Filtering of Mammograms Based on Convolution with Directional Fractal Masks to Enhance Microcalcifications
Abstract
:Featured Application
Abstract
1. Introduction
2. Description of the algorithm
- A preprocessing step to detect and localize microcalcifications.
- The isolation, enhancement and segmentation of a particular microcalcification.
- Zoom the region of interest of the image around the microcalcification clusters. Use simple nearest neighbor zooming without interpolation.
- Use a complete set of fractal filters for pixel diffusion. Convolve each fractal mask with the original ROI. A very good zooming factor is × 8 when a set of fractal masks composed of 5 × 5 pixels is used, with angular increments of 45 degrees from 0 to 315 degrees.
- Duplicate the resulting image, and then calculate the logarithm of the first image and the exponential of the second one.
- Apply the logical operation XOR between both images. After the previous enhance contrast step, the XOR operation produces a grayscale segmented image. This result permits the definition of isolevel regions and a clear delineation of the shape of the selected microcalcification. The logical XOR operation between images is described in the Image user guide [29].
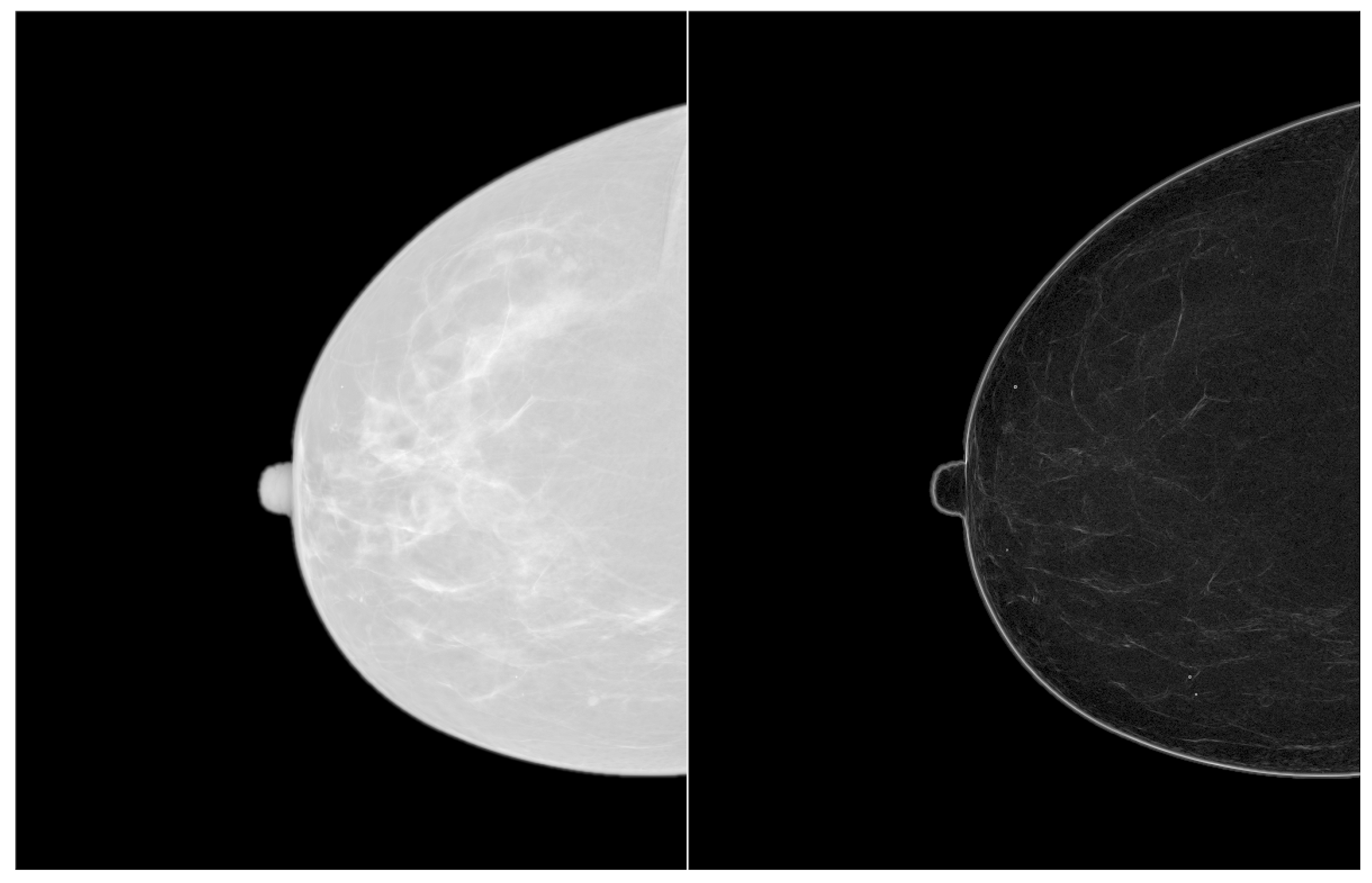
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Sanchez, R.; Garcia, J.A.; Fernandez-Valdivia, J.; Fernandez-Vidal, X.R. How to define the notion of microcalcifications in digitized mammograms. In Proceedings of the 15th International Conference of Pattern Recognition, Barcelona, Spain, 3–7 September 2000; Volume 1, pp. 494–499. [Google Scholar]
- Bazzani, A.; Bevilacqua, A.; Bollini, D.; Brancaccio, R.; Campanini, R.; Lanconelli, N.; Riccardi, A.; Romani, D. An SVM classifier to separate false signals from microcalcifications in digital mammograms. Phys. Med. Biol. 2001, 46, 1651–1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthorpe, R.J.; Worden, K. Classification of multi-site damage using support vector machines. J. Phys. Conf. Ser. 2011, 305, 012059. [Google Scholar] [CrossRef] [Green Version]
- Davies, D.H.; Dance, D.R. Automatic computer detection of clustered calcifications in digital mammograms. Phys. Med. Biol. 1990, 35, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Elter, M.; Held, C. Semiautomatic segmentation for the computer aided diagnosis of clustered microcalcifications. Proc. SPIE 2008. [Google Scholar] [CrossRef]
- Thangavel, K.; Karnan, M. Computer Aided Diagnosis in Digital Mammograms: Detection of Microcalcifications by Meta Heuristics Algorithms. GVIP J. 2005, 5, 41–55. [Google Scholar]
- Jing, H.; Yang, Y.; Nishikawa, R.M. Detection of clustered microcalcifications using spatial point process modeling. Phys. Med. Biol. 2011, 56, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bozek, J.; Mustra, M.; Dela, K.; Grgic, M. A Survey of Image Processing Algorithms in Digital Mammography. In Recent Advances in Multimedia Signal Processing and Communications; Springer: Berlin/Heidelberg, Germany, 2009; pp. 631–657. [Google Scholar]
- Li, H.; Liu, K.J.R.; Lo, S.-C.B. Fractal modelling and segmentation for the enhancement of microcalcifications in digital mammograms. IEEE Trans. Med Imaging 1997, 16, 785–798. [Google Scholar]
- Dudczyk, J.; Kawalec, A. Fractal features of specific emitter identification. Acta Phys. Pol. A 2013, 124, 406–409. [Google Scholar] [CrossRef]
- Dudczyk, J.; Kawalec, A. Identification of emitter sources in the aspect of their fractal features. Bull. Pol. Acad. Sci. 2013, 61, 623–628. [Google Scholar] [CrossRef] [Green Version]
- Sankar, D.; Thomas, T. Fractal Modeling of Mammograms based on Mean and Variance for the Detection of Microcalcifications. In Proceedings of the 2007 International Conference on Computational Intelligence and Multimedia Applications, Sivakasi, Tamil Nadu, India, 13–15 December 2007; pp. 334–338. [Google Scholar]
- Rojas, J.A.; Alpuente, J.; Rojas, I.M.; Vignote, S. Fractal-based image enhancement techniques inspired by differential interference contrast microscopy. J. Opt. A Pure Appl. Opt. 2009, 11, 1–8. [Google Scholar] [CrossRef]
- Sakellaropoulos, P.; Costaridou, L.; Panayiotakis, G. A wavelet-based spatially adaptive method for mammographic contrast enhancement. Phys. Med. Biol. 2003, 48, 787–803. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.; Pina, D.R.; Miranda JR, A.; Duarte, S.B. Application of wavelets to the evaluation of phantom images for mammography quality control. Phys. Med. Biol. 2012, 57, 7177–7190. [Google Scholar] [CrossRef] [PubMed]
- Salvado, J.; Roque, B. Detection of Calcifications in Digital Mammograms using Wavelet Analysis and Contrast Enhancement. In Proceedings of the IEEE International Workshop on Intelligent Signal Processing, Faro, Portugal, 1–3 September 2005; pp. 200–205. [Google Scholar]
- Suckling, J.; Parker, J.; Dance, D.R.; Astley, S.; Hutt, I.; Boggis, C.R.M.; Ricketts, I.; Stamatakis, E.; Cernaez, N.; Kok, S.L.; et al. The Mammographic Image Analysis Society Digital Mammograms Database. In Proceedings of the 2nd International Workshop on Digital Mammography, York, UK, 10–12 July 1994; pp. 375–378. [Google Scholar]
- Juarez, L.C.; Ponomaryov, V.; Sanchez RJ, L. Detection of Microcalcifications in Digital Mammograms Images Using Wavelet Transform. In Proceedings of the Electronics, Robotics and Automotive Mechanics Conference, Cuernavaca, Mexico, 26–29 September 2006; Volume 2, pp. 58–61. [Google Scholar]
- Song, L.; Wang, Q.; Gao, J. Microcalcifications detection using combination of wavelet transform and morphology. In Proceedings of the 8th International Conference on Signal Processing, ICSP 2006, Guilin, China, 16–20 November 2006; Volume 4, pp. 16–20. [Google Scholar]
- Heinlein, P.; Drexl, J.; Scheneider, W. Integrated Wavelets for Enhancement of Microcalcifications in Digital Mammograph. IEEE Trans. Med Imaging 2003, 22, 402–413. [Google Scholar] [CrossRef]
- Mencattini, A.; Salmeri, M.; Lojacono, R.; Frigerio, M.; Caselli, F. Mammographic Images Enhancement and Denoising for Breast Cancer Detection Using Dyadic Wavelet Processing. IEEE Trans. Instrum. Meas. 2008, 57, 1422–1430. [Google Scholar] [CrossRef]
- Tsai, D.Y.; Matsuyama, E.; Chen, H.M. Improving image quality in medical images using a combined method of undecimated wavelet transform and wavelet coefficient mapping. Int. J. Biomed. Imaging 2013, 2013, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jagatheeswari, P.; Suresh Kumar, S.; Mary Linda, M. Quadrant dynamic with automatic plateau limit histogram equalization for image enhancement. Math. Probl. Eng. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Perona, P.; Malik, J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 629–639. [Google Scholar] [CrossRef] [Green Version]
- Bunde, A.; Havlin, S. Fractals in Science; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Weickert, J. Anisotropic Diffusion in Image Processing; Teuber Verlag: Stuttgart, Germany, 1998. [Google Scholar]
- Lindeberg, T. Feature detection with automatic scale selection. Int. J. Comput. Vis. 1998, 30, 77–116. [Google Scholar]
- Lindeberg, T. Generalized axiomatic scale-space theory. Adv. Imaging Electron Phys. 2013, 178, 1–96. [Google Scholar]
- ImageJ User Guide. Available online: https://imagej.nih.gov/ij/docs/guide/ (accessed on 26 February 2019).
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529–555. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Digital Repository. Available online: https://bcdr.eu/information/downloads (accessed on 30 January 2019).
- IMAGEJ PLUGINS Developed at UMRS-INSERM 514 (Reims, France) under the Supervision of Noël BONNET. Available online: https://imagej.nih.gov/ij/plugins/inserm514/#No-threshold (accessed on 25 January 2019).
- Shensa, M.J. Discrete Wavelet Transforms: Wedding the a trous and Mallat algorithms. IEEE Trans. Signal Process. 1992, 40, 2.464–2.482. [Google Scholar]
- Chang, C.H.-M.; Laine, A. Coherence of Multiscale Features for Enhancement of Digital Mammograms. IEEE Trans. Inf. Technol. Biomed. 1999, 3, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.C.; Karayiannis, N.B. Detection of Microcalcifications in Digital Mammograms Using Wavelets. IEEE Trans. Med. Imaging 1998, 17, 498–509. [Google Scholar] [CrossRef] [PubMed]

















| Algorithm | Time (s) |
|---|---|
| Edges | 0.04 |
| Zoom | 0.05 |
| ROI | CII Fractal | CII CLAHE | CII Atrous | CII Haar |
|---|---|---|---|---|
| Mammogram1 | 9.00329385 | 0.78247068 | 2.03116094 | 1.06843971 |
| Mammogram2 | 3.56662498 | 0.79058571 | 0.41786664 | 1.40218904 |
| Mammogram3 | 4.14070347 | 2.14333457 | 0.84633132 | 0.42674917 |
| Mammogram4 | 4.274771 | 1.36151986 | 1.49876914 | 0.68315829 |
| Mammogram5 | 2.67706791 | 1.10035115 | 1.58543392 | 1.56701435 |
| Mammogram6 | 2.758558 | 0.30791769 | 0.24688073 | 0.5524436 |
| Mammogram7 | 3.65402443 | 1.19168866 | 1.5369818 | 1.13065924 |
| Mammogram8 | 5.40648243 | 0.34326964 | 0.89710593 | 1.44899629 |
| Mammogram9 | 2.99186103 | 1.10269593 | 1.20163771 | 2.09266653 |
| Mammogram10 | 4.40374138 | 0.56431707 | 0.35189964 | 0.09538283 |
| Mammogram11 | 3.20109307 | 0.54621531 | 3.05219551 | 2.78978836 |
| Mammogram12 | 3.11400989 | 1.49727518 | 0.56953524 | 0.34750761 |
| Mammogram13 | 4.3998446 | 0.89017546 | 2.98889635 | 2.29192808 |
| Mammogram14 | 4.255415 | 0.05292143 | 0.11102128 | 0.15881968 |
| Mammogram15 | 6.11817365 | 1.19879222 | 0.109 | 0.12262106 |
| Mammogram16 | 7.65287633 | 1.47122771 | 2.58386652 | 4.78291022 |
| Mammogram17 | 4.08819571 | 0.63090961 | 5.12244121 | 8.09071728 |
| Mammogram18 | 5.16468499 | 1.48876198 | 1.61087759 | 2.02798801 |
| Mammogram19 | 10.7184181 | 3.6155906 | 0.52099839 | 0.98244152 |
| Mammogram20 | 11.7366576 | 6.12117844 | 1.28762178 | 1.18855975 |
| Mammogram21 | 23.3671203 | 3.03378728 | 1.44678872 | 6.18991257 |
| Mammogram22 | 58.4529769 | 9.96264465 | 0.60447142 | 6.81814316 |
| Mammogram23 | 12.6516423 | 0.28899773 | 2.4594087 | 12.3941907 |
| Mammogram24 | 1.65498759 | 1.43151168 | 7.15552118 | 7.87362745 |
| Mammogram25 | 2.87451829 | 2.33973211 | 0.70591978 | 0.89812281 |
| Mammogram26 | 3.36468404 | 0.47004935 | 1.66922658 | 1.37948437 |
| Mammogram27 | 8.996256 | 1.22939224 | 0.327697 | 1.13475112 |
| Mammogram28 | 3.88369335 | 0.41867164 | 1.82381339 | 2.77346834 |
| Mammogram29 | 2.89182277 | 3.2923157 | 3.55288931 | 2.11941866 |
| Mammogram30 | 3.80692411 | 1.12374573 | 2.61267326 | 2.68808429 |
| Mammogram31 | 46.0914604 | 11.677673 | 1.00957551 | 0.78720545 |
| Mammogram32 | 7.65678676 | 1.26215444 | 42.9881506 | 28.0206854 |
| Mammogram33 | 16.5575214 | 2.32745222 | 1.7570377 | 1.26522097 |
| Mammogram34 | 15.6562376 | 0.93201257 | 1.69772751 | 0.92729605 |
| Mammogram35 | 3.21988538 | 2.28041558 | 4.08016937 | 3.08891691 |
| Mammogram36 | 2.05907292 | 1.41508463 | 3.48174126 | 2.65394081 |
| Mammogram37 | 3.00040019 | 3.79383083 | 1.04593283 | 0.5417953 |
| Mammogram38 | 3.0048273 | 1.49438715 | 3.78050959 | 1.68271289 |
| Mammogram39 | 4.56890806 | 1.90957747 | 1.55319022 | 0.97922433 |
| Mammogram40 | 6.01854508 | 1.83805163 | 1.63728231 | 0.40482588 |
| ROI | PSNR Original | PSNR Fractal | PSNR CLAHE | PSNR Atrous | PSNR Haar |
|---|---|---|---|---|---|
| Mammogram1 | 14.9520375 | 26.0173658 | 5.29331434 | 19.1798131 | 28.3784897 |
| Mammogram2 | 10.3780269 | 25.7073607 | 3.75265808 | 9.82987038 | 8.113002 |
| Mammogram3 | 6.2968653 | 17.1834711 | 2.74185551 | 7.55762459 | 9.03115773 |
| Mammogram4 | 12.5311327 | 19.8738643 | 4.56125727 | 13.7681427 | 20.5082699 |
| Mammogram5 | 5.02124095 | 21.8352635 | 3.34000548 | 4.58755005 | 5.8595959 |
| Mammogram6 | 7.39672241 | 12.8427779 | 3.22695738 | 7.38436646 | 13.7693594 |
| Mammogram7 | 6.57004815 | 21.9250510 | 3.07412349 | 8.04142869 | 6.91677123 |
| Mammogram8 | 6.39077642 | 18.7973014 | 2.82520137 | 6.0909552 | 10.0848015 |
| Mammogram9 | 9.13698222 | 31.4672378 | 4.64409266 | 11.7880713 | 16.5602278 |
| Mammogram10 | 8.61808019 | 14.7112292 | 3.25784414 | 11.5992335 | 29.5477321 |
| Mammogram11 | 5.77780888 | 34.0763958 | 2.90028926 | 6.91335505 | 8.87043705 |
| Mammogram12 | 4.89290219 | 13.1691046 | 2.36540812 | 5.1120106 | 7.4325419 |
| Mammogram13 | 4.84325384 | 30.2829792 | 3.14601435 | 7.74845119 | 50.0889064 |
| Mammogram14 | 6.12435205 | 25.5351583 | 2.35412286 | 4.75589499 | 11.180581 |
| Mammogram15 | 4.6581734 | 8.83677307 | 2.4930723 | 5.90852575 | 11.0134513 |
| Mammogram16 | 8.18221154 | 10.0063355 | 3.43123805 | 9.03891051 | 0.27177053 |
| Mammogram17 | 5.28929 | 18.080768 | 2.77471237 | 4.61410416 | 21.225357 |
| Mammogram18 | 10.9771912 | 27.0783787 | 3.59516952 | 14.4752557 | 16.5651798 |
| Mammogram19 | 2.98289168 | 14.2268846 | 2.2561202 | 2.47962232 | 10.8961836 |
| Mammogram20 | 5.23672425 | 13.0167117 | 2.13761694 | 6.33414072 | 6.64206507 |
| Mammogram21 | 7.96266149 | 19.1265475 | 3.63517605 | 9.91701614 | 28.8666667 |
| Mammogram22 | 6.07544615 | 26.030846 | 2.62601951 | 6.32458276 | 14.6507345 |
| Mammogram23 | 3.44512195 | 13.6106648 | 1.89943456 | 5.28660093 | 8.60732738 |
| Mammogram24 | 3.70408645 | 16.7877909 | 2.06785039 | 5.77580242 | 8.76845222 |
| Mammogram25 | 4.06758289 | 10.116543 | 2.05662157 | 5.89074025 | 1.7334873 |
| Mammogram26 | 2.71627173 | 8.5 | 1.96199084 | 5.04315432 | 4.74570783 |
| Mammogram27 | 6.0246116 | 17.6884346 | 3.1371602 | 7.05380693 | 7.44753125 |
| Mammogram28 | 4.1505851 | 18.3424969 | 2.21777943 | 3.14137946 | 10.2381395 |
| Mammogram29 | 9.23976876 | 18.5481108 | 2.96087432 | 6.22028179 | 9.17295229 |
| Mammogram30 | 10.2501316 | 17.795401 | 3.8837785 | 13.3533498 | 14.1378149 |
| Mammogram31 | 5.05765431 | 11.6534946 | 2.56873638 | 11.6788344 | 7.8616893 |
| Mammogram32 | 11.3763816 | 27.0865679 | 3.97316532 | 15.7074364 | 13.3525884 |
| Mammogram33 | 9.23906741 | 28.088304 | 3.50745082 | 17.0012467 | 20.5801554 |
| Mammogram34 | 13.2308365 | 27.5411554 | 3.83210849 | 14.1484887 | 17.3376225 |
| Mammogram35 | 11.1090101 | 15.0001036 | 3.7831803 | 15.1415094 | 18.3323799 |
| Mammogram36 | 4.06041894 | 15.2942219 | 2.09879274 | 6.4998754 | 5.28183644 |
| Mammogram37 | 3.71631451 | 0.00761101 | 2.01044365 | 4.7762951 | 6.33515326 |
| Mammogram38 | 4.677277 | 15.3555181 | 3.19396062 | 11.4677325 | 7.46167068 |
| Mammogram39 | 12.2380422 | 23.4266447 | 5.00444973 | 17.1568245 | 16.6131838 |
| Mammogram40 | 3.47104752 | 15.1217554 | 2.3643739 | 5.45404851 | 5.07032952 |
| Mean | 6.92460218 | 18.7448156 | 3.07386053 | 8.85615834 | 13.0874717 |
| ROI | ASNR Original | ASNR Fractal | ASNR CLAHE | ASNR Atrous | ASNR Haar |
|---|---|---|---|---|---|
| Mammogram1 | 1.92138478 | 5.91102842 | 0.50921582 | 4.62321948 | 3.87551487 |
| Mammogram2 | 2.11717295 | 4.04948062 | 0.66931446 | 0.7440891 | 2.91985632 |
| Mammogram3 | 1.11138964 | 2.87870917 | 1.10719469 | 1.4921516 | 1.14638695 |
| Mammogram4 | 1.19000813 | 2.53444474 | 0.75085653 | 1.93138289 | 1.46009451 |
| Mammogram5 | 2.14968053 | 2.08434665 | 1.01530075 | 1.73020568 | 2.37210234 |
| Mammogram6 | 0.81209447 | 0.63797351 | 0.08715843 | 0.51395081 | 2.81033618 |
| Mammogram7 | 1.4074159 | 1.78118013 | 0.54110148 | 1.32290242 | 0.93022445 |
| Mammogram8 | 0.93712233 | 2.40160703 | 0.13560235 | 0.91253874 | 2.36980528 |
| Mammogram9 | 1.69113606 | 4.22512774 | 1.23281362 | 0.82040917 | 2.28332525 |
| Mammogram10 | 0.89741712 | 1.1995997 | 0.16657803 | 1.38046406 | 1.04852321 |
| Mammogram11 | 1.40445102 | 3.65471941 | 0.4385359 | 1.13876221 | 1.28477261 |
| Mammogram12 | 1.22417692 | 1.89515882 | 0.73293645 | 1.08708665 | 1.03798883 |
| Mammogram13 | 0.6209227 | 4.88426383 | 0.52534689 | 1.38448172 | 8.30343562 |
| Mammogram14 | 0.8024838 | 1.45149701 | 0.01418545 | 0.22644391 | 0.85464318 |
| Mammogram15 | 0.28838518 | 0.28987778 | 0.11143834 | 0.19321097 | 0.87504425 |
| Mammogram16 | 0.57740385 | 1.03213381 | 0.28080032 | 0.70804961 | 2.57346265 |
| Mammogram17 | 1.2180696 | 2.01317042 | 0.28141294 | 1.25546664 | 8.75792407 |
| Mammogram18 | 0.99763521 | 2.18082 | 0.58401093 | 2.5930329 | 4.12344746 |
| Mammogram19 | 0.35321101 | 1.97358239 | 0.58705142 | 0.34156223 | 2.67227115 |
| Mammogram20 | 0.08239649 | 0.42903662 | 0.19140577 | 0.4516453 | 0.60373063 |
| Mammogram21 | 0.23385149 | 0.94274415 | 0.22510653 | 0.38111577 | 5.19347826 |
| Mammogram22 | 0.04347826 | 1.46518485 | 0.17659523 | 0.0500683 | 1.24485876 |
| Mammogram23 | 0.22085933 | 1.88823377 | 0.02445453 | 0.1146704 | 1.01824018 |
| Mammogram24 | 0.86318562 | 0.72308279 | 0.46427999 | 1.83528733 | 3.16042413 |
| Mammogram25 | 0.56456352 | 0.6007358 | 0.43309007 | 0.99209694 | 0.72797757 |
| Mammogram26 | 0.62170787 | 1.03764453 | 0.1075992 | 1.14966919 | 1.42266566 |
| Mammogram27 | 0.42524978 | 1.32044393 | 0.23826021 | 0.23486314 | 0.92555878 |
| Mammogram28 | 0.98888188 | 1.66122365 | 0.14326355 | 0.78717024 | 3.30046512 |
| Mammogram29 | 0.62944271 | 0.90394626 | 0.97520851 | 2.47923031 | 2.82704771 |
| Mammogram30 | 1.82259222 | 1.75038596 | 0.67784449 | 4.1556576 | 5.12858568 |
| Mammogram31 | 0.06991747 | 1.1118492 | 0.23888096 | 1.75173575 | 1.26344129 |
| Mammogram32 | 1.26412458 | 5.23676782 | 0.62172245 | 3.72363557 | 2.26123466 |
| Mammogram33 | 0.81605488 | 7.48991722 | 0.66454681 | 3.60964967 | 4.15342514 |
| Mammogram34 | 0.8720686 | 3.81977367 | 0.20177187 | 1.95579345 | 1.41544118 |
| Mammogram35 | 1.58946982 | 1.68700155 | 1.38676962 | 2.71666667 | 3.22685534 |
| Mammogram36 | 1.51692168 | 1.96504786 | 1.17126575 | 3.88831298 | 3.56533882 |
| Mammogram37 | 0.5233848 | 0.00066433 | 0.70270183 | 1.79359303 | 1.77911239 |
| Mammogram38 | 1.48137163 | 1.21644201 | 0.56248339 | 2.81507116 | 1.37146786 |
| Mammogram39 | 2.59148568 | 2.2776691 | 1.69948894 | 4.80868716 | 3.96694375 |
| Mammogram40 | 0.66859479 | 1.92905101 | 0.45997426 | 1.87097142 | 0.64877398 |
| Mean | 1.01244458 | 2.16338918 | 0.52843922 | 1.64912505 | 2.52260565 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Montero, R.; Martinez-Rojas, J.-A.; Lopez-Espi, P.-L.; Nuñez-Martin, L.; Diez-Jimenez, E. Filtering of Mammograms Based on Convolution with Directional Fractal Masks to Enhance Microcalcifications. Appl. Sci. 2019, 9, 1194. https://doi.org/10.3390/app9061194
Sanchez-Montero R, Martinez-Rojas J-A, Lopez-Espi P-L, Nuñez-Martin L, Diez-Jimenez E. Filtering of Mammograms Based on Convolution with Directional Fractal Masks to Enhance Microcalcifications. Applied Sciences. 2019; 9(6):1194. https://doi.org/10.3390/app9061194
Chicago/Turabian StyleSanchez-Montero, Rocio, Juan-Antonio Martinez-Rojas, Pablo-Luis Lopez-Espi, Luis Nuñez-Martin, and Efren Diez-Jimenez. 2019. "Filtering of Mammograms Based on Convolution with Directional Fractal Masks to Enhance Microcalcifications" Applied Sciences 9, no. 6: 1194. https://doi.org/10.3390/app9061194





