS, as a negative control (group 3). The resulting antibody-dependent humoral immune responses against the SEC-purified HA proteins were first evaluated by SDS–PAGE and Western blot (Figure 6a). Five mouse sera collected after the second and third immunizations of each group (artificial H5.c1 protein, native H5TG and PBS) were mixed and used as a primary antibody. Specific IgG antibodies against SEC-purified HA proteins were detected following the second and the third immunizations. After the third immunization, a much stronger band signal was clearly visible compared to that following the second immunization in both the artificial H5.c1 and native H5TG groups. The band signal of the artificial H5.c1 group was significantly stronger than that of the native H5TG group following the second immunization. In the negative control mice group vaccinated with PBS, there was no HA-specific IgG antibody response induced.
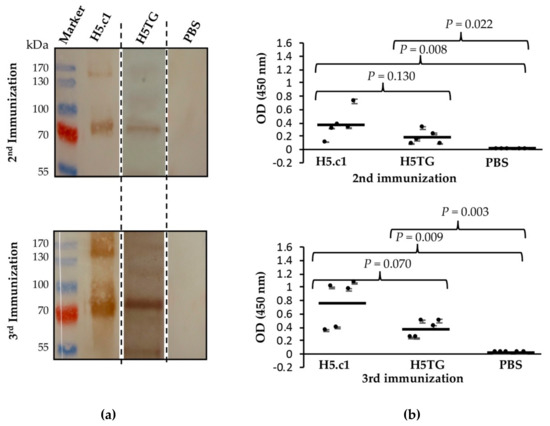
Figure 6.
Western blot to detect haemagglutinin (HA)-specific IgG antibodies in mice sera (a) and an indirect ELISA to detect HA-specific IgG antibodies in mice sera at the 16 × 10-3 serum dilution (b).
ELISA is the most common immunoassay used for detecting and quantifying a target substance (typically an antigen or antibody) in which an enzymatic reaction is used to amplify the signal if the target substance is present [35]. In this study, the immune responses in the sera of five mice of groups 1 and 2 were measured against the SEC-purified HA proteins by indirect ELISA. Five individual sera in each group were characterized and compared by t-test analysis in Sigma Plot software. Figure 6b shows the mean OD450 values of each mouse, with the standard deviations represented as bars, individual values as dots, and geometric mean titres as horizontal bars for each test group. The mean OD450 value of group 1 (artificial H5.c1) mouse sera at the 16 × 10-3 serum dilution was not significantly different from that of group 2 (the native H5TG) following the second immunization (P value = 0.130) and the third immunization (P value = 0.070). No immune response against HA proteins was detected in mouse sera from mice vaccinated with PBS.
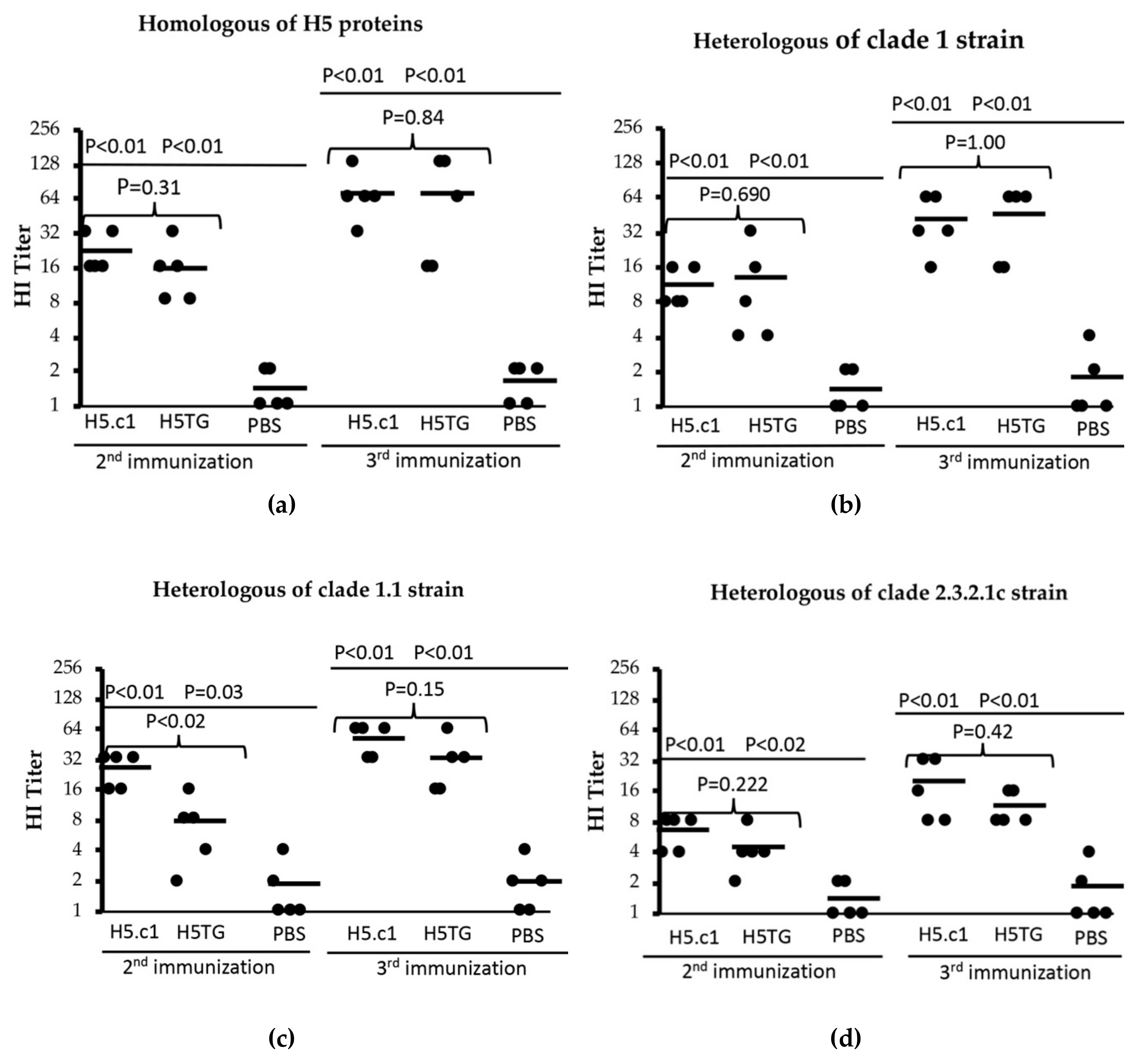
Hemagglutination inhibition (HI) assays are commonly used to measure neutralizing antibodies induced in serum after immunization with HA protein. In this study, four HI assays, including one for homologous artificial HA proteins and three for heterologous strains, were used for testing induced neutralizing antibodies in mouse sera following the second and third immunizations. The results are shown in Figure 7, in which a single dot indicates the HA titre of a single serum sample, and the horizontal bars represent the geometric mean titre of each test group.
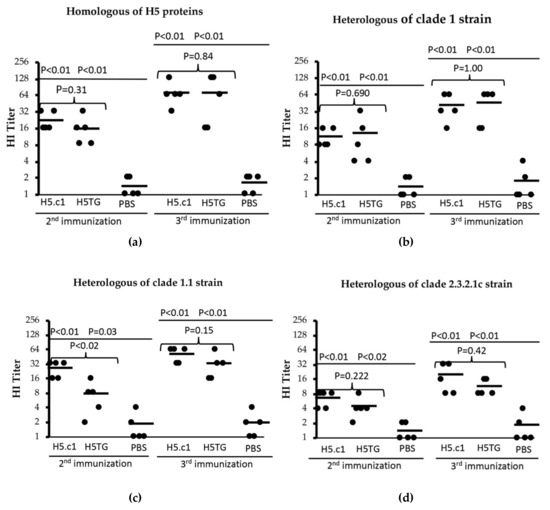
Figure 7.
Haemagglutinin inhibition (HI) titres against homologous HA proteins (a) and heterologous strains (A/Vietnam/1194/2004(H5N1) – clade 1 (b), A/duck/Vietnam/ST0970/2009(H5N1) - clade 1.1 (c) and A/duck/Vietnam/HT2/2014(H5N1) - clade 2.3.2.1c (d)).
In our first HI assay, the SEC-purified artificial H5.c1 and H5TG proteins were used as the homologues for the tested mouse sera. In the mouse group that was vaccinated with the artificial H5.c1 protein, the HI titre ranged from 16–32, and the HI titre mean was 22.4 after the second immunization. Following the third immunization, all of the sera had HI titres from 32–128, and the HI titre mean was 70.4. In the mouse group that was vaccinated with the native H5.c1 protein, the HI titre ranged from 8–32, and the HI titre mean was 16 after the second immunization. Following the third immunization, all of the sera had HI titres from 16–128, and the HI titre mean was 70. However, the difference between these two groups was not statistically significant, with a P value > 0.05 (as shown in Figure 7a).
Second, a heterologous inactivated virus strain whose HA protein derives from A/Vietnam/1194/2004(H5N1) NIBRG–14, clade 1, was used. The haemagglutinin amino acid sequence similarities of this strain compared to that of the artificial H5.c1 and the native H5TG proteins were 98.4% and 99.6%, respectively (see Appendix A Figure A3). All of the mice that were vaccinated with the artificial H5.c1 protein had HI titres from 8–16 with a mean of 11.2 after the 2nd immunization and 16–64 with a mean of 41.6 after the 3rd immunization. All of the mice that were vaccinated the native H5TG protein had HI titres from 4–32 with a mean of 12.8 after the 2nd immunization and 16–64 with a mean of 45 after the 3rd immunization (as shown in Figure 7b).
Third, we used a heterologous inactivated virus strain, A/duck/Vietnam/ST0970/2009(H5N1), clade 1.1, with the haemagglutinin ectodomain amino acid sequence similarity compared to that of the artificial H5.c1 and the native H5TG protein of 98,8% and 97.6%, respectively (Appendix A Figure A3). All of the mice that were vaccinated the artificial H5.c1 protein had HI titres from 16–32 with a mean of 25.6 (the second immunization) and 32–64 with a mean of 51.2 (the third immunization). All of the mice that were vaccinated the native H5TG protein had HI titres from 2–16 with a mean of 7.6 after the second immunization and 16–64 with a mean of 32 after the third immunization (as shown in Figure 7c).
Finally, a heterologous inactivated virus strain, A/duck/Vietnam/HT2/2014(H5N1), clade 2.3.2.1c, was introduced for the HI assay. The amino acid sequence similarity of the haemagglutinin ectodomain of this strain compared to that of the artificial H5.c1 and the native H5TG proteins was 92.5% and 92.9%, respectively (see Appendix A Figure A3). The results are shown in Figure 7d. All of the mice that were vaccinated the artificial H5.c1 protein had HI titres from 4–8 with a mean of 6.4 (the second immunization) and 8–32 with a mean of 19.2 (the third immunization). All of the mice that were vaccinated the native H5TG protein had HI titres from 2–8 with a mean of 4 (the second immunization) and 8–16 with a mean of 11 (the third immunization). In contrast, in all of the HI assays, the PBS negative control serum group had an HI titre of ≤ 4. These HI titre results indicated that the artificial H5.c1 and the native H5TG protein induced neutralizing antibodies against homologous protein and that of A/H5N1 heterologous strain clade 1 and clade 1.1 more than against that of A/H5N1 heterologous strains from clade 2.3.2.1c.
4. Discussion
The HA is a major target antigen for influenza vaccine manufacture and plays a crucial role in the host immune protection against influenza viruses exhibited at neutralizing activity [5]. HA sequence keeps changing and thus the influenza vaccice needs to be updated annually. This has prompted many scientists to study in a new trend, looking for HA sequences that are able to produce a broad and long-lasting immune response [8,9,10,11,12,36,37,38]. Following this trend, in the present study, two novel artificial trimeric haemagglutinin proteins representing A/H5N1 that was detected in poultry from 2005 to 2015 in Vietnam, H5.c1 (representing all of the subclades 1.1, 1.1.1, and 1.1.2) and H5.c2 (representing all of the subclades 2.3.2.1, 2.3.2.1a, 2.3.2.1b, and 2.3.2.1c), were designed. The artificial HA protein sequences of each group (clade 1 and clade 2) were compared to those of the native HA sequences. The amino acid sequence alignment results showed that the sequence identity of artificial H5.c1 and native H5TG sequences was 98.4%, whereas the sequence identity of artificial H5.c2 and native H5Dk sequences was 99.4% at the amino acid level. The differences in some important sites in the amino acid sequence of HA proteins, such as protease cleavage sites and antigenic sites of the HA polypeptide, are shown in Table 1. The amino acid sequence of the artificial H5.c1 protein was not similar to that of the native H5TG sequences at the cleavage site (341G>R) and antigenic site (156Q>K), whereas differences at the amino acid level of artificial H5.c2 and native H5Dk sequences were at antigenic sites (99A>P, 140D>N). When comparing the three-dimensional structures between artificial and native proteins, the H5 protein structures were not distinguishable. However, the level of protein expression of the artificial H5.c2 protein in N. benthamiana was lower than that of the native H5Dk protein, whereas the protein expression level of the artificial H5.c1 protein and that of native H5TG in N. benthamiana was similar. The difference in protein expression level between artificial H5.c2 and native H5Dk protein might be explained by the dissimilarity in amino acid sequence since amino acids act as substrates for protein synthesis as well as protein expression [39]. It has been reported that the high level of amino acid mutation-based HA expression can increase or remain HA-assayed titre and HI-assayed titre of influenza vaccine [40]. The lower level of protein expression of the H5.c2 protein led to a lower HA titre obtained in the haemagglutination assay than that of H5.c1 protein in the tobacco crude extracts. In addition, the small protein expression level of H5.c2 protein in tobacco leaves resulting in difficulty to purify essential H5.c2 protein amount for further experiments.
Biostructural characterization of the artificial H5.c1 protein which highly expressed in plant was performed by cross-linking reactions and SEC. These results confirmed again that the artificial H5.c1 protein structure was a trimeric. Various studies demonstrated the capacity to efficiently induce neutralizing antibodies of oligomeric and trimeric haemagglutinins in vaccinated animals, whereas monomeric haemagglutinins do not have that ability [19,20,21,41,42,43]. Hoang and co-workers demonstrated that 10 µg of plant-made purified trimeric haemagglutinins [21] were highly immunogenic and elicited neutralizing antibodies in mice against homologous inactivated A/H5N1 strains. In this study, with 2.5 µg of IMAC-purified trimeric haemagglutinins inoculated per mouse, the artificial H5.c1 protein-specific IgG antibodies in mouse sera and neutralizing antibodies after vaccination were strongly induced, as described in the indirect ELISA and HI assay results. Notably, the neutralizing antibodies against a homologous artificial H5.c1 protein and those from two heterologous A/H5N1 strains of clade 1 and clade 1.1 were expressed by HI titre, with the mean HI titres after the third immunization all being over 40. For utilization as a vaccine candidate, the recombinant haemagglutinins have to induce strong target-specific antibodies and neutralizing antibodies with a serum HI titre ≥ 40 [44]. So, it indicated that the artificial H5.c1 protein can become a potential influenza vaccine candidate.
Due to viral antigenic drift, the currently available influenza vaccines have become less effective at protection, and formulations must be updated annually. The development of a broadly protective vaccine or universal influenza vaccine with long-term protective immunity against antigenically distant influenza virus strains is urgently needed [16]. Several studies have utilized haemagglutinin (HA) to generate broadly protective vaccines. However, all of them were based on the conserved influenza virus haemagglutinin sequence and the ability to stimulate cross-reactive neutralizing antibody responses to the HA protein [8,9,10,11,12,36,37,38]. In this study, we successfully generated a novel artificial trimeric H5.c1 haemagglutinin and subsequently demonstrated that these artificial H5.c1 proteins induced strong cross-reactive antibody responses against three heterologous strains, A/Vietnam/1194/2004(H5N1) (clade 1), A/duck/Vietnam/ST0970/2009(H5N1) (clade 1.1) and A/duck/Vietnam/HT2/2014(H5N1) (clade 2.3.2.1c). Therefore, the artificial H5.c1 protein can become a promising influenza vaccine candidate against many different A/H5N1 strains in poultry in Vietnam.
5. Conclusions
The production of influenza subunit vaccine candidates using the plant-based transient expression system is an efficient method due to its scalability and potential for rapid production. Two major procedures have been used to efficiently produce influenza vaccine candidates: expression of haemagglutinins and purification of expressed HA proteins. The novel artificial HA proteins were successfully expressed in plants. The protein expression level, function, and structure of the artificial H5.c1 haemagglutinin protein were similar to those of the native H5TG haemagglutinin protein. The purified artificial H5.c1 protein strongly elicited neutralizing antibody responses in mice. Thus, the artificial H5.c1 protein is expected to become a promising influenza vaccine candidate. Further work will be focused on applying the artificial H5.c1 protein in chicken studies and virus challenges to assess the ability to protect animals against different variants of A/H5N1 clades 1.1, 1.1.1, and 1.1.2.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/9/21/4605/s1, Figure S1: Optimization of imidazole concentration for purification of H5.c1 by immobilized metal ion affinity chromatography (IMAC), Figure S2: Purification of H5.c1 and H5TG by size exclusion chromatography (SEC).
Author Contributions
Conceptualization, methodology, data curation, formal analysis; Writing—Original draft preparation, Writing—Review and editing, project administration, funding acquisition, supervision, H.H.C.; Investigation, conceptualization, methodology, data curation, formal analysis, Writing—Original draft preparation and editing, V.T.P.; Investigation, Writing—Original draft preparation and editing, T.T.H. and H.T.P.; conceptualization, methodology, Writing—Review and editing, T.H.L., N.B.P. and U.C.; Supervision, conceptualization, methodology, Writing—Review and editing, T.H.V.
Funding
The study is supported by Ministry of Science and Technology (MOST), Vietnam; the project “Study on production of immunogenic HA from A/H5N1 virus by agroinfiltration in tobacco”, code NĐT.07.GER.15 at IBT, Hanoi, Vietnam and the Federal Ministry of Education and Research (BMBF), Germany, “Plantfluvac” (Production, purification and characterization of plant-based Avian flu antigens), code 031A283 at IPK, Gatersleben, Germany.
Acknowledgments
We would like to thank Do Thi Thao from IBT, Hanoi, Vietnam for animal experiments. We are grateful to Nguyen Trung Nam from IBT, Hanoi, Vietnam and Tran Xuan Hanh from National Veterinary Joint Stock Company (NAVETCO), Vietnam, for providing the inactivated A/H5N1 virus strains for HI assays. This study is also a part of the thesis results of PhD student Van Thi Pham at GUST, VAST, Hanoi, Vietnam.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Four HA sequences, including the clade 1 group (artificial H5.c1 and native H5TG) and the clade 2 group (artificial H5.c2 and native H5Dk), were submitted to http://raptorx.uchicago.edu/ to predict the structure of these proteins. The results are shown in Appendix A Figure A1. All of them were compared at the amino acid level with each other and with two HA sequences (designated NIBRG14 and ST0970, deriving from the native strains A/Vietnam/1194/2004(H5N1) and A/duck/Vietnam/ST0970/2009(H5N1), respectively) using HI assays. The results were expressed by an evolutionary tree using MEGA7 (shown in Appendix A Figure A2) and percent identity using MegAlign/Lasergene7 (shown in Appendix A Figure A3).
Figure A1.
The predicted HA protein structures by RaptorX.
Figure A1.
The predicted HA protein structures by RaptorX.
Figure A2.
Evolutionary relationships of HA proteins
Figure A2.
Evolutionary relationships of HA proteins
Figure A3.
Amino acid sequence identity percentage of HA proteins.
Figure A3.
Amino acid sequence identity percentage of HA proteins.
References
- Yen, H.L.; Webster, R.G. Pandemic Influenza as a Current Threat. In Vaccines for Pandemic Influenza; Compans, R.W., Orenstein, W.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 3–24. [Google Scholar] [CrossRef]
- Office International des Epizooties (OIE)—World Organization for Animal Health. Update on Avian Influenza in Animals. 2019. Available online: http://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/ (accessed on 09 September 2019).
- Wilson, I.A.; Cox, N.J. Structural basis of immune recognition of influenza virus hemagglutinin. Annu. Rev. Immunol. 1990, 8, 737–771. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Marasco, W.A. Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 2011, 1217, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Shoji, Y.; Jones, R.M.; Mett, V.; Chichester, J.A.; Musiychuk, K.; Sun, X.; Tumpey, T.M.; Green, B.J.; Shamloul, M.; Norikane, J.; et al. A plant-produced H1N1 trimeric hemagglutinin protects mice from a lethal influenza virus challenge. Hum. Vaccines Immunother. 2013, 9, 553–560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le, T.H.; Nguyen, N.T.B. Evolutionary dynamics of highly pathogenic avian influenza A/H5N1 HA clades and vaccine implementation in Vietnam. Clin. Exp. Vaccine Res. 2014, 3, 117–127. [Google Scholar] [CrossRef]
- Crevar, C.J.; Ross, T.M. Elicitation of protective immune responses using a bivalent H5N1 VLP vaccine. Virol. J. 2008, 5, 131. [Google Scholar] [CrossRef]
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schults-Cherry, S.; Wiley, C.A.; Ross, T.M. A Computationally Optimized Hemagglutinin Virus-Like Particle Vaccine Elicits Broadly Reactive Antibodies that Protect Nonhuman Primates from H5N1 Infection. J. Infect. Dis. 2012, 205, 1562–1570. [Google Scholar] [CrossRef]
- Giles, B.M.; Bissel, S.J.; Dealmeida, D.R.; Wiley, C.A.; Ross, T.M. Breadth and Protective Efficacy Are Increased by Vaccination with Computationally Optimized Hemagglutinin but Not with Polyvalent Hemagglutinin-Based H5N1 Virus-Like Particle Vaccines. Clin. Vaccine Immunol. 2012, 19, 128–139. [Google Scholar] [CrossRef]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U.; et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef]
- Crevar, C.J.; Carter, D.M.; Lee, K.Y.J.; Ross, T.M. Cocktail of H5N1 COBRA HA vaccines elicit protective antibodies against H5N1 viruses from multiple clades. Hum. Vaccines Immunother. 2015, 11, 572–583. [Google Scholar] [CrossRef]
- Cho, A.; Wrammert, J. Implications of broadly neutralizing antibodies in the development of a universal influenza vaccine. Curr. Opin. Virol. 2016, 17, 110–115. [Google Scholar] [CrossRef]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Karikoó, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Lai, H.; Hurtado, J.; Stahnke, J.; Leuzinger, K.; Dent, M. Agroinfiltration as an Effective and Scalable Strategy of Gene Delivery for Production of Pharmaceutical Proteins. Adv. Tech. Biol. Med. 2013, 1, 10. [Google Scholar] [CrossRef]
- Topp, E.; Irwin, R.; McAllister, T.; Lessard, M.; Joensuu, J.J.; Kolotilin, I.; Conrad, U.; Stöger, E.; Mor, T.; Warzecha, H.; et al. The case for plant-made veterinary immuno therapeutics. Biotechnol. Adv. 2016, 34, 597–604. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, M.A.; Couture, M.M.J.; Charland, N.; Trépanier, S.; Landry, N.; Ors, F.; Vézina, L.P. The production of hemagglutinin-based virus-like particles in plants: A rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 2010, 8, 607–619. [Google Scholar] [CrossRef] [PubMed]
- D’Aoust, M.A.; Lavoie, P.O.; Couture, M.M.J.; Trépanier, S.; Guay, J.M.; Dargis, M.; Mongrand, S.; Landry, N.; Ward, B.J.; Vézina, L.P. Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 2008, 6, 930–940. [Google Scholar] [CrossRef]
- Impagliazzo, A.; Milder, F.; Kuipers, H.; Wagner, M.V.; Zhu, X.; Hoffman, R.M.; Van Meersbergen, R.; Huizingh, J.; Wanningen, P.; Verspuij, J.; et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015, 349, 1301–1306. [Google Scholar] [CrossRef]
- Weldon, W.C.; Wang, B.Z.; Martin, M.P.; Koutsonanos, D.G.; Skountzou, I.; Compans, R.W. Enhanced immunogenicity of stabilized trimeric soluble influenza hemagglutinin. PLoS ONE 2010, 5, e12466. [Google Scholar] [CrossRef]
- Phan, H.T. ELPylated Avian Flu Vaccines from Plants: Improvement of Expression and Development of a New Purification Strategy. Ph.D. Thesis, Martin-Luther-Universität Halle-Wittenberg, Halle (Saale), Germany, 2 October 2012. [Google Scholar]
- Phan, H.T.; Pohl, J.; Floss, D.M.; Rabenstein, F.; Veits, J.; Le, B.T.; Chu, H.H.; Hause, G.; Mettenleiter, T.; Conrad, U. ELPylated haemagglutinins produced in tobacco plants induce potentially neutralizing antibodies against H5N1 viruses in mice. Plant Biotechnol. J. 2013, 11, 582–593. [Google Scholar] [CrossRef]
- Harbury, P.; Zhang, T.; Kim, P.; Alber, T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 1993, 262, 1401–1407. [Google Scholar] [CrossRef]
- Phan, H.T.; Conrad, U. Plant-Based Vaccine Antigen Production. In Vaccine Technologies for Veterinary Viral Diseases: Methods and Protocols; Brun, A., Ed.; Humana Press; Springer: New York, NY, USA, 2016; Volume 1349, pp. 35–47. [Google Scholar] [CrossRef]
- Phan, H.T.; Ho, T.T.; Chu, H.H.; Vu, T.H.; Gresch, U.; Conrad, U. Neutralizing immune responses induced by oligomeric H5N1-hemagglutinins from plants. Vet. Res. 2017, 48, 53. [Google Scholar] [CrossRef][Green Version]
- Sudarshana, M.R.; Plesha, M.A.; Uratsu, S.L.; Falk, B.W.; Dandekar, A.M.; Huang, T.K.; McDonald, K.A. A chemically inducible cucumber mosaic virus amplicon system for expression of heterologous proteins in plant tissues. Plant Biotechnol. J. 2006, 4, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Conley, A.J.; Joensuu, J.J.; Jevnikar, A.M.; Menassa, R.; Brandle, J.E. Optimization of elastin-like polypeptide fusions for expression and purification of recombinant proteins in plants. Biotechnol. Bioeng. 2009, 103, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Gahrtz, M.; Conrad, U. Immunomodulation of plant function by in vitro selected single-chain Fv intrabodies. Methods Mol. Biol. 2009, 483, 289–312. [Google Scholar]
- Pham, N.B.; Ho, T.T.; Nguyen, G.T.; Le, T.T.; Le, N.T.; Chang, H.C.; Pham, M.D.; Conrad, U.; Chu, H.H. Nanodiamond enhances immune responses in mice against recombinant HA/H7N9 protein. J. Nanobiotechnol. 2017, 15, 69. [Google Scholar] [CrossRef][Green Version]
- Office International des Epizooties (OIE). Avian influenza (infection with avian influenza viruses), in Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2019; Chapter 3.3.4. World Organization for Animal Health: Cham, Switzerland; pp. 830–831. Available online: http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/ (accessed on 05 June 2019).
- Phan, H.T.; Hause, B.; Hause, G.; Arcalis, E.; Stoger, E.; Maresch, D.; Altmann, F.; Joensuu, J.; Conrad, U. Influence of elastin-like polypeptide and hydrophobin on recombinant hemagglutinin accumulations in transgenic tobacco plants. PLoS ONE 2014, 9, e99347. [Google Scholar] [CrossRef]
- Bornhorstand, J.A.; Falke, J.J. Purification of proteins using polyhistidine affinity tags. Methods Enzymol. 2000, 326, 245–254. [Google Scholar]
- Davies, G.E.; Stark, G.R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc. Natl. Acad. Sci. USA 1970, 66, 651–656. [Google Scholar] [CrossRef]
- Ruigrok, R.W.; Aitken, A.; Calder, L.J.; Martin, S.R.; Skehel, J.J.; Wharton, S.A.; Weis, W.; Wiley, D.C. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J. Gen. Virol. 1988, 69, 2785–2795. [Google Scholar] [CrossRef]
- O’Connor, T.P. SNAP Assay Technology. Top. Companion Anim. Med. 2015, 30, 132–138. [Google Scholar] [CrossRef][Green Version]
- Anderson, C.S.; Ortega, S.; Chaves, F.A.; Clark, A.M.; Yang, H.; Topham, D.J.; DeDiego, M.L. Natural and directed antigenic drift of the H1 influenza virus hemagglutinin stalk domain. Sci. Rep. 2017, 7, 14614. [Google Scholar] [CrossRef] [PubMed]
- Rajaão, D.S.; Peérez, D.R. Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front. Microbiol. 2018, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Mallajosyula, V.V.; Li, O.T.; Chin, A.W.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016, 6, 22666. [Google Scholar] [CrossRef] [PubMed]
- Kimball, S.R.; Jefferson, L.S. Amino acids as regulators of gene expression. Nutr. Metab. (Lond.) 2004, 1, 3. [Google Scholar] [CrossRef]
- Barman, S.; Krylov, P.S.; Turner, J.C.; Franks, J.; Webster, R.G.; Husain, M.; Webby, R.J. Manipulation of neuraminidase packaging signals and hemagglutinin residues improves the growth of A/Anhui/1/2013 (H7N9) influenza vaccine virus yield in eggs. Vaccine 2017, 35, 1424–1430. [Google Scholar] [CrossRef]
- Bosch, B.J.; Bodewes, R.; De Vries, R.P.; Kreijtz, J.H.; Bartelink, W.; Van Amerongen, G.; Rimmelzwaan, G.F.; De Haan, C.A.; Osterhaus, A.D.; Rottier, P.J. Recombinant soluble, multimeric HA and NA exhibit distinctive types of protection against pandemic swine-origin 2009 A(H1N1) influenza virus infection in ferrets. J. Virol. 2010, 84, 10366–10374. [Google Scholar] [CrossRef]
- Cornelissen, L.A.H.M.; De Vries, R.P.; De Boer-Luijtze, E.A.; Rigter, A.; Rottier, P.J.M.; De Haan, C.A.M.; Tripp, R. A single immunization with soluble recombinant trimeric hemagglutinin protects chickens against highly pathogenic avian influenza virus H5N1. PLoS ONE 2010, 5, e10645. [Google Scholar] [CrossRef]
- Wei, C.J.; Xu, L.; Kong, W.P.; Shi, W.; Canis, K.; Stevens, J.; Yang, Z.Y.; Dell, A.; Haslam, S.M.; Wilson, I.A.; et al. Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J. Virol. 2008, 82, 6200–6208. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. Epidemiol. Infect. 1972, 70, 767–777. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).