Lamination, Borders, and Thalamic Projections of the Primary Visual Cortex in Human, Non-Human Primate, and Rodent Brains
Abstract
1. Introduction
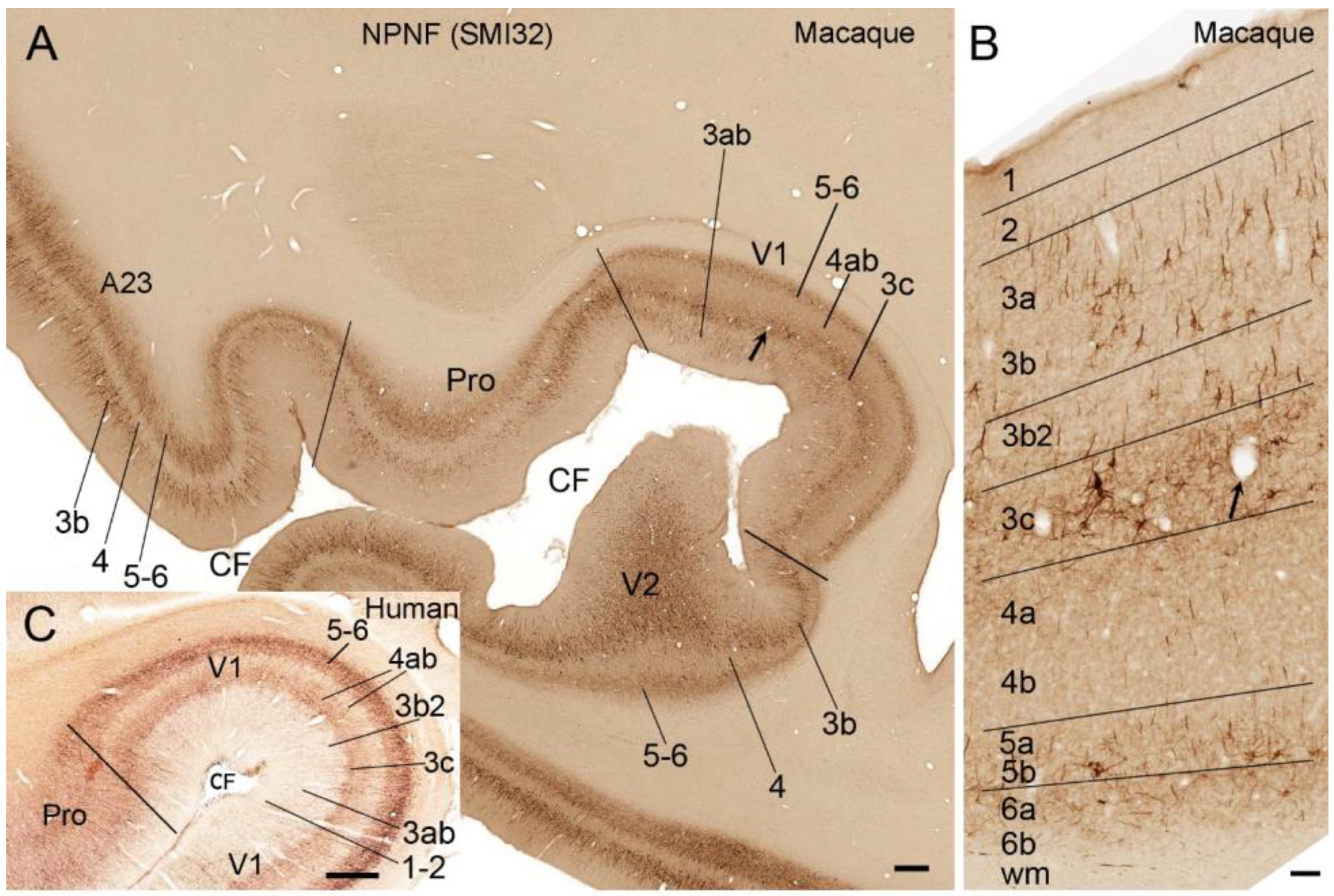
2. Laminar Organization of V1 in Humans and NHPs
3. Laminar Organization of V1 in Rodents
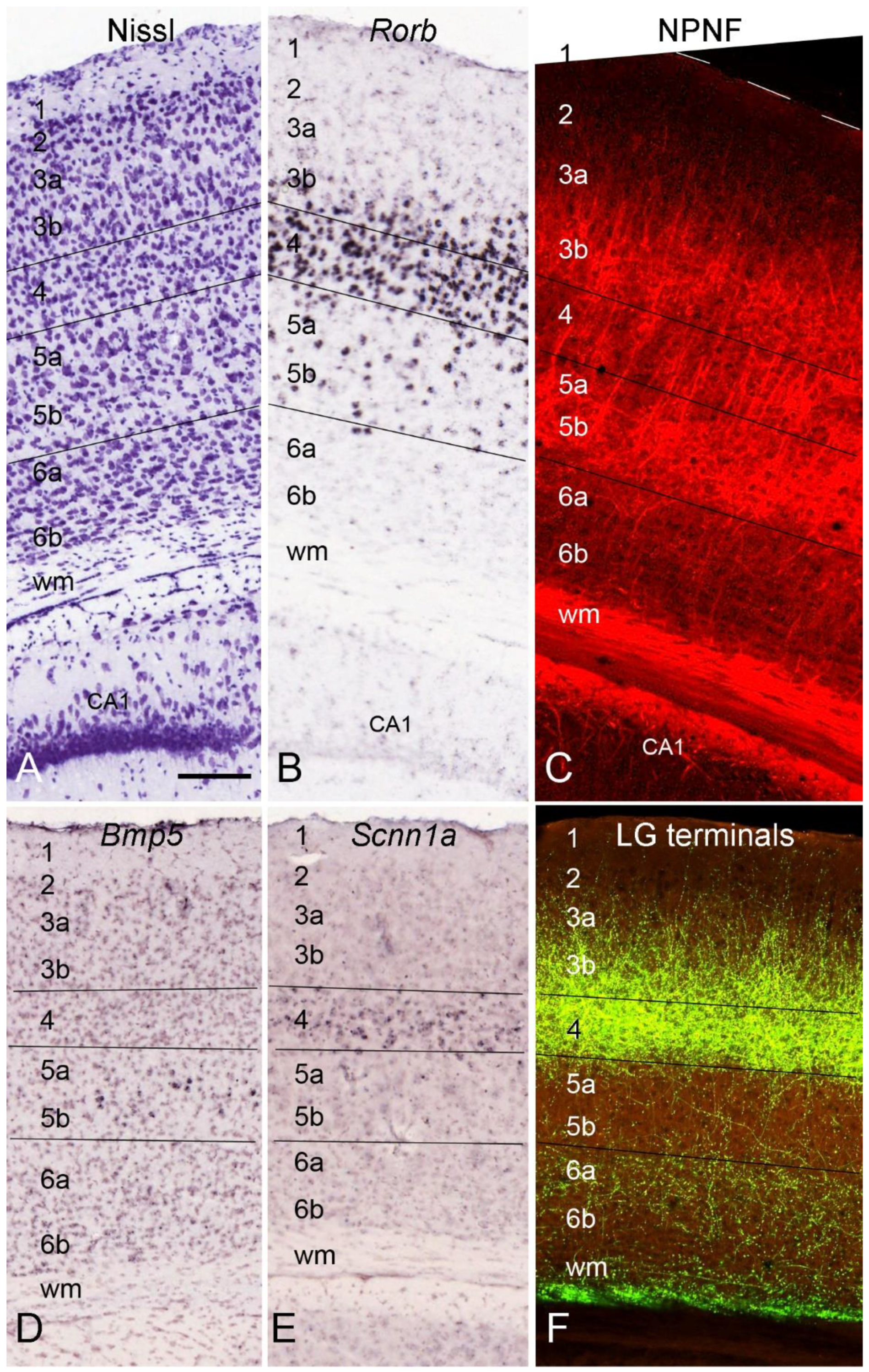
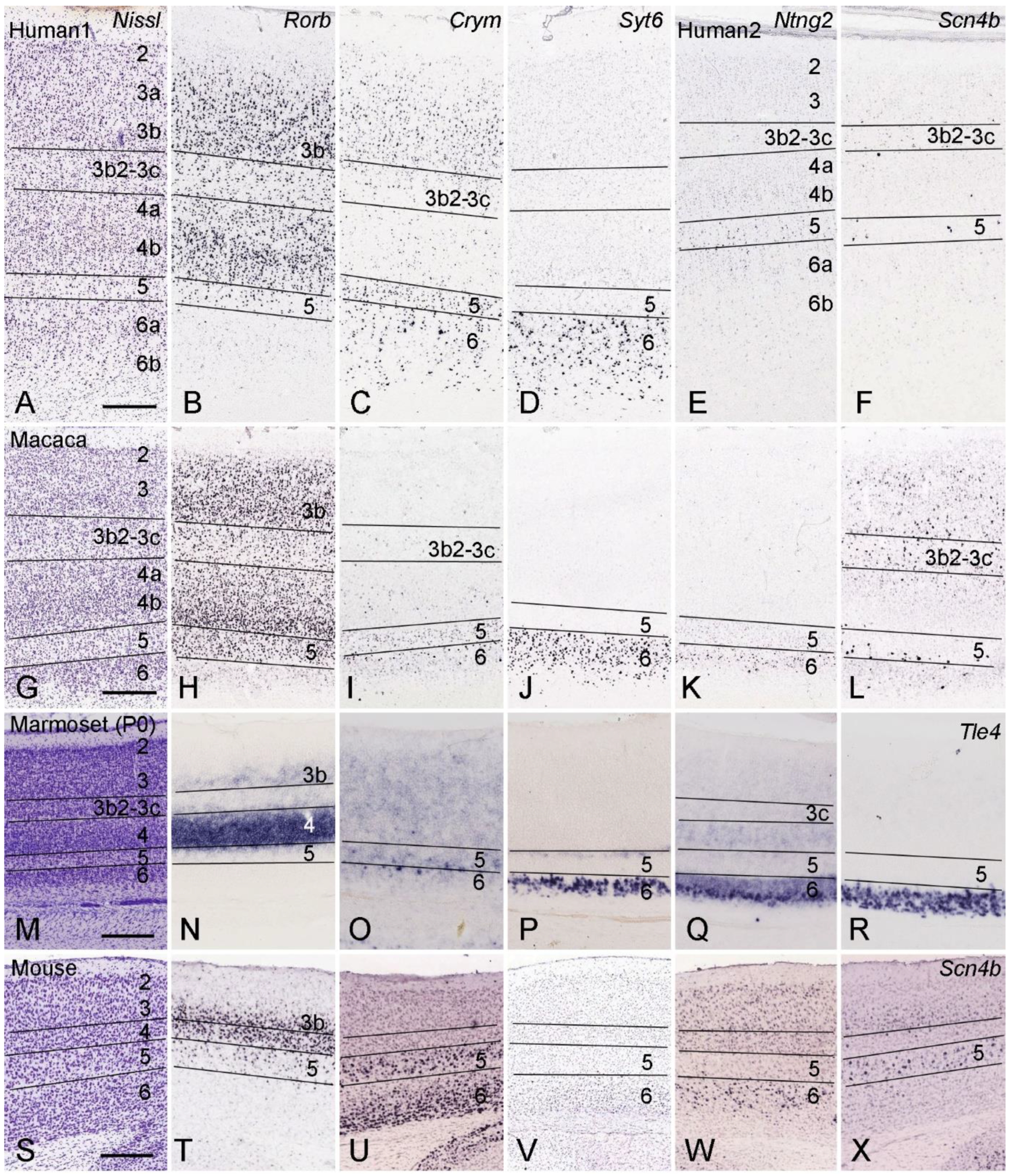
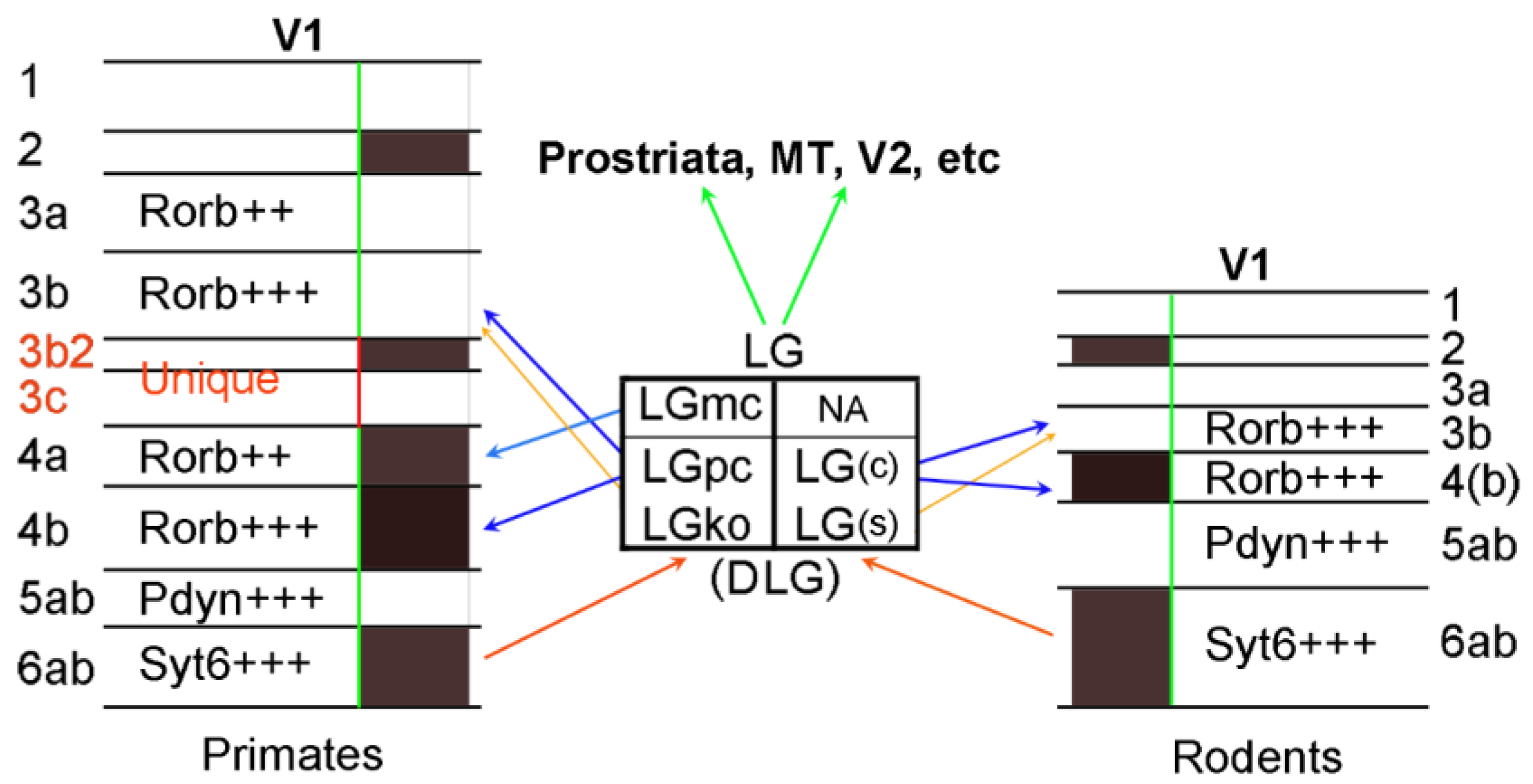
4. Layer-Specific Molecular Markers for V1 across Species
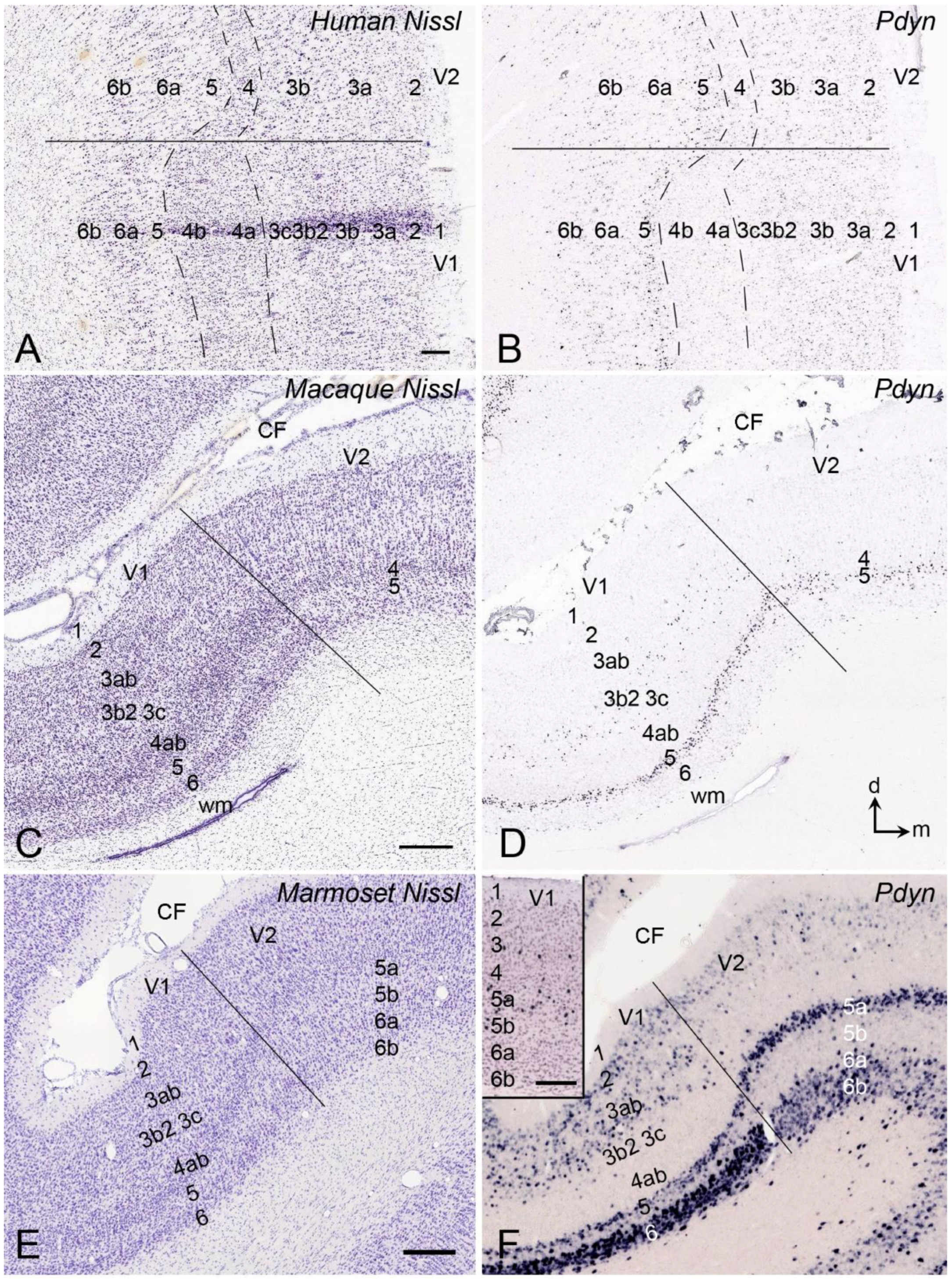
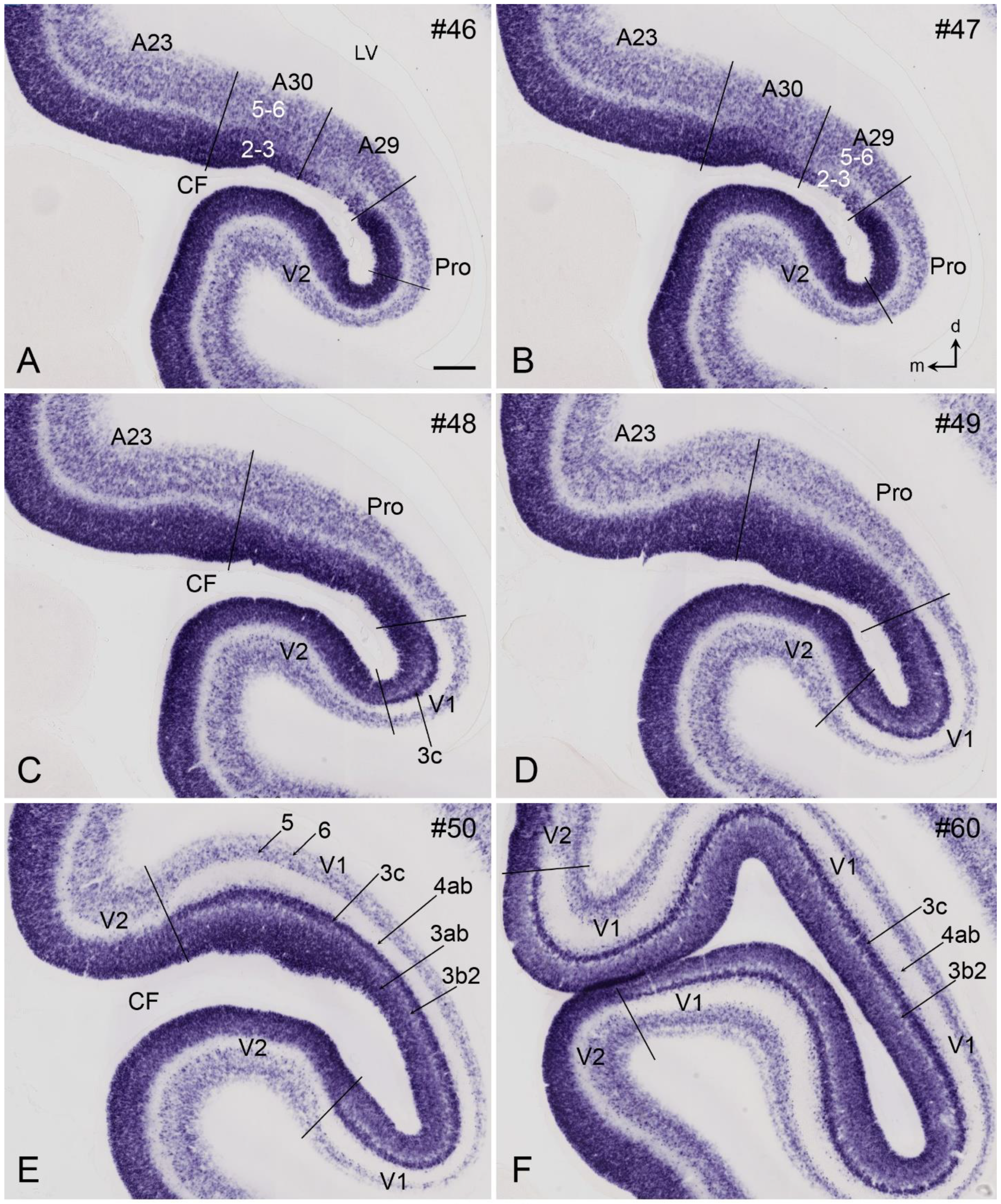
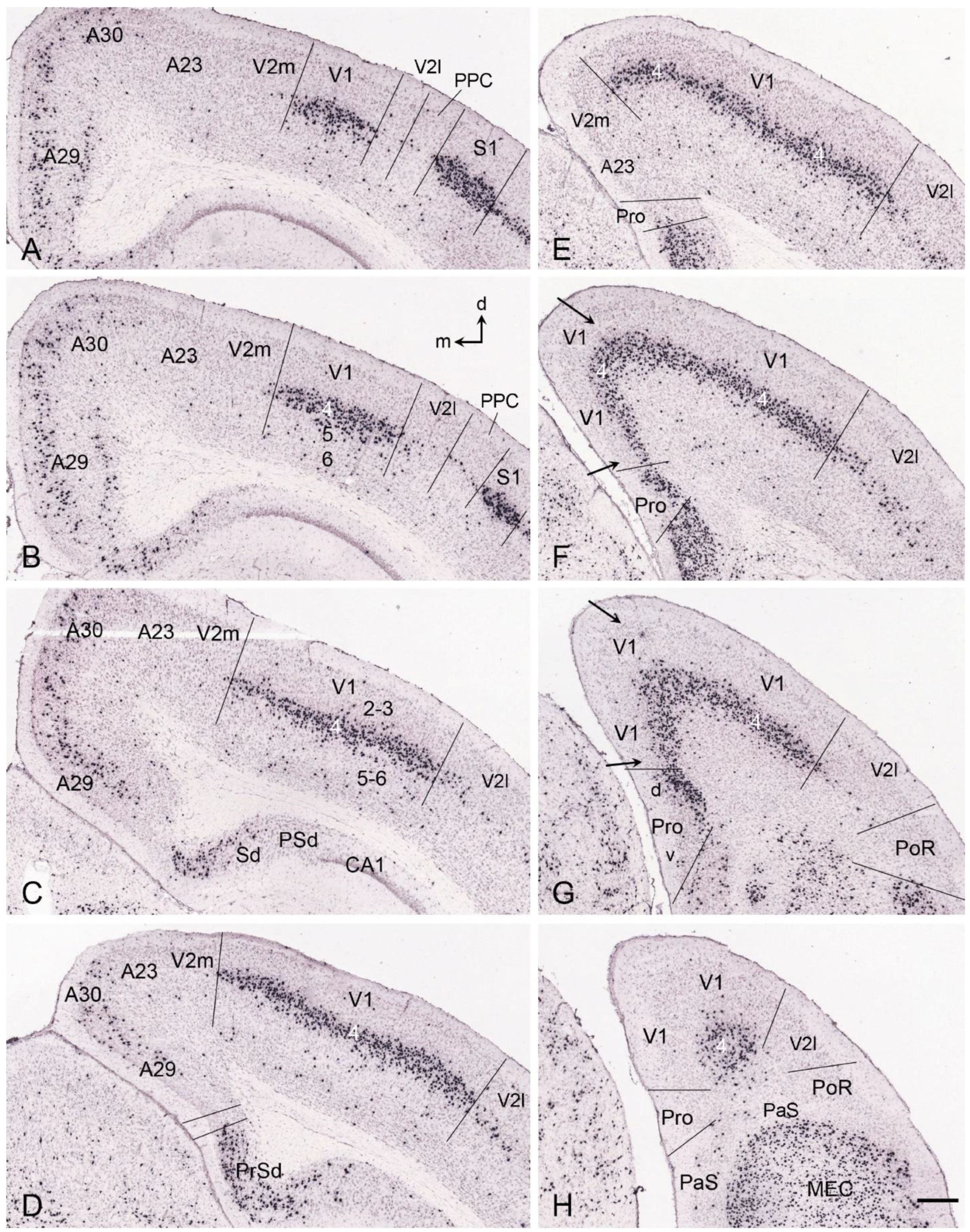
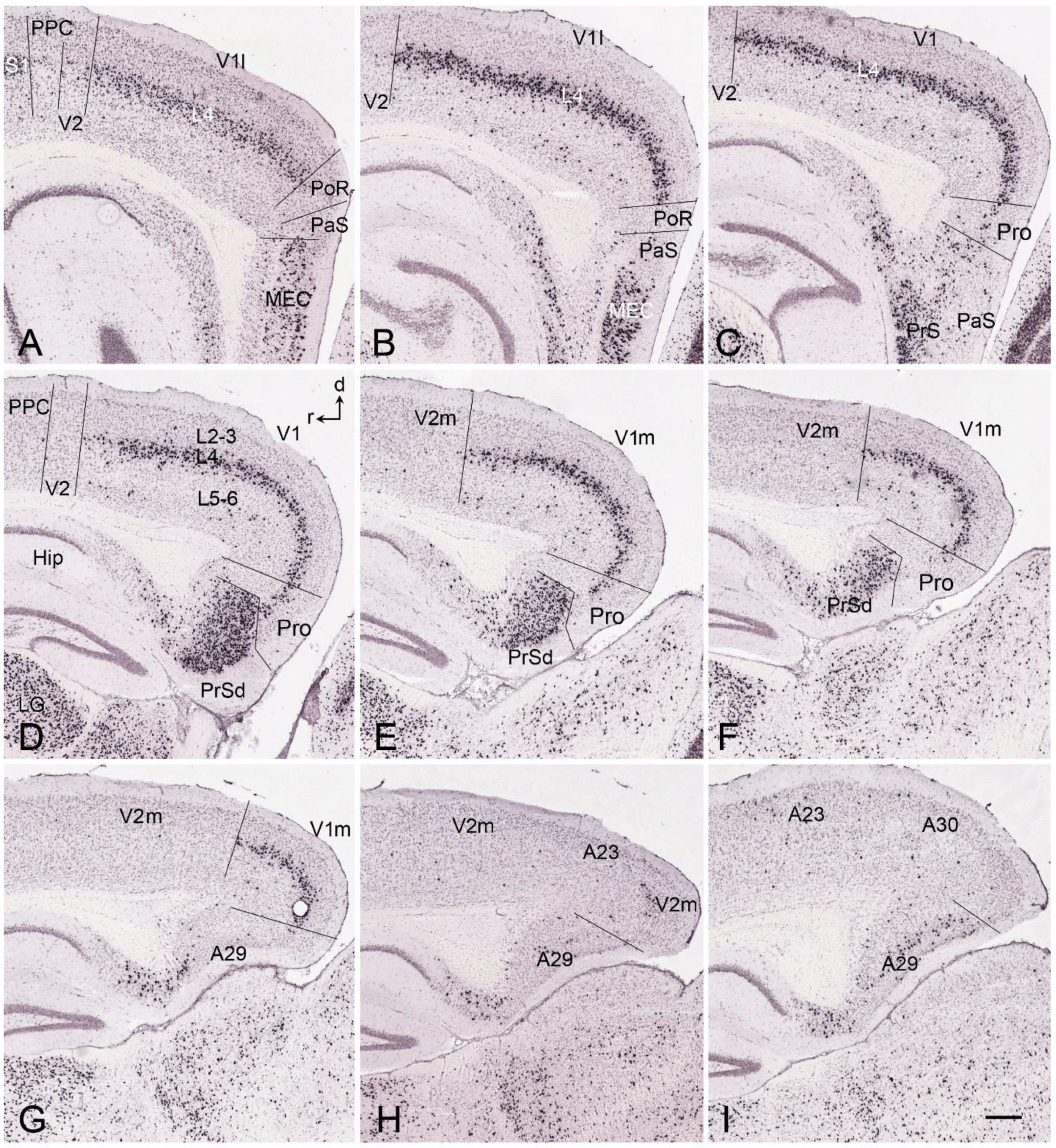
5. V1 Borders with V2 and Prostriata in Adult Humans and NHPs
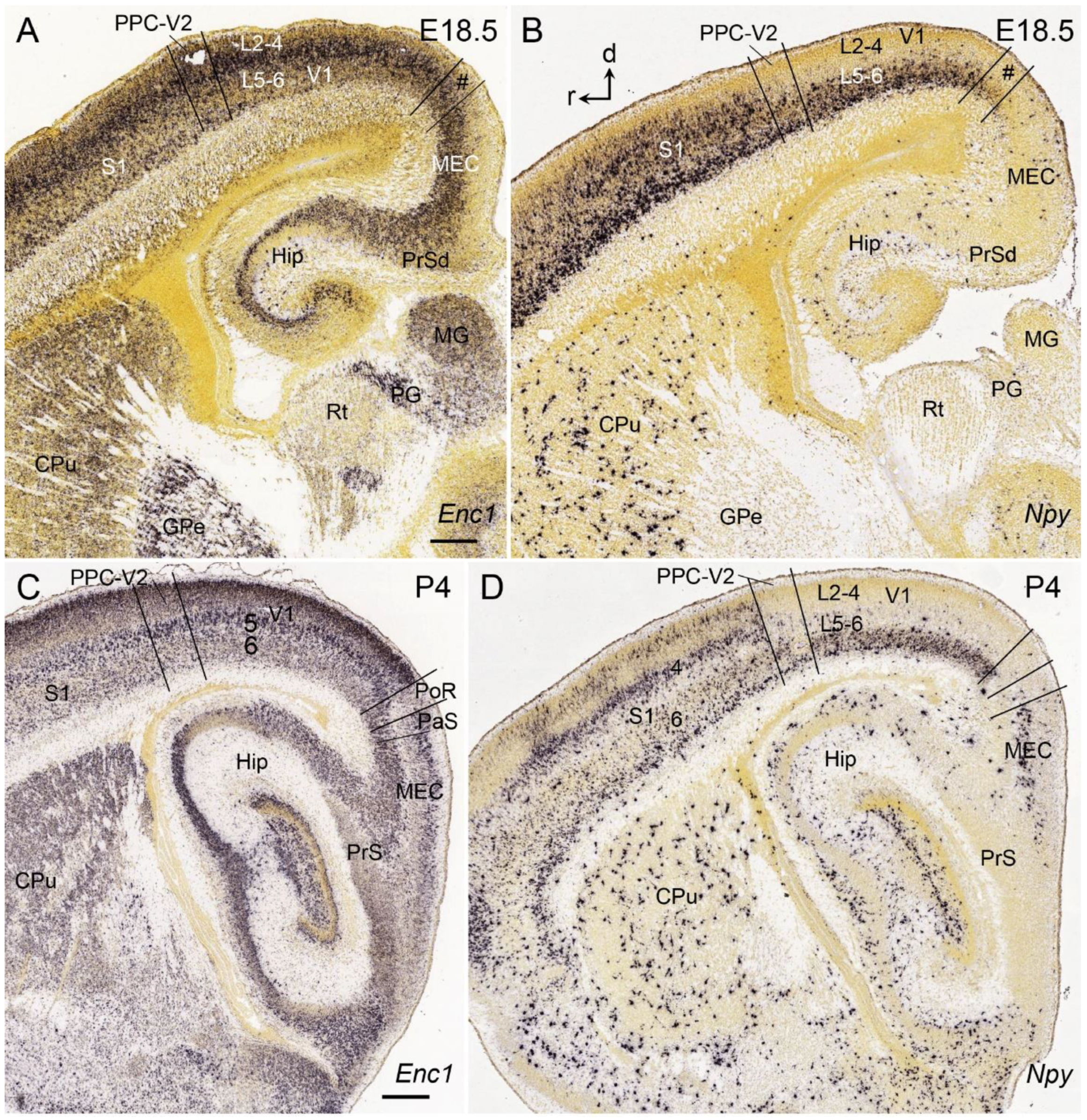
6. Boundary Determination of V1 in Prenatal Humans
7. Boundary Determination of V1 in Prenatal Macaque Monkeys
8. Boundary Determination of V1 in Newborn Marmosets
9. Boundary Determination of V1 in Developing Mice
10. Boundary Determination of V1 in Adult Mice
11. Thalamocortical Projections of V1 in Human and NHP Brains
12. Corticothalamic Projections of V1 in Human and NHP Brains
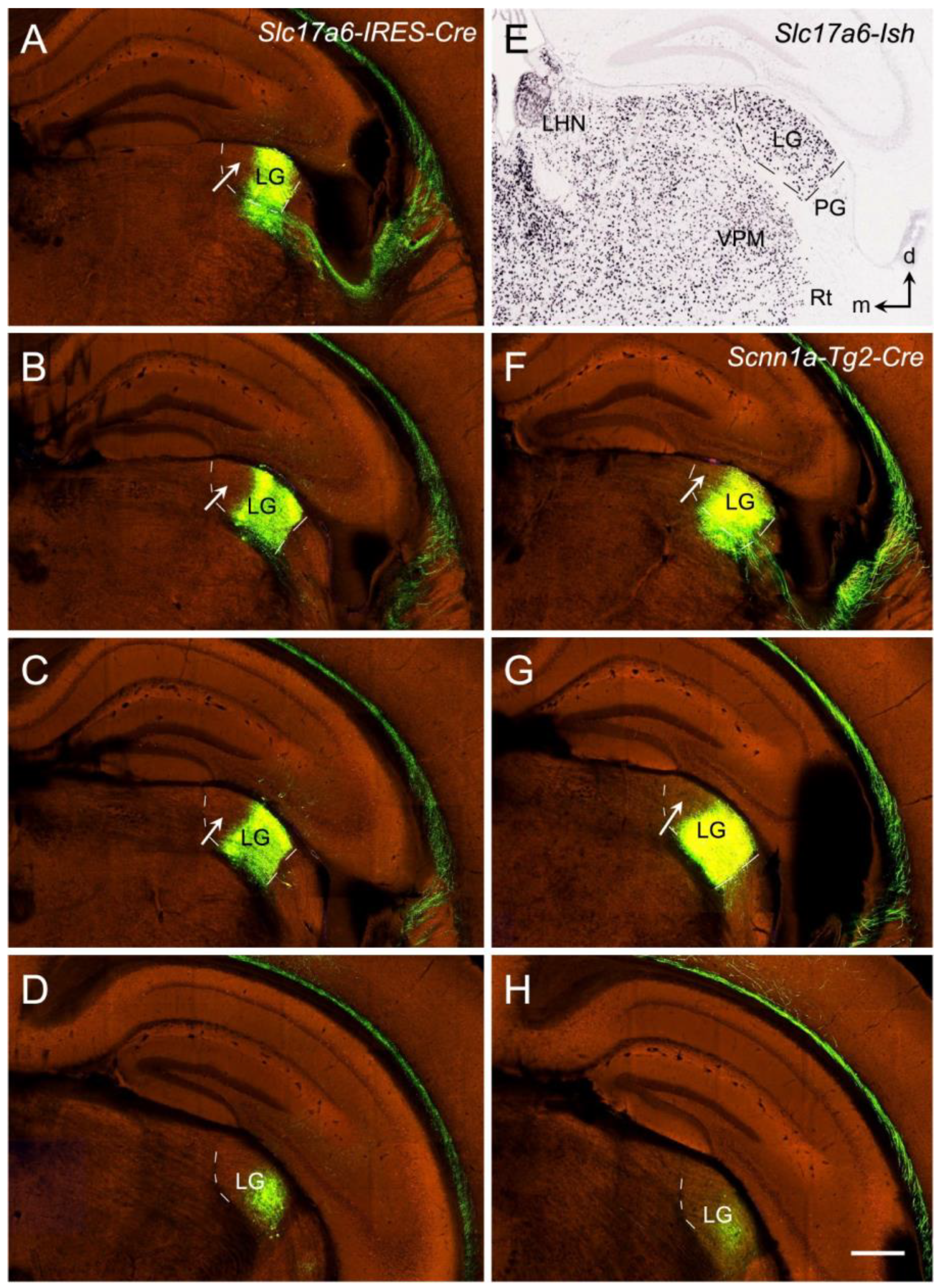
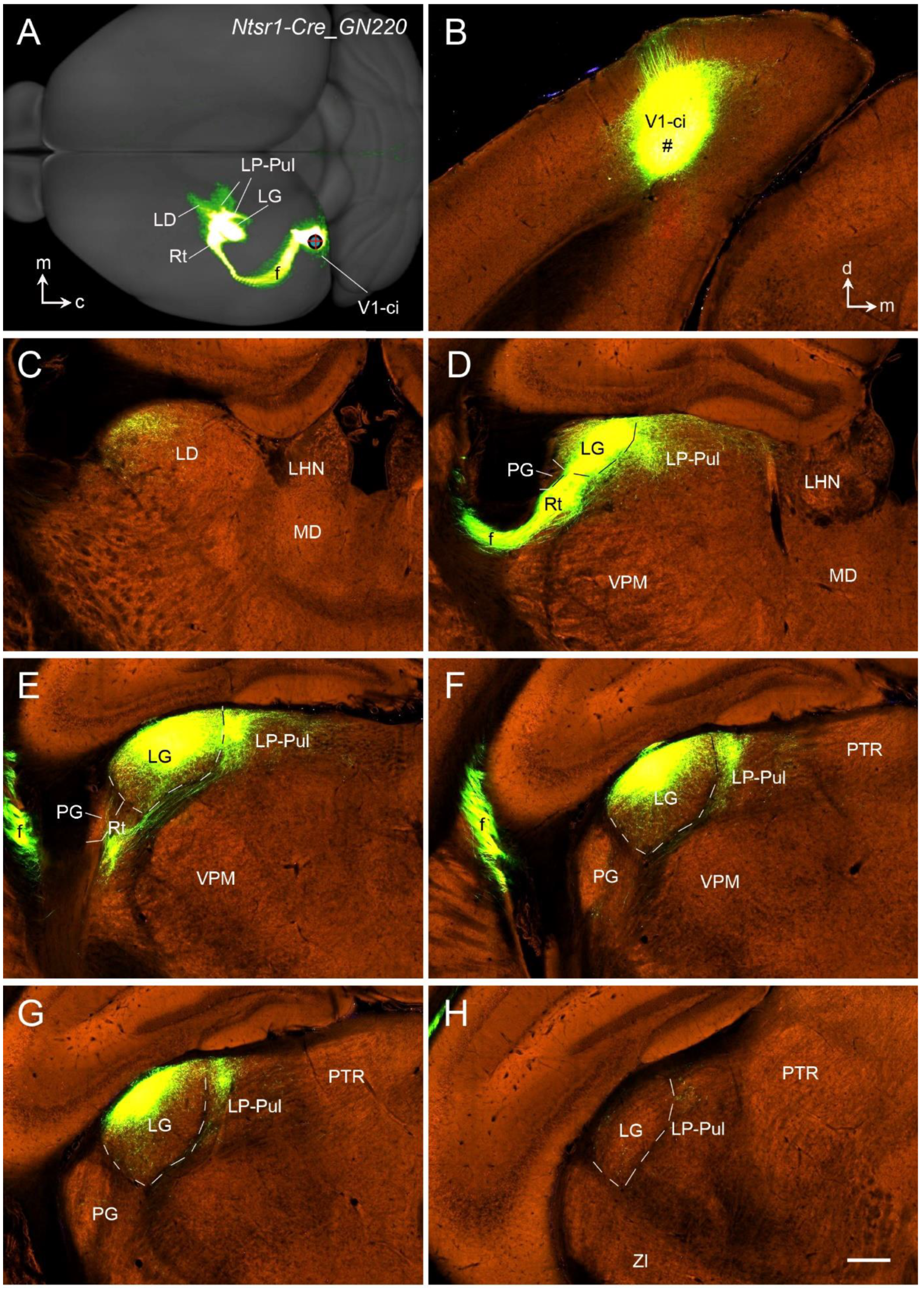
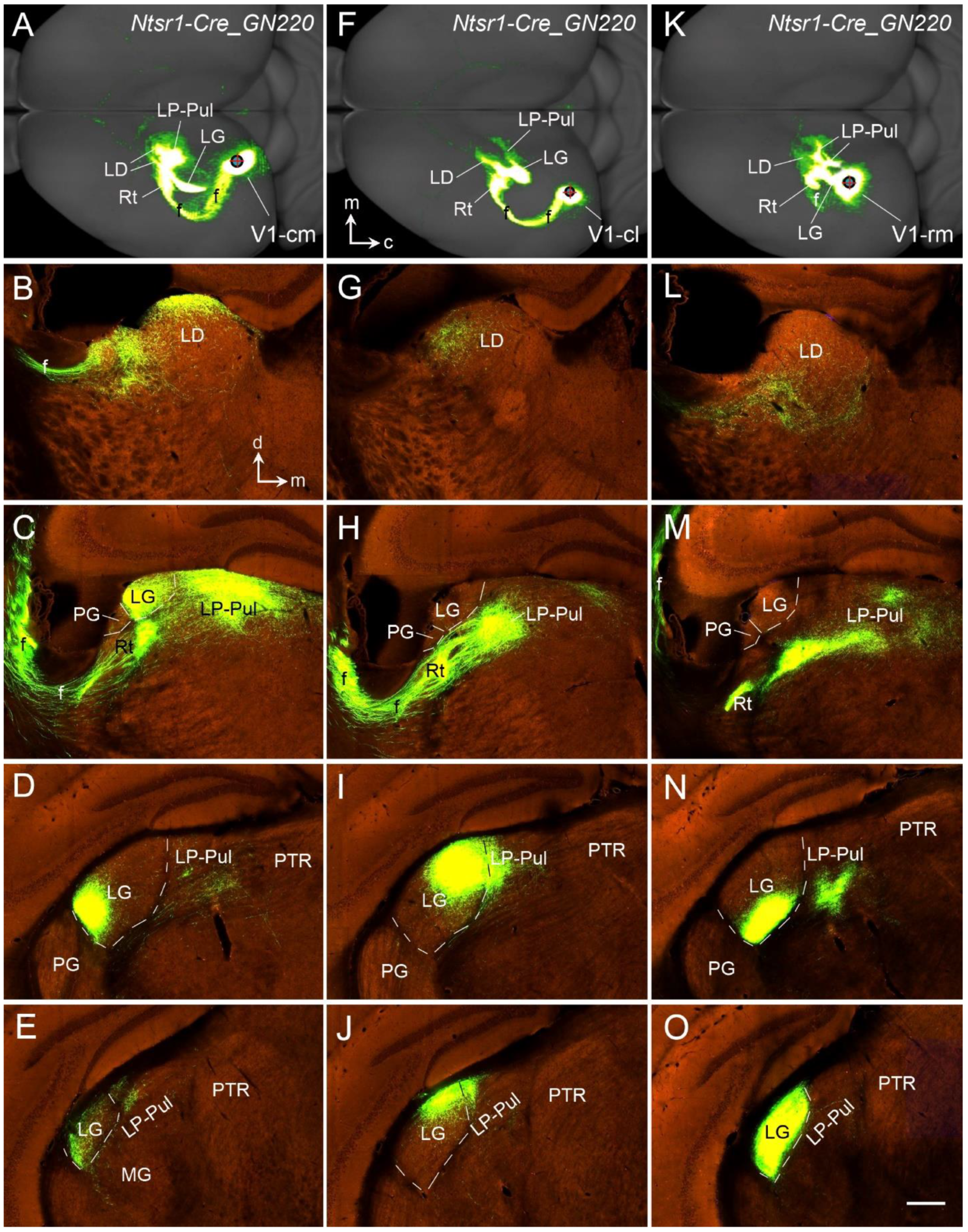
13. Thalamocortical Projections of the Rodent V1
14. Corticothalamic Projections of the Rodent V1
15. Possible Neural Circuits Underlying Blindsight
16. Laminar Development and Vision-Related Functional Maturation and Disorders
17. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hawrylycz, M.J.; Lein, E.S.; Guillozet-Bongaarts, A.L.; Shen, E.H.; Ng, L.; Miller, J.A.; Van De Lagemaat, L.N.; Smith, K.A.; Ebbert, A.; Riley, Z.L.; et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012, 489, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Royall, J.J.; Sunkin, S.M.; Ng, L.; Facer, B.A.; Lesnar, P.; Guillozet-Bongaarts, A.; McMurray, B.; Szafer, A.; Dolbeare, T.A.; et al. Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol. 2016, 524, 3127–3481. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Ding, S.-L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional landscape of the prenatal human brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Royall, J.J.; Lesnar, P.; Facer, B.A.; Smith, K.A.; Wei, Y.; Brouner, K.; Dalley, R.A.; Dee, N.; Dolbeare, T.A.; et al. Cellular resolution anatomical and molecular atlases for prenatal human brains. J. Comp. Neurol. 2022, 530, 6–503. [Google Scholar] [CrossRef]
- Bernard, A.; Lubbers, L.S.; Tanis, K.Q.; Luo, R.; Podtelezhnikov, A.A.; Finney, E.M.; McWhorter, M.M.; Serikawa, K.; Lemon, T.; Morgan, R.; et al. Transcriptional architecture of the primate neocortex. Neuron 2012, 73, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Bakken, T.E.; Miller, J.A.; Ding, S.-L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Dalley, R.A.; Royall, J.J.; Lemon, T.; et al. A comprehensive transcriptional map of primate brain development. Nature 2016, 535, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.G.; Stone, J.M.; Karten, H.J. High-resolution digital brain atlases: A Hubble telescope for the brain. Ann. New York Acad. Sci. 2011, 1225, E147–E159. [Google Scholar] [CrossRef]
- Mikula, S.; Trotts, I.; Stone, J.M.; Jones, E.G. Internet-enabled high-resolution brain mapping and virtual microscopy. NeuroImage 2007, 35, 9–15. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.W.; Harris, J.A.; Ng, L.; Winslow, B.; Cain, N.; Mihalas, S.; Wang, Q.; Lau, C.; Kuan, L.; Henry, A.M.; et al. A mesoscale connectome of the mouse brain. Nature 2014, 508, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Shimogori, T.; Abe, A.; Go, Y.; Hashikawa, T.; Kishi, N.; Kikuchi, S.S.; Kita, Y.; Niimi, K.; Nishibe, H.; Okuno, M.; et al. Digital gene atlas of neonate common marmoset brain. Neurosci. Res. 2018, 128, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kita, Y.; Nishibe, H.; Wang, Y.; Hashikawa, T.; Kikuchi, S.S.; U, M.; Yoshida, A.C.; Yoshida, C.; Kawase, T.; Ishii, S.; et al. Cellular-resolution gene expression profiling in the neonatal marmoset brain reveals dynamic species- and region-specific differences. Proc. Natl. Acad. Sci. USA 2021, 118, e2020125118. [Google Scholar] [CrossRef] [PubMed]
- Brodmann, K. Vergleichende Lokalisationslehre der Gro Hirnrinde; Verlag von Johann Ambrosius Barth: Leipzig, Germany, 1909. [Google Scholar]
- Hevner, R.; Wong-Riley, M. Regulation of cytochrome oxidase protein levels by functional activity in the macaque monkey visual system. J. Neurosci. 1990, 10, 1331–1340. [Google Scholar] [CrossRef]
- Yao, S.; Zhou, Q.; Li, S.; Takahata, T. Immunoreactivity of vesicular glutamate transporter 2 corresponds to cytochrome oxidase-rich subcompartments in the visual cortex of squirrel monkeys. Front. Neuroanat. 2021, 15, 629473. [Google Scholar] [CrossRef] [PubMed]
- Bryant, K.L.; Suwyn, C.; Reding, K.M.; Smiley, J.F.; Hackett, T.A.; Preuss, T.M. Evidence for ape and human specializations in geniculostriate projections from VGLUT2 immunohistochemistry. Brain Behav. Evol. 2012, 80, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Marin, V.; Ahmed, T.H.; Afzal, Y.C.; Hawken, M.J. Distribution of vesicular glutamate transporter 2 (VGluT2) in the primary visual cortex of the macaque and human. J. Comp. Neurol. 2013, 521, 130–151. [Google Scholar] [CrossRef]
- Hässler, R. Comparative Anatomy of the Central Visual System in Day- and Night-Active Primates; Hässler, R., Thieme, S.S., Eds.; Evolution of the Forebrain: Stuttgart, Germany, 1967. [Google Scholar]
- Fitzpatrick, D.; Itoh, K.; Diamond, I. The laminar organization of the lateral geniculate body and the striate cortex in the squirrel monkey (Saimiri sciureus). J. Neurosci. 1983, 3, 673–702. [Google Scholar] [CrossRef] [PubMed]
- Elston, G.N.; Rosa, M.G. The occipitoparietal pathway of the macaque monkey: Comparison of pyramidal cell morphology in layer III of functionally related cortical visual areas. Cereb. Cortex 1997, 7, 432–452. [Google Scholar] [CrossRef]
- Takahata, T.; Shukla, R.; Yamamori, T.; Kaas, J.H. Differential expression patterns of striate-cortex-enriched genes among Old World, New World and prosimian primates. Cereb. Cortex 2012, 22, 2313–2321. [Google Scholar] [CrossRef] [PubMed]
- Balaram, P.; Kaas, J.H. Towards a unified scheme of cortical lamination for primary visual cortex across primates: Insights from NeuN and VGLUT2 immunoreactivity. Front. Neuroanat. 2014, 8, 81. [Google Scholar] [CrossRef]
- Spatz, W.B. Topographically organized reciprocal connections between areas 17 and MT (visual area of superior temporal sulcus) in the marmoset Callithrix jacchus. Exp. Brain Res. 1977, 27, 559–572. [Google Scholar] [CrossRef]
- Palmer, S.M.; Rosa, M.G.P. A distinct anatomical network of cortical areas for analysis of motion in far peripheral vision. Eur. J. Neurosci. 2006, 24, 2389–2405. [Google Scholar] [CrossRef] [PubMed]
- Diamond, I.T.; Conley, M.; Itoh, K.; Fitzpatrick, D. Laminar organization of geniculocortical projections in Galago senegalensis and Aotus trivirgatus. J. Comp. Neurol. 1985, 242, 584–610. [Google Scholar] [CrossRef] [PubMed]
- Bourne, J.A.; Rosa, M.G.P. Neurofilament protein expression in the geniculostriate pathway of a New World monkey (Callithrix jacchus). Exp. Brain Res. 2003, 150, 19–24. [Google Scholar] [CrossRef]
- Blasdel, G.; Lund, J. Termination of afferent axons in macaque striate cortex. J. Neurosci. 1983, 3, 1389–1413. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.; Feldman, M.L. The projection of the lateral geniculate nucleus to area 17 of the rat cerebral cortex. I. General description. J. Neurocytol. 1976, 5, 63–84. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-L.; Elberger, A.J. Postnatal development of biotinylated dextran amine-labeled corpus callosum axons projecting from the visual and auditory cortices to the visual cortex of the rat. Exp. Brain Res. 2001, 136, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ding, S.-L.; Li, Y.; Royall, J.; Feng, D.; Lesnar, P.; Graddis, N.; Naeemi, M.; Facer, B.; Ho, A.; et al. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell 2020, 181, 936–953.e20. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.; Kaas, J.H. Architectonic Subdivisions of Neocortex in the Gray Squirrel (Sciurus carolinensis). Neuroscience 2008, 291, 1301–1333. [Google Scholar] [CrossRef] [PubMed]
- Morecraft, R.J.; Rockland, K.S.; Van Hoesen, G.W. Localization of area prostriata and its projection to the cingulate motor cortex in the rhesus monkey. Cereb. Cortex 2000, 10, 192–203. [Google Scholar] [CrossRef][Green Version]
- Ding, S.; Morecraft, R.J.; Van Hoesen, G.W. Topography, cytoarchitecture, and cellular phenotypes of cortical areas that form the cingulo-parahippocampal isthmus and adjoining retrocalcarine areas in the monkey. J. Comp. Neurol. 2003, 456, 184–201. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson CPetrides, M.; Rosa, M.; Tokuno, H. The Marmoset Brain in Stereotaxic Coordinates; Academic Press: San Diego, CA, USA, 2012. [Google Scholar]
- Zeng, H.; Shen, E.H.; Hohmann, J.G.; Oh, S.W.; Bernard, A.; Royall, J.J.; Glattfelder, K.J.; Sunkin, S.M.; Morris, J.A.; Guillozet-Bongaarts, A.L.; et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 2012, 149, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Jorstad, N.L.; Close, J.; Johansen, N.; Yanny, A.M.; Barkan, E.R.; Travaglini, K.J.; Bertagnolli, D.; Campos, J.; Casper, T.; Crichton, K.; et al. Transcriptomic cytoarchitecture reveals principles of human neocortex organization. Science 2023, 382, eadf6812. [Google Scholar] [CrossRef]
- Tochitani, S.; Liang, F.; Watakabe, A.; Hashikawa, T.; Yamamori, T. The occ1 gene is preferentially expressed in the primary visual cortex in an activity-dependent manner: A pattern of gene expression related to the cytoarchitectonic area in adult macaque neocortex. Eur. J. Neurosci. 2001, 13, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Takahata, T.; Komatsu, Y.; Watakabe, A.; Hashikawa, T.; Tochitani, S.; Yamamori, T. Differential expression patterns of occ1-related genes in adult monkey visual cortex. Cereb. Cortex 2009, 19, 1937–1951. [Google Scholar] [CrossRef] [PubMed]
- Watakabe, A.; Komatsu, Y.; Sadakane, O.; Shimegi, S.; Takahata, T.; Higo, N.; Tochitani, S.; Hashikawa, T.; Naito, T.; Osaki, H.; et al. Enriched expression of serotonin 1B and 2A receptor genes in macaque visual cortex and their bidirectional modulatory effects on neuronal responses. Cereb. Cortex 2009, 19, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Takahata, T.; Komatsu, Y.; Watakabe, A.; Hashikawa, T.; Tochitani, S.; Yamamori, T. Activity-dependent expression of occ1 in excitatory neurons is a characteristic feature of the primate visual cortex. Cereb. Cortex 2006, 16, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zheng, D.; Garey, L. Prenatal development of GABA-immunoreactive neurons in the human striate cortex. Dev. Brain Res. 1992, 65, 191–204. [Google Scholar] [CrossRef]
- Bayer, S.A.; Altman, J. The Human Brain during the Third Trimester; CRC Press LLC: Boca Raton, FL, USA, 2004. [Google Scholar]
- Šestan, N.; Rakic, P.; Donoghue, M.J. Independent parcellation of the embryonic visual cortex and thalamus revealed by combinatorial Eph/ephrin gene expression. Curr. Biol. 2001, 11, 39–43. [Google Scholar] [CrossRef][Green Version]
- Mashiko, H.; Yoshida, A.C.; Kikuchi, S.S.; Niimi, K.; Takahashi, E.; Aruga, J.; Okano, H.; Shimogori, T. Comparative anatomy of marmoset and mouse cortex from genomic expression. J. Neurosci. 2012, 32, 5039–5053. [Google Scholar] [CrossRef]
- Chen, S.; Chen, C.; Xiang, X.; Zhang, S.; Ding, S. Chemoarchitecture of area prostriata in adult and developing mice: Comparison with presubiculum and parasubiculum. J. Comp. Neurol. 2022, 530, 2486–2517. [Google Scholar] [CrossRef]
- Chen, C.-H.; Hu, J.-M.; Zhang, S.-Y.; Xiang, X.-J.; Chen, S.-Q.; Ding, S.-L. Rodent area prostriata converges multimodal hierarchical inputs and projects to the structures important for visuomotor behaviors. Front. Neurosci. 2021, 15, 772016. [Google Scholar] [CrossRef]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates; Academic Press: San Diego, CA, USA, 2013. [Google Scholar]
- Swanson, L.W. Brain Maps: Structure of the Rat Brain, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2004. [Google Scholar]
- Bienkowski, M.S.; Benavidez, N.L.; Wu, K.; Gou, L.; Becerra, M.; Dong, H. Extrastriate connectivity of the mouse dorsal lateral geniculate thalamic nucleus. J. Comp. Neurol. 2019, 527, 1419–1442. [Google Scholar] [CrossRef]
- Xiang, X.-J.; Chen, S.-Q.; Zhang, X.-Q.; Chen, C.-H.; Zhang, S.-Y.; Cai, H.-R.; Ding, S.-L. Possible rodent equivalent of the posterior cingulate cortex (area 23) interconnects with multimodal cortical and subcortical regions. Front. Neurosci. 2023, 17, 1194299. [Google Scholar] [CrossRef]
- Lu, W.; Chen, S.; Chen, X.; Hu, J.; Xuan, A.; Ding, S. Localization of area prostriata and its connections with primary visual cortex in rodent. J. Comp. Neurol. 2020, 528, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, D.; Starck, G.; Ljungberg, M.; Ribbelin, S.; Jönsson, L.; Malmgren, K.; Rydenhag, B. Intersubject variability in the anterior extent of the optic radiation assessed by tractography. Epilepsy Res. 2007, 77, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Sherbondy, A.J.; Dougherty, R.F.; Napel, S.; Wandell, B.A. Identifying the human optic radiation using diffusion imaging and fiber tractography. J. Vis. 2008, 8, 12. [Google Scholar] [CrossRef]
- Wu, W.; Rigolo, L.; O’Donnell, L.J.; Norton, I.; Shriver, S.; Golby, A.J. Visual pathway study using in vivo diffusion tensor imaging tractography to complement classic anatomy. Neurosurgery 2012, 70, ons145–ons156. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Worthy, K.H.; Liu, C.; Rosa, M.G.; Atapour, N. Parvalbumin as a neurochemical marker of the primate optic radiation. iScience 2023, 26, 106608. [Google Scholar] [CrossRef] [PubMed]
- Norden, J.J.; Kaas, J.H. The identification of relay neurons in the dorsal lateral geniculate nucleus of monkeys using horseradish peroxidase. J. Comp. Neurol. 1978, 182, 707–725. [Google Scholar] [CrossRef]
- Henderickson, A.E.; Wilson, J.R.; Ogren, M.P. The neurological organization of pathways between the dorsal lateral geniculate nucleus and visual cortex in old world and new world primates. J. Comp. Neurol. 1978, 182, 123–136. [Google Scholar] [CrossRef]
- Rezak, M.; Benevento, L. A comparison of the organization of the projections of the dorsal lateral geniculate nucleus, the inferior pulvinar and adjacent lateral pulvinar to primary visual cortex (area 17) in the macaque monkey. Brain Res. 1979, 167, 19–40. [Google Scholar] [CrossRef]
- Spatz, W.B. The retino-geniculo-cortical pathway in Callithrix. II. The geniculo-cortical projection. Exp. Brain Res. 1979, 36, 401–410. [Google Scholar] [CrossRef]
- DeBruyn, E.; Casagrande, V. Demonstration of ocular dominance columns in a New World primate by means of monocular deprivation. Brain Res. 1981, 207, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Dick, A.; Kaske, A.; Creutzfeldt, O.D. Topographical and topological organization of the thalamocortical projection to the striate and prestriate cortex in the marmoset (Callithrix jacchus). Exp. Brain Res. 1991, 84, 233–253. [Google Scholar] [CrossRef]
- Benevento, L.A.; Yoshida, K. The afferent and efferent organization of the lateral geniculo-prestriate pathways in the macaque monkey. J. Comp. Neurol. 1981, 203, 455–474. [Google Scholar] [CrossRef]
- Fries, W. The projection from the lateral geniculate nucleus to the prestriate cortex of the macaque monkey. Proc. R Soc. Lond. Ser. B Biol. Sci. 1981, 213, 73–80. [Google Scholar] [CrossRef]
- Yukie, M.; Iwai, E. Direct projection from the dorsal lateral geniculate nucleus to the prestriate cortex in macaque monkeys. J. Comp. Neurol. 1981, 201, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Bullier, J.; Kennedy, H. Projection of the lateral geniculate nucleus onto cortical area V2 in the macaque monkey. Exp. Brain Res. 1983, 53, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Sincich, L.C.; Park, K.F.; Wohlgemuth, M.J.; Horton, J.C. Bypassing V1: A direct geniculate input to area MT. Nat. Neurosci. 2004, 7, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Warner, C.E.; Goldshmit, Y.; Bourne, J.A. Retinal afferents synapse with relay cells targeting the middle temporal area in the pulvinar and lateral geniculate nuclei. Front. Neuroanat. 2010, 4, 8. [Google Scholar] [CrossRef]
- Malpeli, J.G.; Baker, F.H. The representation of the visual field in the lateral geniculate nucleus of Macaca mulatta. J. Comp. Neurol. 1975, 161, 569–594. [Google Scholar] [CrossRef] [PubMed]
- Perkel, D.J.; Bullier, J.; Kennedy, H. Topography of the afferent connectivity of area 17 in the macaque monkey: A double-labelling study. J. Comp. Neurol. 1986, 253, 374–402. [Google Scholar] [CrossRef] [PubMed]
- Tigges, J.; Tigges, M.; Perachio, A.A. Complementary laminar terminations of afferents to area 17 originating in area 18 and in the lateral geniculate nucleus in squirrel monkey. J. Comp. Neurol. 1977, 176, 87–100. [Google Scholar] [CrossRef]
- Weber, J.T.; Huerta, M.F.; Kaas, J.H.; Harting, J.K. The projections of the lateral geniculate nucleus of the squirrel monkey: Studies of the interlaminar zones and the S layers. J. Comp. Neurol. 1983, 213, 135–145. [Google Scholar] [CrossRef]
- Casagrande, V.; Yazar, F.; Jones, K.; Ding, Y. The morphology of the koniocellular axon pathway in the macaque monkey. Cereb. Cortex 2007, 17, 2334–2345. [Google Scholar] [CrossRef]
- Solomon, S.G. Striate cortex in dichromatic and trichromatic marmosets: Neurochemical compartmentalization and geniculate input. J. Comp. Neurol. 2002, 450, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Evrard, H.C.; Shapcott, K.A.; Haverkamp, S.; Logothetis, N.K.; Schmid, M.C. Cell-targeted optogenetics and electrical microstimulation reveal the primate koniocellular projection to supra-granular visual cortex. Neuron 2016, 90, 143–151. [Google Scholar] [CrossRef]
- Boyd, J.D.; Mavity-Hudson, J.A.; Casagrande, V.A. The connections of layer 4 subdivisions in the primary visual cortex (V1) of the owl monkey. Cereb. Cortex 2000, 10, 644–662. [Google Scholar] [CrossRef]
- Burkhalter, A.; Bernardo, K.L. Organization of corticocortical connections in human visual cortex. Proc. Natl. Acad. Sci. USA 1989, 86, 1071–1075. [Google Scholar] [CrossRef]
- Glendenning, K.K.; Kofron, E.; Diamond, I.T. Laminar organization of projections of the lateral geniculate nucleus to the striate cortex inGalago. Brain Res. 1976, 105, 538–546. [Google Scholar] [CrossRef]
- Spatz, W.; Erdmann, G. Striate cortex projections to the lateral geniculate and other thalamic nuclei; a study using degeneration and autoradiographic tracing methods in the marmosetCallithrix. Brain Res. 1974, 82, 91–108. [Google Scholar] [CrossRef]
- Graham, J. Some topographical connections of the striate cortex with subcortical structures in Macaca fascicularis. Exp. Brain Res. 1982, 47, 1–14. [Google Scholar] [CrossRef]
- Lund, J.S.; Lund, R.D.; Hendrickson, A.E.; Bunt, A.H.; Fuchs, A.F. The origin of efferent pathways from the primary visual cortex, area 17, of the macaque monkey as shown by retrograde transport of horseradish peroxidase. J. Comp. Neurol. 1975, 164, 287–303. [Google Scholar] [CrossRef]
- Fitzpatrick, D.; Usrey, W.M.; Schofield, B.R.; Einstein, G. The sublaminar organization of corticogeniculate neurons in layer 6 of macaque striate cortex. Vis. Neurosci. 1994, 11, 307–315. [Google Scholar] [CrossRef]
- Ichida, J.M.; Casagrande, V.A. Organization of the feedback pathway from striate cortex (V1) to the lateral geniculate nucleus (LGN) in the owl monkey (Aotus trivirgatus). J. Comp. Neurol. 2002, 454, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Fries, W.; Keizer, K.; Kuypers, H.G.J.M. Large layer VI cells in macaque striate cortex (Meynert cells) project to both superior colliculus and prestriate visual area V5. Exp. Brain Res. 1985, 58, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Nhan, H.L.; Callaway, E.M. Morphology of superior colliculus- and middle temporal area-projecting neurons in primate primary visual cortex. J. Comp. Neurol. 2012, 520, 52–80. [Google Scholar] [CrossRef] [PubMed]
- Harting, J.K.; Huerta, M.F. The geniculostriate projection in the grey squirrel: Preliminary autoradiographic data. Brain Res. 1983, 272, 341–349. [Google Scholar] [CrossRef]
- Kaas, J.H.; Hall, W.C.; Diamond, I.T. Visual cortex of the grey squirrel (Sciurus carolinensis): Architectonic subdivisions and connections from the visual thalamus. J. Comp. Neurol. 1972, 145, 273–305. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Burkhalter, A. Evidence for excitatory amino acid neurotransmitters in the geniculo-cortical pathway and local projections within rat primary visual cortex. Exp. Brain Res. 1992, 89, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Leiwe, M.N.; Hendry, A.C.; Bard, A.D.; Eglen, S.J.; Lowe, A.S.; Thompson, I.D. Geniculo-cortical projection diversity revealed within the mouse visual thalamus. PLoS ONE 2016, 11, e0144846. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ribak, C.E.; Peters, A. An autoradiographic study of the projections from the lateral geniculate body of the rat. Brain Res. 1975, 92, 341–368. [Google Scholar] [CrossRef] [PubMed]
- Kurzawski, J.W.; Mikellidou, K.; Morrone, M.C.; Pestilli, F. The visual white matter connecting human area prostriata and the thalamus is retinotopically organized. Brain Struct. Funct. 2020, 225, 1839–1853. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, J.; Deschênes, M. Corticothalamic projections from the primary visual cortex in rats: A single fiber study using biocytin as an anterograde tracer. Neuroscience 1995, 66, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Froudarakis, E.; Fahey, P.G.; Reimer, J.; Smirnakis, S.M.; Tehovnik, E.J.; Tolias, A.S. The visual cortex in context. Annu. Rev. Vis. Sci. 2019, 5, 317–339. [Google Scholar] [CrossRef]
- Roger, M.; Arnault, P. Anatomical study of the connections of the primary auditory area in the rat. J. Comp. Neurol. 1989, 287, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Hazama, M.; Kimura, A.; Donishi, T.; Sakoda, T.; Tamai, Y. Topography of corticothalamic projections from the auditory cortex of the rat. Neuroscience 2004, 124, 655–667. [Google Scholar] [CrossRef]
- Bourassa, J.; Pinault, D.; Deschênes, M. Corticothalamic projections from the cortical barrel field to the somatosensory thalamus in rats: A single-fibre study using biocytin as an anterograde tracer. Eur. J. Neurosci. 1995, 7, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Killackey, H.P.; Sherman, S.M. Corticothalamic projections from the rat primary somatosensory cortex. J. Neurosci. 2003, 23, 7381–7384. [Google Scholar] [CrossRef]
- Ding, S.-L.; Yao, Z.; Hirokawa, K.E.; Nguyen, T.N.; Graybuck, L.T.; Fong, O.; Bohn, P.; Ngo, K.; Smith, K.A.; Koch, C.; et al. Distinct transcriptomic cell types and neural circuits of the subiculum and prosubiculum along the dorsal-ventral axis. Cell Rep. 2020, 31, 107648. [Google Scholar] [CrossRef]
- Kinoshita, M.; Kato, R.; Isa, K.; Kobayashi, K.; Kobayashi, K.; Onoe, H.; Isa, T. Dissecting the circuit for blindsight to reveal the critical role of pulvinar and superior colliculus. Nat. Commun. 2019, 10, 135. [Google Scholar] [CrossRef] [PubMed]
- Takakuwa, N.; Isa, K.; Onoe, H.; Takahashi, J.; Isa, T. Contribution of the Pulvinar and Lateral Geniculate Nucleus to the Control of Visually Guided Saccades in Blindsight Monkeys. J. Neurosci. 2021, 41, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.C.; Mrowka, S.W.; Turchi, J.; Saunders, R.C.; Wilke, M.; Peters, A.J.; Ye, F.Q.; Leopold, D.A. Blindsight depends on the lateral geniculate nucleus. Nature 2010, 466, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.P.G.; Sumner, P.; Chambers, C.D. The timing and neuroanatomy of conscious vision as revealed by TMS-induced blindsight. J. Cogn. Neurosci. 2014, 26, 1507–1518. [Google Scholar] [CrossRef]
- Atapour, N.; Worthy, K.H.; Lui, L.L.; Yu, H.-H.; Rosa, M.G.P. Neuronal degeneration in the dorsal lateral geniculate nucleus following lesions of primary visual cortex: Comparison of young adult and geriatric marmoset monkeys. Brain Struct. Funct. 2017, 222, 3283–3293. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-H.; Atapour, N.; Chaplin, T.A.; Worthy, K.H.; Rosa, M.G. Robust visual responses and normal retinotopy in primate lateral geniculate nucleus following long-term lesions of striate cortex. J. Neurosci. 2018, 38, 3955–3970. [Google Scholar] [CrossRef]
- Atapour, N.; Worthy, K.H.; Rosa, M.G.P. Neurochemical changes in the primate lateral geniculate nucleus following lesions of striate cortex in infancy and adulthood: Implications for residual vision and blindsight. Brain Struct. Funct. 2021, 226, 2763–2775. [Google Scholar] [CrossRef]
- Atapour, N.; Worthy, K.H.; Rosa, M.G.P. Remodeling of lateral geniculate nucleus projections to extrastriate area MT following long-term lesions of striate cortex. Proc. Natl. Acad. Sci. USA 2022, 119, e2117137119. [Google Scholar] [CrossRef] [PubMed]
- Cowey, A.; Stoerig, P. Projection patterns of surviving neurons in the dorsal lateral geniculate nucleus following discrete lesions of striate cortex: Implications for residual vision. Exp. Brain Res. 1989, 75, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Isa, T.; Yoshida, M. Neural Mechanism of Blindsight in a Macaque Model. Neuroscience 2021, 469, 138–161. [Google Scholar] [CrossRef] [PubMed]
- Bridge, H.; Bell, A.H.; Ainsworth, M.; Sallet, J.; Premereur, E.; Ahmed, B.; Mitchell, A.S.; Schüffelgen, U.; Buckley, M.; Tendler, B.C.; et al. Preserved extrastriate visual network in a monkey with substantial, naturally occurring damage to primary visual cortex. eLife 2019, 8, e42325. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-M.; Chen, C.-H.; Chen, S.-Q.; Ding, S.-L. Afferent Projections to Area Prostriata of the Mouse. Front. Neuroanat. 2020, 14, 605021. [Google Scholar] [CrossRef]
- Chen, C.-H.; Hu, J.-M.; Chen, S.-Q.; Liu, S.-M.; Ding, S.-L. Homotopic commissural projections of area prostriata in rat and mouse: Comparison with presubiculum and parasubiculum. Front. Neural Circuits 2020, 14, 605332. [Google Scholar] [CrossRef] [PubMed]
- Barone, P.; Dehay, C.; Berland, M.; Bullier, J.; Kennedy, H. Developmental remodeling of primate visual cortical pathways. Cereb. Cortex 1995, 5, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, C.P.D.; Khalil, R. Postnatal Development of Visual Cortical Function in the Mammalian Brain. Front. Syst. Neurosci. 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.Y.; A Pinsk, M.; Kastner, S. Neural Basis of Biased Competition in Development: Sensory Competition in Visual Cortex of School-Aged Children. Cereb. Cortex 2021, 31, 3107–3121. [Google Scholar] [CrossRef]
- Pucilowska, J.; Vithayathil, J.; Tavares, E.J.; Kelly, C.; Karlo, J.C.; Landreth, G.E. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J. Neurosci. 2015, 35, 3190–3200. [Google Scholar] [CrossRef] [PubMed]
- Adler, W.T.; Platt, M.P.; Mehlhorn, A.J.; Haight, J.L.; Currier, T.A.; Etchegaray, M.A.; Galaburda, A.M.; Rosen, G.D. Position of Neocortical Neurons Transfected at Different Gestational Ages with shRNA Targeted against Candidate Dyslexia Susceptibility Genes. PLoS ONE 2013, 8, e65179. [Google Scholar] [CrossRef]
- Platt, M.; Adler, W.; Mehlhorn, A.; Johnson, G.; Wright, K.; Choi, R.; Tsang, W.; Poon, M.; Yeung, S.; Waye, M.; et al. Embryonic disruption of the candidate dyslexia susceptibility gene homologue Kiaa0319-like results in neuronal migration disorders. Neuroscience 2013, 248, 585–593. [Google Scholar] [CrossRef]
- Farah, R.; Ionta, S.; Horowitz-Kraus, T. Neuro-Behavioral Correlates of Executive Dysfunctions in Dyslexia Over Development from Childhood to Adulthood. Front. Psychol. 2021, 12, 708863. [Google Scholar] [CrossRef] [PubMed]
- Stein, J. The current status of the magnocellular theory of developmental dyslexia. Neuropsychologia 2019, 130, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Stein, J. Dyslexia: The Role of Vision and Visual Attention. Curr. Dev. Disord. Rep. 2014, 1, 267–280. [Google Scholar] [CrossRef]
- Livingstone, M.S.; Rosen, G.D.; Drislane, F.W.; Galaburda, A.M. Physiological and anatomical evidence for a magnocellular defect in developmental dyslexia. Proc. Natl. Acad. Sci. USA 1991, 88, 7943–7947. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.-L. Lamination, Borders, and Thalamic Projections of the Primary Visual Cortex in Human, Non-Human Primate, and Rodent Brains. Brain Sci. 2024, 14, 372. https://doi.org/10.3390/brainsci14040372
Ding S-L. Lamination, Borders, and Thalamic Projections of the Primary Visual Cortex in Human, Non-Human Primate, and Rodent Brains. Brain Sciences. 2024; 14(4):372. https://doi.org/10.3390/brainsci14040372
Chicago/Turabian StyleDing, Song-Lin. 2024. "Lamination, Borders, and Thalamic Projections of the Primary Visual Cortex in Human, Non-Human Primate, and Rodent Brains" Brain Sciences 14, no. 4: 372. https://doi.org/10.3390/brainsci14040372
APA StyleDing, S.-L. (2024). Lamination, Borders, and Thalamic Projections of the Primary Visual Cortex in Human, Non-Human Primate, and Rodent Brains. Brain Sciences, 14(4), 372. https://doi.org/10.3390/brainsci14040372




