Insights into the Mechanisms of Lactobacillus acidophilus Activity against Entamoeba histolytica by Using Thiol Redox Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. E. histolytica and L. acidophilus Culture
2.2. Reagents
2.3. Ferrous Oxidation-Xylenol Orange (FOX) Assay
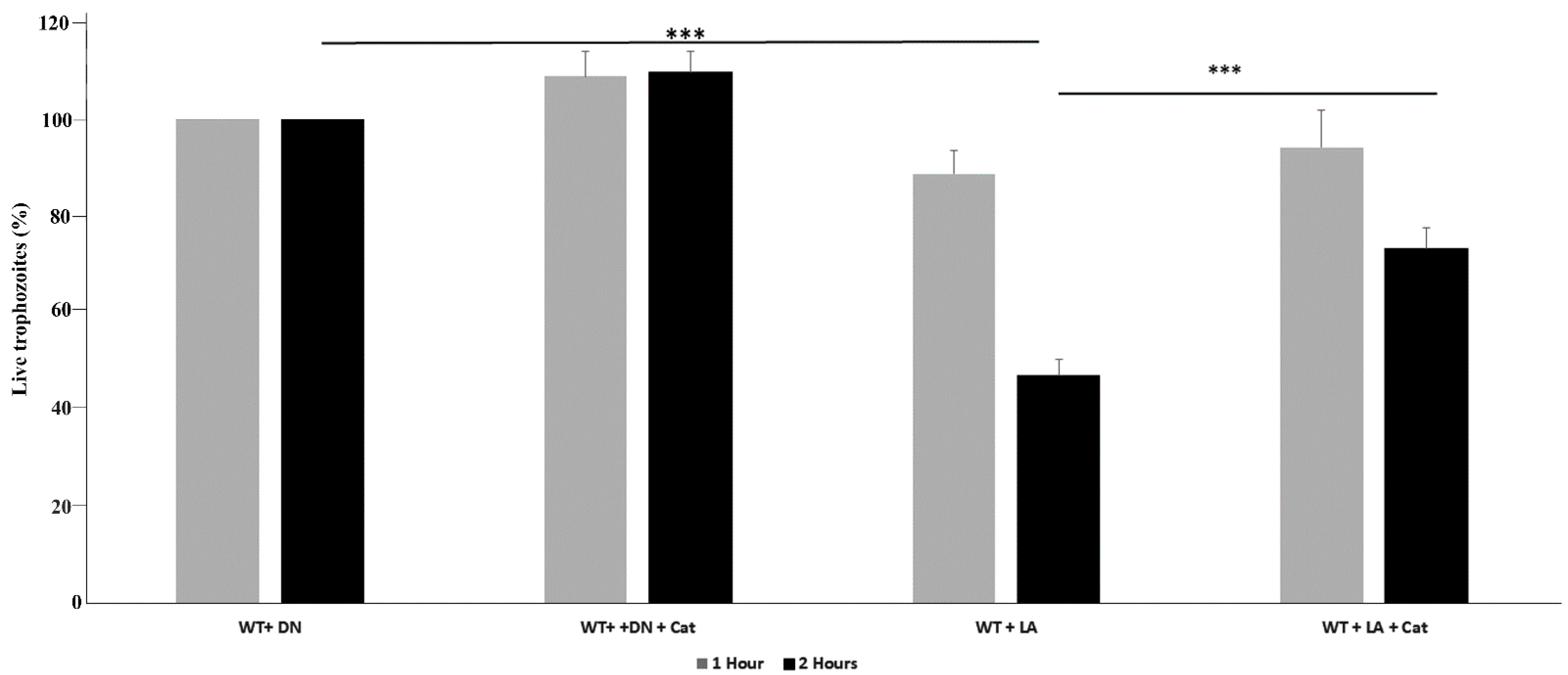
2.4. Viability of E. histolytica Trophozoites
2.5. Detection of Oxidized Proteins (OXs) by Resin-Assisted Capture RAC (OX–RAC)
2.6. In-Gel Proteolysis and MS Analysis
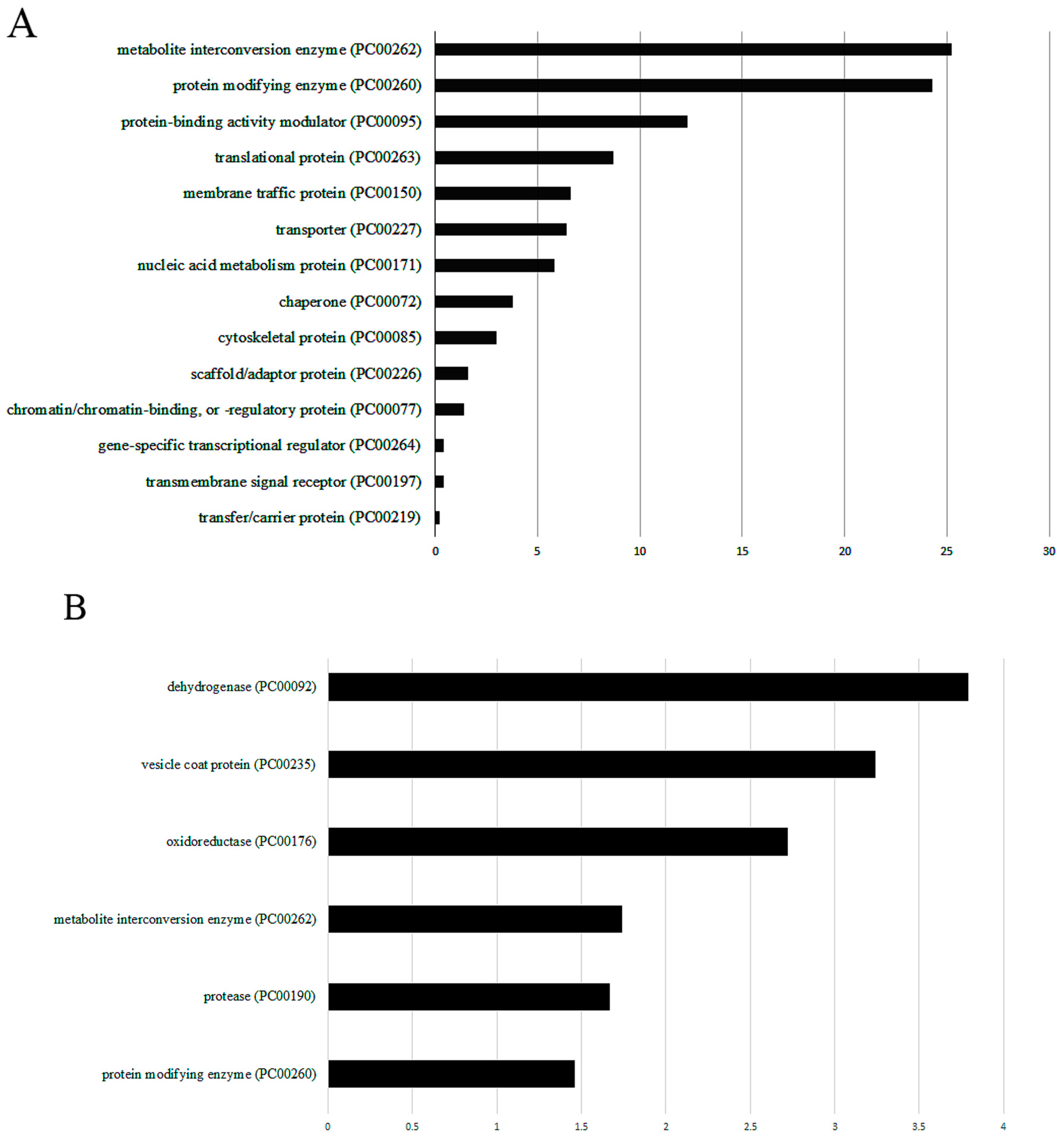
2.7. Classification of OXs According to Their Protein Class and Statistical Overrepresentation Test
2.8. Measurement of Cysteine Proteases (CPs) Activity
2.9. Adhesion Assay
3. Results and Discussion
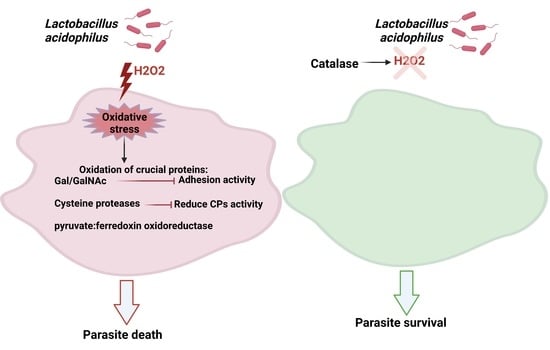
3.1. L. acidophilus Amebicide Activity Depends on the Formation of H2O2
3.2. Resin-Assisted Capture (RAC) of Oxidized Proteins (OX) Coupled to Mass Spectrometry (OX–RAC) Analysis of E. histolytica Trophozoites Exposed to L. acidophilus
3.3. E. histolytica CP Activity Is Impaired by L. acidophilus
3.4. Adhesion of E. histolytica Trophozoites to HeLa Cells Is Impaired by L. acidophilus
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marie, C.; Petri, W.A., Jr. Regulation of Virulence of Entamoeba histolytica. Annu. Rev. Microbiol. 2014, 68, 493–520. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.J.; Ganguly, N.K.; Mahajan, R.C.; Dilawari, J.B. Antibody dependent cellular cytotoxicity in experimental intestinal & hepatic amoebiasis. Indian J. Med. Res. 1988, 87, 170–175. [Google Scholar] [PubMed]
- Schulz, T.F.; Kollaritsch, H.; Hengster, P.; Stemberger, H.; Scheiner, O.; Wiedemann, G.; Dierich, M.P. Molecular weight analysis of Entamoeba histolytica antigens recognized by IgG and IgM antibodies in the sera of patients with amoebiasis. Trop. Med. Parasitol. 1987, 38, 149–152. [Google Scholar] [PubMed]
- Rastew, E.; Vicente, J.B.; Singh, U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int. J. Parasitol. 2012, 42, 1007–1015. [Google Scholar] [CrossRef] [Green Version]
- Santi-Rocca, J.; Smith, S.; Weber, C.; Pineda, E.; Hon, C.-C.; Saavedra, E.; Olivos-García, A.; Rousseau, S.; Dillies, M.-A.; Coppée, J.-Y.; et al. Endoplasmic reticulum stress-sensing mechanism is activated in Entamoeba histolytica upon treatment with nitric oxide. PLoS ONE 2012, 7, e31777. [Google Scholar] [CrossRef] [Green Version]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Alterzon, S.B.; Hertz, R.; Methling, K.; Lalk, M.; Ankri, S. Proteomic Identification of Oxidized Proteins in Entamoeba histolytica by Resin-Assisted Capture: Insights into the Role of Arginase in Resistance to Oxidative Stress. PLoS Negl. Trop. Dis. 2016, 10, e0004340. [Google Scholar] [CrossRef] [Green Version]
- Hertz, R.; Ben Lulu, S.; Shahi, P.; Trebicz-Geffen, M.; Benhar, M.; Ankri, S. Proteomic Identification of S-Nitrosylated Proteins in the Parasite Entamoeba histolytica by Resin-Assisted Capture: Insights into the Regulation of the Gal/GalNAc Lectin by Nitric Oxide. PLoS ONE. 2014, 9, e91518. [Google Scholar] [CrossRef] [Green Version]
- Roe, F.J. Metronidazole: Review of uses and toxicity. J. Antimicrob. Chemother. 1977, 3, 205–212. [Google Scholar] [CrossRef]
- Andersson, K.E. Pharmacokinetics of Nitroimidazoles—Spectrum of Adverse Reactions. Scand. J. Infect. Dis. 1981, 26, 60–67. [Google Scholar]
- Bansal, D.; Sehgal, R.; Chawla, Y.; Mahajan, R.C.; Malla, N. In vitro activity of antiamoebic drugs against clinical isolates of Entamoeba histolytica and Entamoeba dispar. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 27. [Google Scholar] [CrossRef] [Green Version]
- Iyer, L.R.B.N.; Naik, S.; Paul, J. Antioxidant enzyme profile of two clinical isolates of Entamoeba histolytica varying in sensitivity to antiamoebic drugs. World J. Clin. Infect. Dis. 2017, 7, 21–23. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travers, M.A.; Florent, I.; Kohl, L.; Grellier, P. Probiotics for the control of parasites: An overview. J. Parasitol. Res. 2011, 2011, 610769. [Google Scholar] [CrossRef] [PubMed]
- Bar, A.K.; Phukan, N.; Pinheiro, J.; Simoes-Barbosa, A. The Interplay of Host Microbiota and Parasitic Protozoans at Mucosal Interfaces: Implications for the Outcomes of Infections and Diseases. PLoS Negl. Trop. Dis. 2015, 9, e0004176. [Google Scholar] [CrossRef]
- Goyal, N.; Tiwari, R.P.; Shukla, G. Lactobacillus rhamnosus GG as an Effective Probiotic for Murine Giardiasis. Interdiscip. Perspect. Infect. Dis. 2011, 2011, 795219. [Google Scholar] [CrossRef] [Green Version]
- Sarjapuram, N.; Mekala, N.; Singh, M.; Tatu, U. The Potential of Lactobacillus casei and Entercoccus faecium Combination as a Preventive Probiotic Against Entamoeba. Probiotics Antimicrob. Proteins 2017, 9, 142–149. [Google Scholar] [CrossRef]
- Rigothier, M.C.; Maccario, J.; Gayral, P. Inhibitory activity of saccharomyces yeasts on the adhesion of Entamoeba histolytica trophozoites to human erythrocytes in vitro. Parasitol. Res. 1994, 80, 10–15. [Google Scholar] [CrossRef]
- Mansour-Ghanaei, F.; Dehbashi, N.; Yazdanparast, K.; Shafaghi, A. Efficacy of saccharomyces boulardii with antibiotics in acute amoebiasis. World J. Gastroenterol. 2003, 9, 1832–1833. [Google Scholar] [CrossRef]
- Varet, H.; Shaulov, Y.; Sismeiro, O.; Trebicz-Geffen, M.; Legendre, R.; Coppée, J.-Y.; Ankri, S.; Guillen, N. Enteric bacteria boost defences against oxidative stress in Entamoeba histolytica. Sci. Rep. 2018, 8, 9042. [Google Scholar] [CrossRef]
- Collins, E.B.; Aramaki, K. Production of Hydrogen peroxide by Lactobacillus acidophilus. J. Dairy Sci. 1980, 63, 353–357. [Google Scholar] [CrossRef]
- Hertzberger, R.; Arents, J.; Dekker, H.L.; Pridmore, R.D.; Gysler, C.; Kleerebezem, M.; de Mattos, M.J.T. H2O2 production in species of the Lactobacillus acidophilus group: A central role for a novel NADH-dependent flavin reductase. Appl. Environ. Microbiol. 2014, 80, 2229–2239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamond, L.S.; Harlow, D.R.; Cunnick, C.C. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978, 72, 431–432. [Google Scholar] [CrossRef]
- DeLong, J.M.; Prange, R.K.; Hodges, D.M.; Forney, C.; Bishop, M.C.; Quilliam, M. Using a modified ferrous oxidation-xylenol orange (FOX) assay for detection of lipid hydroperoxides in plant tissue. J. Agric. Food Chem. 2002, 50, 248–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version. 16, a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Ankri, S.; Stolarsky, T.; Mirelman, D. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 1998, 28, 777–785. [Google Scholar] [CrossRef]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Hertz, R.; Alterzon-Baumel, S.; Methling, K.; Lalk, M.; Mazumder, M.; Samudrala, G.; Ankri, S. N-acetyl ornithine deacetylase is a moonlighting protein and is involved in the adaptation of Entamoeba histolytica to nitrosative stress. Sci. Rep. 2016, 6, 36323. [Google Scholar] [CrossRef] [Green Version]
- Dinev, T.B.G.; Denev, S.; Dermendzhieva, D.; Tzanova, M.; Valkova, E. Antimicrobial activity of Lactobacillus acidophilus against pathogenic and food spoilage microorganisms: A review. Agric. Sci. Technol. 2017, 91, 3–9. [Google Scholar] [CrossRef]
- Weiss, G.; Rasmussen, S.; Zeuthen, L.H.; Nielsen, B.N.; Jarmer, H.; Jespersen, L.; Frøkiaer, H. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology 2010, 131, 268–281. [Google Scholar] [CrossRef]
- Salari, S.; Ghasemi Nejad Almani, P. Antifungal effects of Lactobacillus acidophilus and Lactobacillus plantarum against different oral Candida species isolated from HIV/AIDS patients: An in vitro study. J. Oral Microbiol. 2020, 12, 1769386. [Google Scholar] [CrossRef]
- Aween, M.M.; Hassan, Z.; Muhialdin, B.J.; Eljamel, Y.A.; Al-Mabrok, A.S.W.; Lani, M.N. Antibacterial activity of Lactobacillus acidophilus strains isolated from honey marketed in Malaysia against selected multiple antibiotic resistant (MAR) Gram-positive bacteria. J. Food Sci. 2012, 77, M364–M371. [Google Scholar] [CrossRef] [PubMed]
- Al-Megrin, W.A.; Mohamed, S.H.; Saleh, M.M.; Yehia, H.M. Preventive role of probiotic bacteria against gastrointestinal diseases in mice caused by Giardia lamblia. Biosci. Rep. 2021, 41, BSR20204114. [Google Scholar] [CrossRef] [PubMed]
- Cadore, P.S.; Walcher, D.L.; de Sousa, N.F.G.C.; Martins, L.H.R.; da Hora, V.P.; Von Groll, A.; de Moura, M.Q.; Berne, M.E.A.; Avila, L.F.D.C.D.; Scaini, C.J. Protective effect of the probiotic Lactobacillus acidophilus ATCC. 4356 in BALB/c mice infected with Toxocara canis. Rev. Inst. Med. Trop. São Paulo 2021, 63, e9. [Google Scholar] [CrossRef] [PubMed]
- Farrag, H.M.; Huseein, E.A.; Abd El-Rady, N.M.; Mostafa, F.A.; Mohamed, S.S.; Gaber, M. The protective effect of Lactobacillus acidophilus on experimental animals challenged with Trichinella spiralis; new insights on their feasibility as prophylaxis in Trichinella spiralis endemic area. Ann. Parasitol. 2021, 67, 195–202. [Google Scholar]
- Alak, J.I.; Wolf, B.W.; Mdurvwa, E.G.; EPimentel-Smith, G.; Kolavala, S.; Abdelrahman, H.; Suppiramaniam, V. Supplementation with Lactobacillus reuteri or L. acidophilus reduced intestinal shedding of cryptosporidium parvum oocysts in immunodeficient C57BL/6 mice. Cell. Mol. Biol. (Noisy-le-grand) 1999, 45, 855–863. [Google Scholar]
- Loew, O. A New Enzyme of General Occurrence in Organismis. Science 1900, 11, 701–702. [Google Scholar] [CrossRef] [Green Version]
- Shaulov, Y.; Sarid, L.; Trebicz-Geffen, M.; Ankri, S. Entamoeba histolytica Adaption to Auranofin: A Phenotypic and Multi-Omics Characterization. Antioxidants 2021, 10, 1240. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; Dezonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Fahey, R.C.; Newton, G.L.; Arrick, B.; Overdank-Bogart, T.; Aley, S.B. Entamoeba histolytica: A eukaryote without glutathione metabolism. Science 1984, 224, 70–72. [Google Scholar] [CrossRef]
- Jeelani, G.; Nozaki, T. Entamoeba thiol-based redox metabolism: A potential target for drug development. Mol. Biochem. Parasitol. 2016, 206, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Pan, G.; Poncz, M.; Wei, J.; Ran, M.; Zhou, Z. Serpin functions in host-pathogen interactions. PeerJ 2018, 6, e4557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riahi, Y.; Siman-Tov, R.; Ankri, S. Molecular cloning, expression and characterization of a serine proteinase inhibitor gene from Entamoeba histolytica. Mol. Biochem. Parasitol. 2004, 133, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.C.; Dafforn, T.R.; Forsyth, S.L.; Missen, M.A.; Horvath, A.J.; Hampson, L.; Hampson, I.N.; Currie, G.; Carrell, R.W.; Coughlin, P.B. Murine serpin. 2A is a redox-sensitive intracellular protein. Biochem. J. 2003, 371, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Stief, T.W.; Aab, A.; Heimburger, N. Oxidative inactivation of purified human alpha-2-antiplasmin, antithrombin III, and C1-inhibitor. Thromb. Res. 1988, 49, 581–589. [Google Scholar] [CrossRef]
- Mangan, M.S.; Bird, C.H.; Kaiserman, D.; Matthews, A.Y.; Hitchen, C.; Steer, D.L.; Thompson, P.E.; Bird, P.I. A Novel Serpin Regulatory Mechanism: SerpinB9 Is Reversibly Inhibited by vicinal disulfide bond formation in the reactive center loop. J. Biol. Chem. 2016, 291, 3626–3638. [Google Scholar] [CrossRef] [Green Version]
- Coudrier, E.; Amblard, F.; Zimmer, C.; Roux, P.; Olivo--Marin, J.C.; Rigothier, M.C.; Guillén, N. Myosin II and the Gal-GalNAc lectin play a crucial role in tissue invasion by Entamoeba histolytica. Cell. Microbiol. 2005, 7, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Bosch, D.E.; Siderovski, D.P. G protein signaling in the parasite Entamoeba histolytica. Exp. Mol. Med. 2013, 45, e15. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, R.; Sharma, S.; Janhawi Arya, R.; Bhattacharya, S.; Bhattacharya, A. EhRho1 regulates phagocytosis by modulating actin dynamics through EhFormin1 and EhProfilin1 in Entamoeba histolytica. Cell. Microbiol. 2018, 20, e12851. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Zhou, B.; Cox, A.D.; Campbell, S.L. Rho GTPases, oxidation, and cell redox control. Small GTPases 2014, 5, e28579. [Google Scholar] [CrossRef] [Green Version]
- Olson, M.F. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases 2018, 9, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Kovacic, H.N.; Irani, K.; Goldschmidt-Clermont, P.J. Redox regulation of human Rac1 stability by the proteasome in human aortic endothelial cells. J. Biol. Chem. 2001, 276, 45856–45861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, R.E.; Warren, L.G.; Susskind, B.; Lo, H.S. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J. Biol. Chem. 1977, 252, 726–731. [Google Scholar] [CrossRef]
- Shaulov, Y.; Nagaraja, S.; Sarid, L.; Trebicz-Geffen, M.; Ankri, S. Formation of oxidised (OX) proteins in Entamoeba histolytica exposed to auranofin and consequences on the parasite virulence. Cell. Microbiol. 2020, 22, e13174. [Google Scholar] [CrossRef]
- Pineda, E.; Encalada, R.; Rodríguez-Zavala, J.S.; Olivos-García, A.; Moreno-Sánchez, R.; Saavedra, E. Pyruvate:ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010, 277, 3382–3395. [Google Scholar] [CrossRef]
- Nowak, N.; Lotter, H.; Tannich, E.; Bruchhaus, I. Resistance of Entamoeba histolytica to the cysteine proteinase inhibitor E64 is associated with secretion of pro-enzymes and reduced pathogenicity. J. Biol. Chem. 2004, 279, 38260–38266. [Google Scholar] [CrossRef] [Green Version]
- Matthiesen, J.; Bär, A.-K.; Bartels, A.-K.; Marien, D.; Ofori, S.; Biller, L.; Tannich, E.; Lotter, H.; Bruchhaus, I. Overexpression of specific cysteine peptidases confers pathogenicity to a nonpathogenic Entamoeba histolytica clone. mBio 2013, 4, e00072-13. [Google Scholar] [CrossRef] [Green Version]
- Irmer, H.; Tillack, M.; Biller, L.; Handal, G.; Leippe, M.; Roeder, T.; Tannich, E.; Bruchhaus, I. Major cysteine peptidases of Entamoeba histolytica are required for aggregation and digestion of erythrocytes but are dispensable for phagocytosis and cytopathogenicity. Mol. Microbiol. 2009, 72, 658–667. [Google Scholar] [CrossRef]
- Hernandez-Cuevas, N.A.; Weber, C.; Hon, C.C.; Guillen, N. Gene expression profiling in Entamoeba histolytica identifies key components in iron uptake and metabolism. PLoS ONE 2014, 9, e107102. [Google Scholar] [CrossRef]
- Lalmanach, G.; Saidi, A.; Bigot, P.; Chazeirat, T.; Lecaille, F.; Wartenberg, M. Regulation of the Proteolytic Activity of Cysteine Cathepsins by Oxidants. Int. J. Mol. Sci. 2020, 21, 1944. [Google Scholar] [CrossRef] [Green Version]
- Bruchhaus, I.; Loftus, B.J.; Hall, N.; Tannich, E. The intestinal protozoan parasite Entamoeba histolytica contains. 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2003, 2, 501–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herraiz, T.; Guillen, H.; Gonzalez-Pena, D.; Aran, V.J. Antimalarial Quinoline Drugs Inhibit beta-Hematin and Increase Free Hemin Catalyzing Peroxidative Reactions and Inhibition of Cysteine Proteases. Sci. Rep. 2019, 9, 15398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirelman, D.; Kobiler, D. Adhesion properties of Entamoeba histolytica. In Adhesion and Microorganism Pathogenicity; Pittman Med London: London, UK, 1981; Volume 80, pp. 17–35. [Google Scholar]
- Aguirre Garcia, M.; Gutierrez-Kobeh, L.; Lopez Vancell, R. Entamoeba histolytica: Adhesins and lectins in the trophozoite surface. Molecules 2015, 20, 2802–2815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petri, W.A., Jr.; Haque, R.; Mann, B.J. The bittersweet interface of parasite and host: Lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 2002, 56, 39–64. [Google Scholar] [CrossRef] [PubMed]
- Vines, R.R.; Ramakrishnan, G.; Rogers, J.B.; Lockhart, L.A.; Mann, B.J.; Petri, W.A., Jr. Regulation of adherence and virulence by the Entamoeba histolytica lectin cytoplasmic domain, which contains a beta2 integrin motif. Mol. Biol. Cell 1998, 9, 2069–2079. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarid, L.; Zanditenas, E.; Ye, J.; Trebicz-Geffen, M.; Ankri, S. Insights into the Mechanisms of Lactobacillus acidophilus Activity against Entamoeba histolytica by Using Thiol Redox Proteomics. Antioxidants 2022, 11, 814. https://doi.org/10.3390/antiox11050814
Sarid L, Zanditenas E, Ye J, Trebicz-Geffen M, Ankri S. Insights into the Mechanisms of Lactobacillus acidophilus Activity against Entamoeba histolytica by Using Thiol Redox Proteomics. Antioxidants. 2022; 11(5):814. https://doi.org/10.3390/antiox11050814
Chicago/Turabian StyleSarid, Lotem, Eva Zanditenas, Jun Ye, Meirav Trebicz-Geffen, and Serge Ankri. 2022. "Insights into the Mechanisms of Lactobacillus acidophilus Activity against Entamoeba histolytica by Using Thiol Redox Proteomics" Antioxidants 11, no. 5: 814. https://doi.org/10.3390/antiox11050814
APA StyleSarid, L., Zanditenas, E., Ye, J., Trebicz-Geffen, M., & Ankri, S. (2022). Insights into the Mechanisms of Lactobacillus acidophilus Activity against Entamoeba histolytica by Using Thiol Redox Proteomics. Antioxidants, 11(5), 814. https://doi.org/10.3390/antiox11050814