Transcriptome Analysis of Peritoneal Cells Reveals the Early Immune Response of Flounder (Paralichthys olivaceus) to Inactivated Vibrio anguillarum Immunization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Fish
2.3. Inactivated Bacteria Immunization and Sample Collection
2.4. RNA Extraction, Library Construction, and Sequencing
2.5. Transcript Assembly and Correlation Analysis of Samples
2.6. Differentially Expressed Genes and WGCNA Analysis
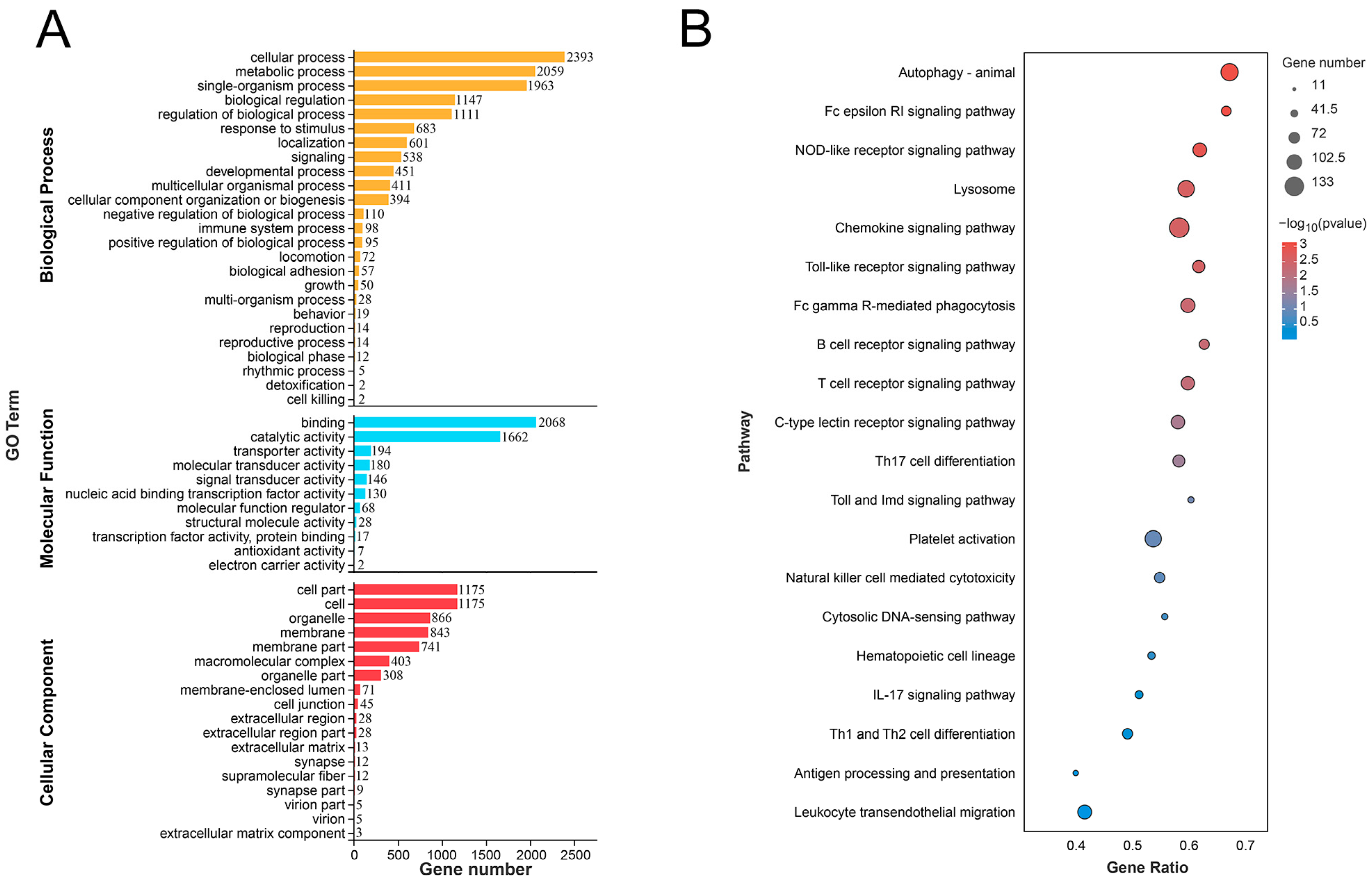
2.7. GO and KEGG Enrichment Analysis
2.8. Quantitative Real-Time PCR (qPCR)
2.9. Statistical Analysis
3. Results
3.1. Transcriptome Sequencing Quality
3.2. Differentially Expressed Genes at Different Times after Immunization
3.3. Gene Co-Expression Modules Post Immunization
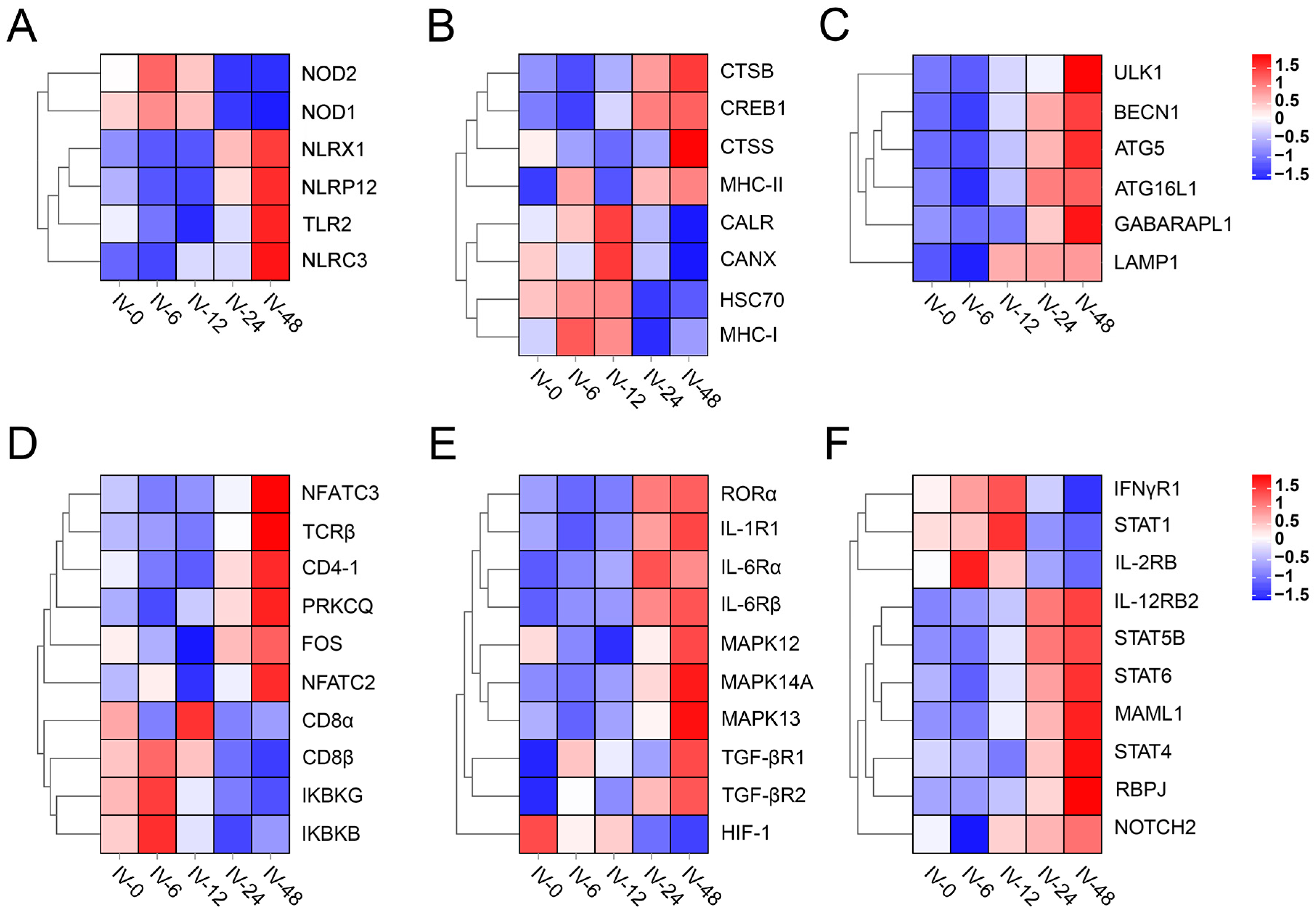
3.4. Enrich Classification Hub Modules Genes
3.5. Expression of T Helper (Th) Cell Differentiation-Related Genes
3.6. Validation of Gene Expression Profiles by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seikai, T. Flounder Culture and Its Challenges in Asia. Rev. Fish. Sci. 2002, 10, 421–432. [Google Scholar] [CrossRef]
- Sohn, H.; Kim, J.; Jin, C.; Lee, J. Identification of Vibrio Species Isolated from Cultured Olive Flounder (Paralichthys olivaceus) in Jeju Island, South Korea. Fish. Aquatic. Sci. 2019, 22, 14. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.; Lievens, B.; Rediers, H. Vibrio anguillarum as a Fish Pathogen: Virulence Factors, Diagnosis and Prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef] [PubMed]
- Toranzo, A.E.; Magariños, B.; Romalde, J.L. A Review of the Main Bacterial Fish Diseases in Mariculture Systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Ning, X.; Sun, L. Gene Network Analysis Reveals a Core Set of Genes Involved in the Immune Response of Japanese Flounder (Paralichthys olivaceus) against Vibrio anguillarum Infection. Fish Shellfish Immunol. 2020, 98, 800–809. [Google Scholar] [CrossRef]
- Sepulcre, M.P.; Sarropoulou, E.; Kotoulas, G.; Meseguer, J.; Mulero, V. Vibrio anguillarum evades the immune response of the bony fish sea bass (Dicentrarchus labrax L.) through the inhibition of leukocyte respiratory burst and down-regulation of apoptotic caspases. Mol. Immunol 2007, 44, 3751–3757. [Google Scholar] [CrossRef]
- Xing, J.; Xu, H.; Tang, X.; Sheng, X.; Zhan, W. A DNA Vaccine Encoding the VAA Gene of Vibrio anguillarum Induces a Protective Immune Response in Flounder. Front. Immunol. 2019, 10, 499. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Y.; Sun, Q.; Li, X.; Sun, L. An Inactivated Bivalent Vaccine Effectively Protects Turbot (Scophthalmus maximus) against Vibrio anguillarum and Vibrio harveyi Infection. Aquaculture 2021, 544, 737158. [Google Scholar] [CrossRef]
- Chakraborty, S.; Cao, T.; Hossain, A.; Gnanagobal, H.; Vasquez, I.; Boyce, D.; Santander, J. Vibrogen-2 Vaccine Trial in Lumpfish (Cyclopterus lumpus) against Vibrio anguillarum. J. Fish Dis. 2019, 42, 1057–1064. [Google Scholar] [CrossRef]
- De Magistris, M.T. Mucosal Delivery of Vaccine Antigens and Its Advantages in Pediatrics. Adv. Drug Deliv. Rev. 2006, 58, 52–67. [Google Scholar] [CrossRef]
- Ma, J.; Bruce, T.J.; Jones, E.M.; Cain, K.D. A Review of Fish Vaccine Development Strategies: Conventional Methods and Modern Biotechnological Approaches. Microorganisms 2019, 7, 569. [Google Scholar] [CrossRef] [PubMed]
- Tlaxca, J.L.; Ellis, S.; Remmele, R.L., Jr. Live Attenuated and Inactivated Viral Vaccine Formulation and Nasal Delivery: Potential and Challenges. Adv. Drug Deliv. Rev. 2015, 93, 56–78. [Google Scholar] [CrossRef] [PubMed]
- Madonia, A.; Melchiorri, C.; Bonamano, S.; Marcelli, M.; Bulfon, C.; Castiglione, F.; Galeotti, M.; Volpatti, D.; Mosca, F.; Tiscar, P.-G.; et al. Computational modeling of immune system of the fish for a more effective vaccination in aquaculture. Bioinformatics 2017, 33, 3065–3071. [Google Scholar] [CrossRef]
- Shi, X.; Chi, H.; Sun, Y.; Tang, X.; Xing, J.; Sheng, X.; Zhan, W. The Early Peritoneal Cavity Immune Response to Vibrio anguillarum Infection and to Inactivated Bacterium in Olive Flounder (Paralichthys olivaceus). Microorganisms 2022, 10, 2175. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Corey, D.R. RNA Sequencing: Platform Selection, Experimental Design, and Data Interpretation. Nucleic Acid Ther. 2012, 22, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Hong, S. Transcriptome Analysis in the Head Kidney of Rainbow Trout (Oncorhynchus mykiss) Immunized with a Combined Vaccine of Formalin-Inactivated Aeromonas salmonicida and Vibrio anguillarum. Vaccines 2021, 9, 1234. [Google Scholar] [CrossRef]
- Fu, Q.; Li, Y.; Zhang, H.; Cao, M.; Zhang, L.; Gao, C.; Cai, X.; Chen, D.; Yang, Z.; Li, J. Comparative Transcriptome Analysis of Spleen Reveals Potential Regulation of Genes and Immune Pathways Following Administration of Aeromonas Salmonicida Subsp. Masoucida Vaccine in Atlantic Salmon (Salmo salar). Mar. Biotechnol. 2022, 24, 97–115. [Google Scholar] [CrossRef]
- Gan, Q.; Chi, H.; Dalmo, R.A.; Meng, X.; Tang, X.; Xing, J.; Sheng, X.; Zhan, W. Characterization of Myeloperoxidase and Its Contribution to Antimicrobial Effect on Extracellular Traps in Flounder (Paralichthys olivaceus). Front. Immunol. 2023, 14, 1124813. [Google Scholar] [CrossRef]
- Li, Q.; Chi, H.; Shi, X.; Gan, Q.; Dalmo, R.A.; Sun, Y.-y.; Tang, X.; Xing, J.; Sheng, X.; Zhan, W. Vaccine Adjuvants Induce Formation of Intraperitoneal Extracellular Traps in Flounder (Paralichthys olivaceus). Front. Cell. Infect. Microbiol. 2022, 12, 875409. [Google Scholar] [CrossRef]
- Li, Q.; Zhan, W.; Xing, J.; Sheng, X. Production, characterisation and applicability of monoclonal antibodies to immunoglobulin of Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2007, 23, 982–990. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C., Jr. Innate Immunity. N. Engl. J. Med. 2000, 343, 338–344. [Google Scholar] [CrossRef]
- Saitoh, K.; Hayashizaki, K.; Yokoyama, Y.; Asahida, T.; Toyohara, H.; Yamashita, Y. Complete Nucleotide Sequence of Japanese Flounder (Paralichthys olivaceus) Mitochondrial Genome: Structural Properties and Cue for Resolving Teleostean Relationship. J. Hered. 2000, 91, 271–278. [Google Scholar] [CrossRef]
- Chu, Q.; Song, W.; Cui, J.; Xu, T. Genome-Guided Transcriptome Analysis of Miiuy Croaker Provides Insights into Pattern Recognition Receptors and Cytokines in Response to Vibrio anguillarum. Dev. Comp. Immunol. 2017, 73, 72–78. [Google Scholar] [CrossRef]
- Bi, D.; Wang, Y.; Gao, Y.; Li, X.; Chu, Q.; Cui, J.; Xu, T. Recognition of Lipopolysaccharide and Activation of NF-kB by Cytosolic Sensor NOD1 in Teleost Fish. Front. Immunol. 2018, 9, 1413. [Google Scholar] [CrossRef]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. NOD2 Is a General Sensor of Peptidoglycan Through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef] [PubMed]
- Tuladhar, S.; Kanneganti, T.-D. NLRP12 in Innate Immunity and Inflammation. Mol. Aspects Med. 2020, 76, 100887. [Google Scholar] [CrossRef] [PubMed]
- Hacker, H.; Karin, M. Regulation and Function of IKK and IKK-Related Kinases. Sci. STKE 2006, 2006, re13. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Swain, B.; Sahoo, B.R.; Maiti, N.K.; Samanta, M. Induction of Toll-Like Receptor (TLR) 2, And Myd88-Dependent TLR-Signaling in Response to Ligand Stimulation and Bacterial Infections in the Indian Major Carp, Mrigal (Cirrhinus mrigala). Mol. Biol. Rep. 2012, 39, 6015–6028. [Google Scholar] [CrossRef] [PubMed]
- Samanta, M.; Swain, B.; Basu, M.; Panda, P.; Mohapatra, G.B.; Sahoo, B.R.; Maiti, N.K. Molecular Characterization of Toll-Like Receptor 2 (Tlr2), Analysis of Its Inductive Expression and Associated Down-Stream Signaling Molecules Following Ligands Exposure and Bacterial Infection in the Indian Major Carp, Rohu (Labeo rohita). Fish Shellfish Immunol. 2012, 32, 411–425. [Google Scholar] [CrossRef]
- Wei, Y.C.; Pan, T.S.; Chang, M.X.; Huang, B.; Xu, Z.; Luo, T.R.; Nie, P. Cloning and Expression of Toll-Like Receptors 1 and 2 from a Teleost Fish, The Orange-Spotted Grouper Epinephelus coioides. Vet. Immunol. Immunopathol. 2011, 141, 173–182. [Google Scholar] [CrossRef]
- Wilson, A.B. MHC and Adaptive Immunity in Teleost Fishes. Immunogenetics 2017, 69, 521–528. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, N.; Hashimoto, K.; Xia, C.; Dijkstra, J.M. Structural Comparison between MHC Classes I and II.; In Evolution, a Class-II-Like Molecule Probably Came First. Front. Immunol. 2021, 12, 621153. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Dijkstra, J.M. Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish. Cells 2019, 8, 378. [Google Scholar] [CrossRef]
- Ahn, D.-H.; Kang, S.; Park, H. Transcriptome Analysis of Immune Response Genes Induced by Pathogen Agonists in The Antarctic Bullhead Notothen Notothenia coriiceps. Fish Shellfish Immunol. 2016, 55, 315–322. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, H.; Xiao, J.; Wang, Q.; Liu, Q.; Zhang, Y. Immune Responses of Zebrafish (Danio rerio) Induced by Bath-Vaccination with a Live Attenuated Vibrio anguillarum Vaccine Candidate. Fish Shellfish Immunol. 2012, 33, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I. Molecular Definitions of Autophagy and Related Processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Münz, C. Non-Canonical Functions of Autophagy Proteins in Immunity and Infection. Mol. Aspects Med. 2021, 82, 100987. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.C.; Dikic, I. Autophagy in Antimicrobial Immunity. Mol. Cell 2014, 54, 224–233. [Google Scholar] [CrossRef]
- Lee, H.K.; Mattei, L.M.; Steinberg, B.E.; Alberts, P.; Lee, Y.H.; Chervonsky, A.; Mizushima, N.; Grinstein, S.; Iwasaki, A. In Vivo Requirement for Atg5 in Antigen Presentation by Dendritic Cells. Immunity 2010, 32, 227–239. [Google Scholar] [CrossRef]
- Fletcher, K.; Ulferts, R.; Jacquin, E.; Veith, T.; Gammoh, N.; Arasteh, J.M.; Mayer, U.; Carding, S.R.; Wileman, T.; Beale, R. The WD 40 Domain of ATG 16L1 Is Required for Its Non-Canonical Role in Lipidation of LC 3 At Single Membranes. EMBO J. 2018, 37, e97840. [Google Scholar] [CrossRef]
- Masud, S.; Prajsnar, T.K.; Torraca, V.; Lamers, G.E.; Benning, M.; Van Der Vaart, M.; Meijer, A.H. Macrophages Target Salmonella by Lc3-Associated Phagocytosis in a Systemic Infection Model. Autophagy 2019, 15, 796–812. [Google Scholar] [CrossRef]
- Saito, S.; Nakashima, A.; Shima, T.; Ito, M. Th1/Th2/Th17 and Regulatory T Cell Paradigm in Pregnancy. Am. J. Reprod. Immunol. 2010, 63, 601–610. [Google Scholar] [CrossRef]
- Robertsen, B. The Interferon System of Teleost Fish. Fish Shellfish Immunol. 2006, 20, 172–191. [Google Scholar] [CrossRef] [PubMed]
- Yoshiura, Y.; Kiryu, I.; Fujiwara, A.; Suetake, H.; Suzuki, Y.; Nakanishi, T.; Ototake, M. Identification and Characterization of Fugu Orthologues of Mammalian Interleukin-12 Subunits. Immunogenetics 2003, 55, 296–306. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, H.; Soliman, H.; Saleh, M.; El-Matbouli, M. CD4: A Vital Player in the Teleost Fish Immune System. Vet. Res. 2019, 50, 1–11. [Google Scholar] [CrossRef]
- Luckheeram, R.V.; Zhou, R.; Verma, A.D.; Xia, B. CD4+ T Cells: Differentiation and Functions. Clin. Dev. Immunol. 2012, 2012, 925135. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Shao, J.Z.; Xiang, L.X.; Wen, Y. Cloning, Characterization and Expression Analysis of Pufferfish Interleukin-4 cDNA: The First Evidence of Th2-Type Cytokine in Fish. Mol. Immunol. 2007, 44, 2078–2086. [Google Scholar] [CrossRef]
- Yagi, R.; Zhu, J.; Paul, W.E. An Updated View on Transcription Factor GATA3-Mediated Regulation of Th1 and Th2 Cell Differentiation. Int. Immunol. 2011, 23, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of Effector CD4 T Cell Populations. Annu. Rev. Immunol. 2009, 28, 445–489. [Google Scholar] [CrossRef]




| Name | Sequence (5′-3′) | Accession Number | Amplicon Length (bp) |
|---|---|---|---|
| NOD2-F | TGGTAGGTAATGGTGTAGGGAATG | XM_020079852.1 | 138 |
| NOD2-R | CCAGGGCTTGAACCAGACTTT | ||
| TLR2-F | CATGGAAACAGAGTAGCTGGGATT | XM_020112938.1 | 151 |
| TLR2-R | TGTGGAGCAGGTTGAGACGC | ||
| MHC-II-F | CTATCACTATTGTGGGCTGCTTTG | XM_020093263.1 | 190 |
| MHC-II-R | TGCTCTGCTTTCTTGACACCTTT | ||
| ATG5-F | CCTCCACTGTCCGTCCAACT | XM_020093489.1 | 256 |
| ATG5-R | CGGTCTATCACTCATCGTCTGG | ||
| GABARAPL1-F | TGTGCTTCCTCATCCGTCAG | XM_020093139.1 | 126 |
| GABARAPL1-R | CCTCTTCATGGTGCTCCTCATA | ||
| TCR β-F | CCCCACTACATCTCAAGGTTTCC | XM_020105957.1 | 151 |
| TCR β-R | CAAAGTTTACACTGCTGCCCAC | ||
| CD4-1-F | CCAGTGGTCCCCACCTAAAA | XM_020093150.1 | 82 |
| CD4-1-R | CACTTCTGGGACGGTGAGATG | ||
| STAT4-F | CCAGCAAAGTCCATCCATACA | XM_020099666.1 | 151 |
| STAT4-R | TCGAAGCACAGATGCTCGTTT | ||
| RORα-F | CCTTACTGCTCCTTCACCAACG | XM_020079419.1 | 252 |
| RORα-R | GGCGAACTCCACCACATACTG | ||
| β-actin-F | GAGGGAAATCGTGCGTGACAT | AF135499.1 | 142 |
| β-actin-R | ATTGCCGATGGTGATGACCTG |
| Sample | Raw Data (bp) | Clean Data (bp) | Q20 (%) | Q30 (%) | GC (%) | Unique Mapped (%) | Total Mapped (%) |
|---|---|---|---|---|---|---|---|
| IV-0-1 | 7,885,244,100 | 7,787,759,502 | 97.16 | 92.38 | 47.89 | 89.53 | 91.34 |
| IV-0-2 | 6,677,778,000 | 6,600,187,229 | 97.55 | 93.22 | 47.92 | 90.05 | 91.83 |
| IV-0-3 | 7,790,959,800 | 7,684,573,272 | 97.45 | 93.05 | 47.90 | 89.62 | 91.43 |
| IV-6-1 | 8,263,775,700 | 8,155,916,011 | 97.35 | 92.88 | 47.61 | 89.33 | 91.15 |
| IV-6-2 | 8,468,135,100 | 8,371,786,696 | 97.65 | 93.46 | 47.64 | 89.64 | 91.53 |
| IV-6-3 | 7,501,817,700 | 7,405,558,922 | 97.45 | 93.10 | 47.60 | 89.40 | 91.19 |
| IV-12-1 | 9,283,881,900 | 9,172,219,940 | 97.47 | 93.13 | 47.93 | 89.96 | 91.83 |
| IV-12-2 | 7,436,369,400 | 7,352,695,274 | 97.36 | 92.87 | 47.75 | 89.56 | 91.41 |
| IV-12-3 | 8,982,513,000 | 8,889,939,324 | 97.55 | 93.19 | 47.79 | 89.85 | 91.7 |
| IV-24-1 | 10,557,306,600 | 10,459,182,090 | 97.38 | 92.91 | 48.01 | 90.01 | 91.89 |
| IV-24-2 | 8,221,355,100 | 8,141,427,164 | 97.56 | 93.38 | 47.61 | 90.33 | 92.12 |
| IV-24-3 | 7,957,429,800 | 7,864,449,269 | 97.20 | 92.46 | 47.98 | 90.16 | 91.97 |
| IV-48-1 | 6,749,397,000 | 6,677,417,329 | 97.46 | 93.12 | 47.70 | 90.04 | 91.82 |
| IV-48-2 | 8,024,550,900 | 7,930,624,583 | 97.22 | 92.59 | 47.40 | 89.59 | 91.36 |
| IV-48-3 | 7,079,832,300 | 7,002,516,806 | 97.28 | 92.62 | 47.41 | 89.94 | 91.66 |
| Module | Gene Number | Module | Gene Number | Module | Gene Number |
|---|---|---|---|---|---|
| Blue | 11,616 | Magenta | 359 | Royal blue | 238 |
| Brown | 2093 | Purple | 354 | Dark red | 237 |
| Red | 1520 | Green–yellow | 329 | Dark green | 230 |
| Green | 693 | Salmon | 300 | Dark grey | 194 |
| Dark turquoise | 506 | Cyan | 290 | Dark orange | 180 |
| Black | 498 | Grey60 | 260 | White | 178 |
| Pink | 385 | Light green | 259 | Saddle brown | 95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, X.; Chi, H.; Zhang, Z.; Li, Q.; Sheng, X.; Tang, X.; Xing, J.; Zhan, W. Transcriptome Analysis of Peritoneal Cells Reveals the Early Immune Response of Flounder (Paralichthys olivaceus) to Inactivated Vibrio anguillarum Immunization. Vaccines 2023, 11, 1603. https://doi.org/10.3390/vaccines11101603
Meng X, Chi H, Zhang Z, Li Q, Sheng X, Tang X, Xing J, Zhan W. Transcriptome Analysis of Peritoneal Cells Reveals the Early Immune Response of Flounder (Paralichthys olivaceus) to Inactivated Vibrio anguillarum Immunization. Vaccines. 2023; 11(10):1603. https://doi.org/10.3390/vaccines11101603
Chicago/Turabian StyleMeng, Xianghu, Heng Chi, Zuobing Zhang, Qian Li, Xiuzhen Sheng, Xiaoqian Tang, Jing Xing, and Wenbin Zhan. 2023. "Transcriptome Analysis of Peritoneal Cells Reveals the Early Immune Response of Flounder (Paralichthys olivaceus) to Inactivated Vibrio anguillarum Immunization" Vaccines 11, no. 10: 1603. https://doi.org/10.3390/vaccines11101603
APA StyleMeng, X., Chi, H., Zhang, Z., Li, Q., Sheng, X., Tang, X., Xing, J., & Zhan, W. (2023). Transcriptome Analysis of Peritoneal Cells Reveals the Early Immune Response of Flounder (Paralichthys olivaceus) to Inactivated Vibrio anguillarum Immunization. Vaccines, 11(10), 1603. https://doi.org/10.3390/vaccines11101603






