Visualization and Characterization of the Brain Regional Heterogeneity of Astrocyte–Astrocyte Structural Interactions by Using Improved Iontophoresis with Dual-Fluorescent Dyes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals
2.3. AAV Microinjections
2.4. Immunohistochemistry (IHC)
2.5. Iontophoresis with Fluorescent Dyes and 3D Reconstruction
2.6. Photostability of Fluorescent Dyes
2.7. Statistical Analyses
3. Results
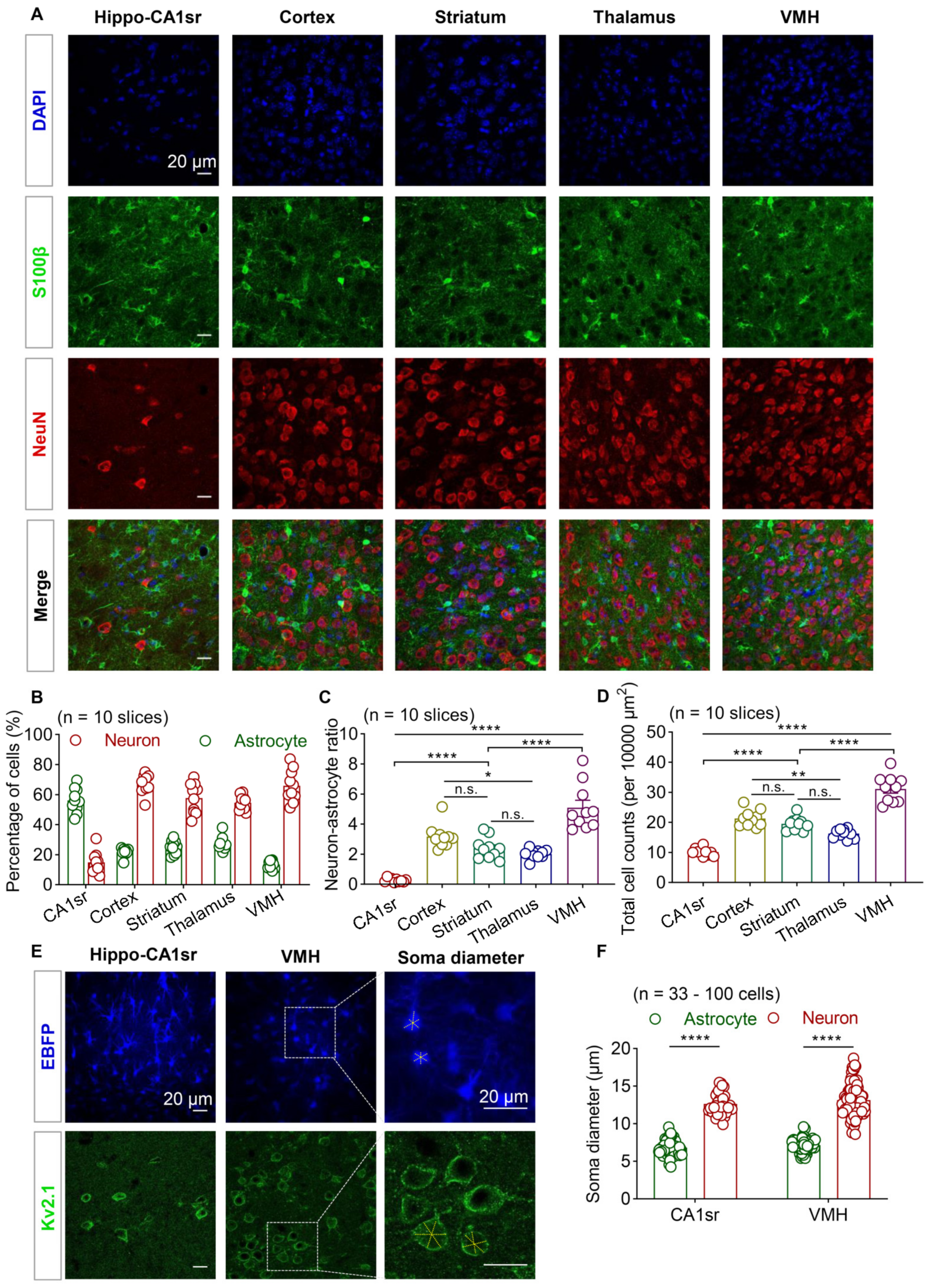
3.1. Brain Regional Heterogeneity of Cell Density and Astrocyte–Neuron Ratio
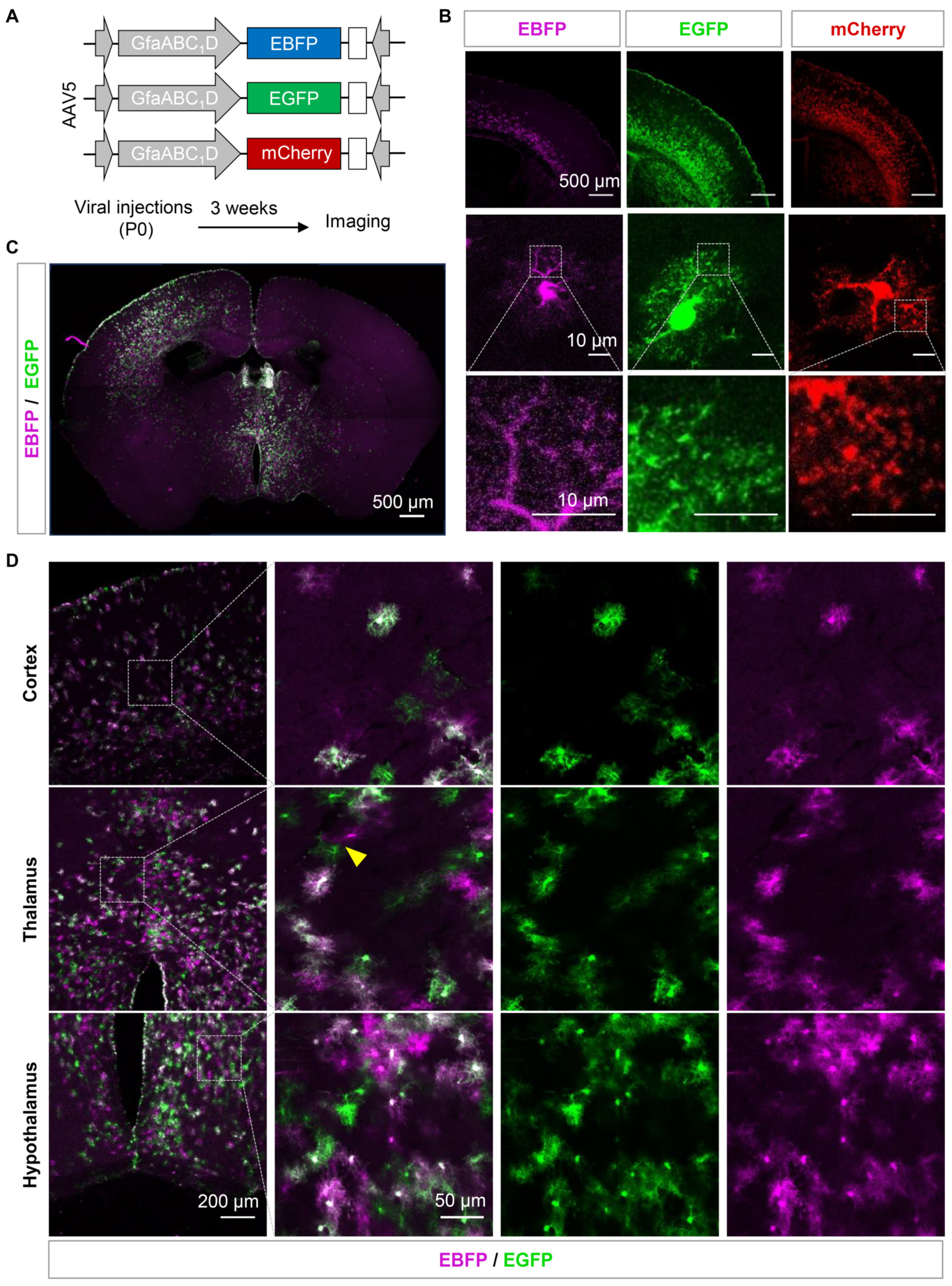
3.2. Sparse Labeling of the Adjacent Astrocytes by Using AAVs Expressing Fluorescent Proteins Exhibited Low Efficiency
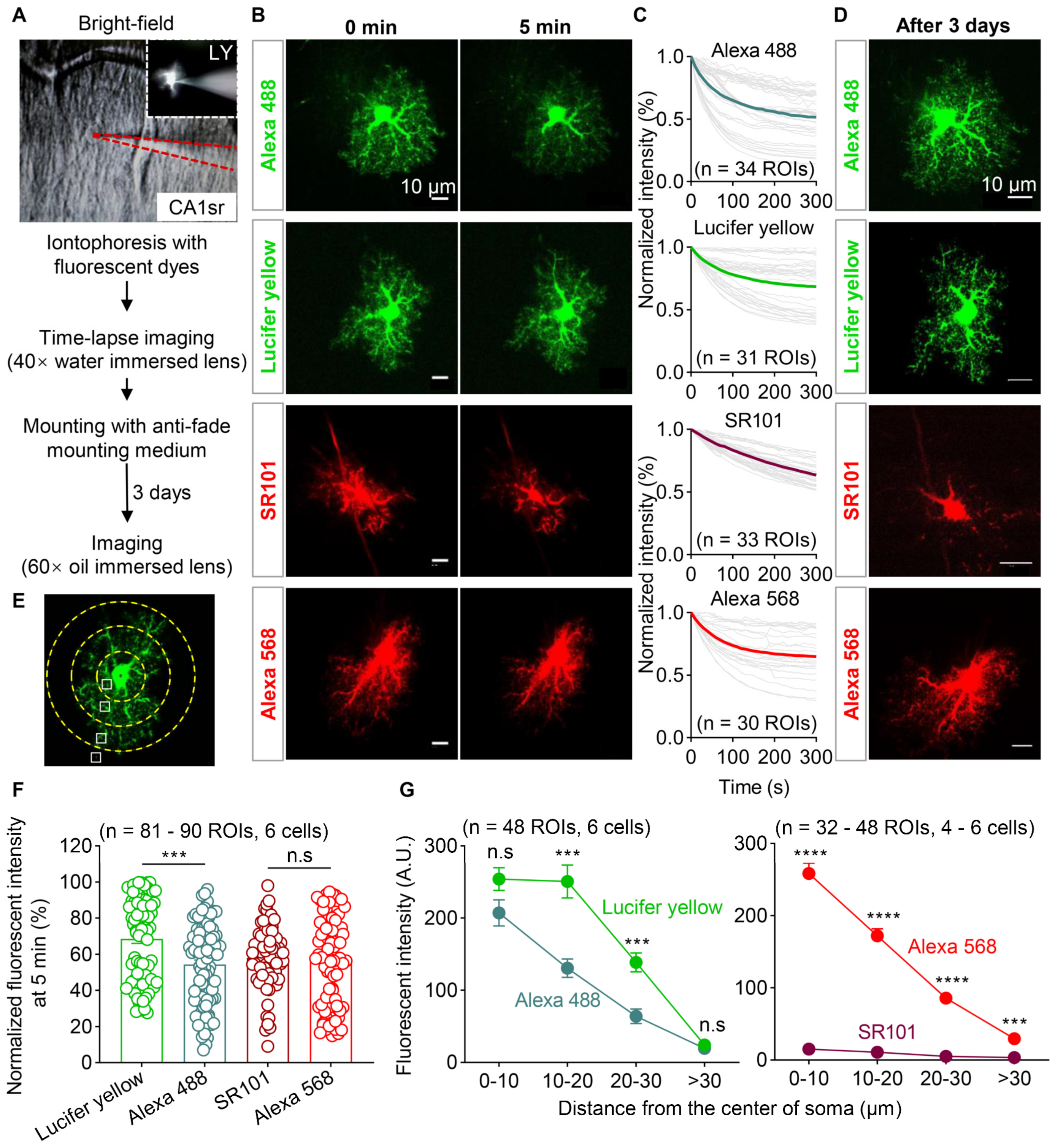
3.3. Photostability and Labeling Efficiency of Four Types of Fluorescent Dyes
3.4. Astrocytes Showed Overlapped Territories in the VMH
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Verkhratsky, A.; Nedergaard, M. Physiology of Astroglia. Physiol. Rev. 2018, 98, 239–389. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Duan, S. Novel Insight into Glial Biology and Diseases. Neurosci. Bull. 2023, 39, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Farhy-Tselnicker, I.; Allen, N.J. Astrocytes, neurons, synapses: A tripartite view on cortical circuit development. Neural Dev. 2018, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Bushong, E.A.; Martone, M.E.; Jones, Y.Z.; Ellisman, M.H. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002, 22, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zuo, Y.X.; Jiang, R.T. Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. CNS Neurosci. Ther. 2019, 25, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Bushong, E.A.; Martone, M.E.; Ellisman, M.H. Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. Int. J. Dev. Neurosci. 2004, 22, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.R. Specification and morphogenesis of astrocytes. Science 2010, 330, 774–778. [Google Scholar] [CrossRef]
- Torres-Ceja, B.; Olsen, M.L. A closer look at astrocyte morphology: Development, heterogeneity, and plasticity at astrocyte leaflets. Curr. Opin. Neurobiol. 2022, 74, 102550. [Google Scholar] [CrossRef]
- Baldwin, K.T.; Tan, C.X.; Strader, S.T.; Jiang, C.; Savage, J.T.; Elorza-Vidal, X.; Contreras, X.; Rülicke, T.; Hippenmeyer, S.; Estévez, R.; et al. HepaCAM controls astrocyte self-organization and coupling. Neuron 2021, 109, 2427–2442.e10. [Google Scholar] [CrossRef]
- Oberheim, N.A.; Wang, X.; Goldman, S.; Nedergaard, M. Astrocytic complexity distinguishes the human brain. Trends Neurosci. 2006, 29, 547–553. [Google Scholar] [CrossRef]
- Stork, T.; Sheehan, A.; Tasdemir-Yilmaz, O.E.; Freeman, M.R. Neuron-glia interactions through the Heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron 2014, 83, 388–403. [Google Scholar] [CrossRef] [PubMed]
- López-Hidalgo, M.; Hoover, W.B.; Schummers, J. Spatial organization of astrocytes in ferret visual cortex. J. Comp. Neurol. 2016, 524, 3561–3576. [Google Scholar] [CrossRef] [PubMed]
- Livet, J.; Weissman, T.A.; Kang, H.; Draft, R.W.; Lu, J.; Bennis, R.A.; Sanes, J.R.; Lichtman, J.W. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 2007, 450, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Endo, F.; Kasai, A.; Soto, J.S.; Yu, X.; Qu, Z.; Hashimoto, H.; Gradinaru, V.; Kawaguchi, R.; Khakh, B.S. Molecular basis of astrocyte diversity and morphology across the CNS in health and disease. Science 2022, 378, eadc9020. [Google Scholar] [CrossRef] [PubMed]
- Karpf, J.; Unichenko, P.; Chalmers, N.; Beyer, F.; Wittmann, M.T.; Schneider, J.; Fidan, E.; Reis, A.; Beckervordersandforth, J.; Brandner, S.; et al. Dentate gyrus astrocytes exhibit layer-specific molecular, morphological and physiological features. Nat. Neurosci. 2022, 25, 1626–1638. [Google Scholar] [CrossRef] [PubMed]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Welle, A.; Kasakow, C.V.; Jungmann, A.M.; Gobbo, D.; Stopper, L.; Nordström, K.; Salhab, A.; Gasparoni, G.; Scheller, A.; Kirchhoff, F.; et al. Epigenetic control of region-specific transcriptional programs in mouse cerebellar and cortical astrocytes. Glia 2021, 69, 2160–2177. [Google Scholar] [CrossRef]
- Lanjakornsiripan, D.; Pior, B.J.; Kawaguchi, D.; Furutachi, S.; Tahara, T.; Katsuyama, Y.; Suzuki, Y.; Fukazawa, Y.; Gotoh, Y. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Matyash, V.; Kettenmann, H. Heterogeneity in astrocyte morphology and physiology. Brain Res. Rev. 2010, 63, 2–10. [Google Scholar] [CrossRef]
- Reeves, A.M.; Shigetomi, E.; Khakh, B.S. Bulk loading of calcium indicator dyes to study astrocyte physiology: Key limitations and improvements using morphological maps. J. Neurosci. 2011, 31, 9353–9358. [Google Scholar] [CrossRef]
- Veldman, M.B.; Park, C.S.; Eyermann, C.M.; Zhang, J.Y.; Zuniga-Sanchez, E.; Hirano, A.A.; Daigle, T.L.; Foster, N.N.; Zhu, M.; Langfelder, P.; et al. Brainwide Genetic Sparse Cell Labeling to Illuminate the Morphology of Neurons and Glia with Cre-Dependent MORF Mice. Neuron 2020, 108, 111–127.e6. [Google Scholar] [CrossRef] [PubMed]
- Zong, H.; Espinosa, J.S.; Su, H.H.; Muzumdar, M.D.; Luo, L. Mosaic analysis with double markers in mice. Cell 2005, 121, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Grunke, S.D.; Levites, Y.; Golde, T.E.; Jankowsky, J.L. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J. Vis. Exp. 2014, 91, 51863. [Google Scholar] [CrossRef]
- Moye, S.L.; Diaz-Castro, B.; Gangwani, M.R.; Khakh, B.S. Visualizing Astrocyte Morphology Using Lucifer Yellow Iontophoresis. J. Vis. Exp. 2019, 151, e60225. [Google Scholar] [CrossRef]
- Webster, V.L.; Mahajan, R.P. Transient hyperaemic response to assess vascular reactivity of skin; effect of locally iontophoresed sodium nitroprusside. Br. J. Anaesth. 2002, 89, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Octeau, J.C.; Chai, H.; Jiang, R.; Bonanno, S.L.; Martin, K.C.; Khakh, B.S. An Optical Neuron-Astrocyte Proximity Assay at Synaptic Distance Scales. Neuron 2018, 98, 49–66.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; He, W.; Tang, T.; Qiu, M. Immunological Markers for Central Nervous System Glia. Neurosci. Bull. 2023, 39, 379–392. [Google Scholar] [CrossRef]
- Ogata, K.; Kosaka, T. Structural and quantitative analysis of astrocytes in the mouse hippocampus. Neuroscience 2002, 113, 221–233. [Google Scholar] [CrossRef]
- Savchenko, V.L.; McKanna, J.A.; Nikonenko, I.R.; Skibo, G.G. Microglia and astrocytes in the adult rat brain: Comparative immunocytochemical analysis demonstrates the efficacy of lipocortin 1 immunoreactivity. Neuroscience 2000, 96, 195–203. [Google Scholar] [CrossRef]
- Trimmer, J.S. Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc. Natl. Acad. Sci. USA 1991, 88, 10764–10768. [Google Scholar] [CrossRef]
- Hwang, P.M.; Fotuhi, M.; Bredt, D.S.; Cunningham, A.M.; Snyder, S.H. Contrasting immunohistochemical localizations in rat brain of two novel K+ channels of the Shab subfamily. J. Neurosci. 1993, 13, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Schulze, W.; Hayata-Takano, A.; Kamo, T.; Nakazawa, T.; Nagayasu, K.; Kasai, A.; Seiriki, K.; Shintani, N.; Ago, Y.; Farfan, C.; et al. Simultaneous neuron- and astrocyte-specific fluorescent marking. Biochem. Biophys. Res. Commun. 2015, 459, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.Y.; Jang, M.J.; Yoo, B.B.; Greenbaum, A.; Ravi, N.; Wu, W.L.; Sánchez-Guardado, L.; Lois, C.; Mazmanian, S.K.; Deverman, B.E.; et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017, 20, 1172–1179. [Google Scholar] [CrossRef] [PubMed]
- Challis, R.C.; Ravindra Kumar, S.; Chan, K.Y.; Challis, C.; Beadle, K.; Jang, M.J.; Kim, H.M.; Rajendran, P.S.; Tompkins, J.D.; Shivkumar, K.; et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019, 14, 379–414, Erratum in Nat. Protoc. 2019, 14, 2597. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Chauhan, B.C. In vivo imaging of adeno-associated viral vector labelled retinal ganglion cells. Sci. Rep. 2018, 8, 1490. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Chen, L.; Liao, P.; Huang, L.; Chen, Z.; Liao, D.; Yang, L.; Wang, J.; Yu, G.; Wang, L.; et al. Astroglial dysfunctions drive aberrant synaptogenesis and social behavioral deficits in mice with neonatal exposure to lengthy general anesthesia. PLoS Biol. 2019, 17, e3000086. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lin, M.Z.; McKeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008, 5, 545–551. [Google Scholar] [CrossRef]
- Panchuk-Voloshina, N.; Haugland, R.P.; Bishop-Stewart, J.; Bhalgat, M.K.; Millard, P.J.; Mao, F.; Leung, W.Y.; Haugland, R.P. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem. 1999, 47, 1179–1188. [Google Scholar] [CrossRef]
- Rai, S.; Bhardwaj, U.; Misra, A.; Singh, S.; Gupta, R. Comparison between photostability of Alexa Fluor 448 and Alexa Fluor 647 with conventional dyes FITC and APC by flow cytometry. Int. J. Lab. Hematol. 2018, 40, e52–e54. [Google Scholar] [CrossRef]
- Abdeladim, L.; Matho, K.S.; Clavreul, S.; Mahou, P.; Sintes, J.M.; Solinas, X.; Arganda-Carreras, I.; Turney, S.G.; Lichtman, J.W.; Chessel, A.; et al. Multicolor multiscale brain imaging with chromatic multiphoton serial microscopy. Nat. Commun. 2019, 10, 1662, Erratum in Nat. Commun. 2019, 10, 2160. [Google Scholar] [CrossRef]
- Clavreul, S.; Abdeladim, L.; Hernández-Garzón, E.; Niculescu, D.; Durand, J.; Ieng, S.H.; Barry, R.; Bonvento, G.; Beaurepaire, E.; Livet, J.; et al. Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nat. Commun. 2019, 10, 4884. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhou, B.; Su, M.; Liao, P.; Lei, F.; Li, X.; Liao, D.; Zhang, X.; Jiang, R. Visualization and Characterization of the Brain Regional Heterogeneity of Astrocyte–Astrocyte Structural Interactions by Using Improved Iontophoresis with Dual-Fluorescent Dyes. Brain Sci. 2023, 13, 1644. https://doi.org/10.3390/brainsci13121644
Li Q, Zhou B, Su M, Liao P, Lei F, Li X, Liao D, Zhang X, Jiang R. Visualization and Characterization of the Brain Regional Heterogeneity of Astrocyte–Astrocyte Structural Interactions by Using Improved Iontophoresis with Dual-Fluorescent Dyes. Brain Sciences. 2023; 13(12):1644. https://doi.org/10.3390/brainsci13121644
Chicago/Turabian StyleLi, Qingran, Bin Zhou, Mengchan Su, Ping Liao, Fan Lei, Xin Li, Daqing Liao, Xia Zhang, and Ruotian Jiang. 2023. "Visualization and Characterization of the Brain Regional Heterogeneity of Astrocyte–Astrocyte Structural Interactions by Using Improved Iontophoresis with Dual-Fluorescent Dyes" Brain Sciences 13, no. 12: 1644. https://doi.org/10.3390/brainsci13121644
APA StyleLi, Q., Zhou, B., Su, M., Liao, P., Lei, F., Li, X., Liao, D., Zhang, X., & Jiang, R. (2023). Visualization and Characterization of the Brain Regional Heterogeneity of Astrocyte–Astrocyte Structural Interactions by Using Improved Iontophoresis with Dual-Fluorescent Dyes. Brain Sciences, 13(12), 1644. https://doi.org/10.3390/brainsci13121644





