A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections
Abstract
1. Introduction
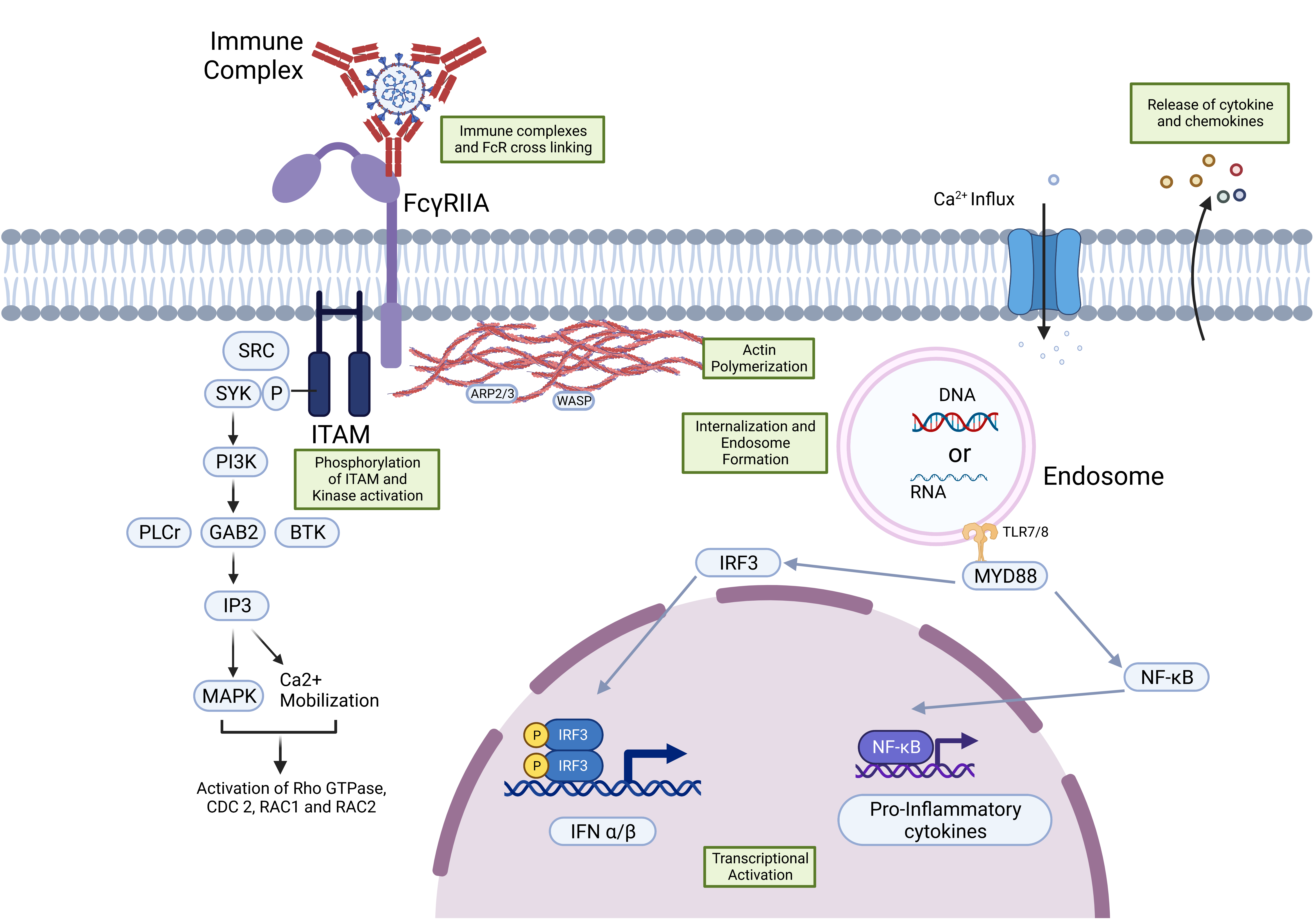
2. FcγR Signaling Pathways and ADE
FcR-Mediated ADE in SARS-CoV-2
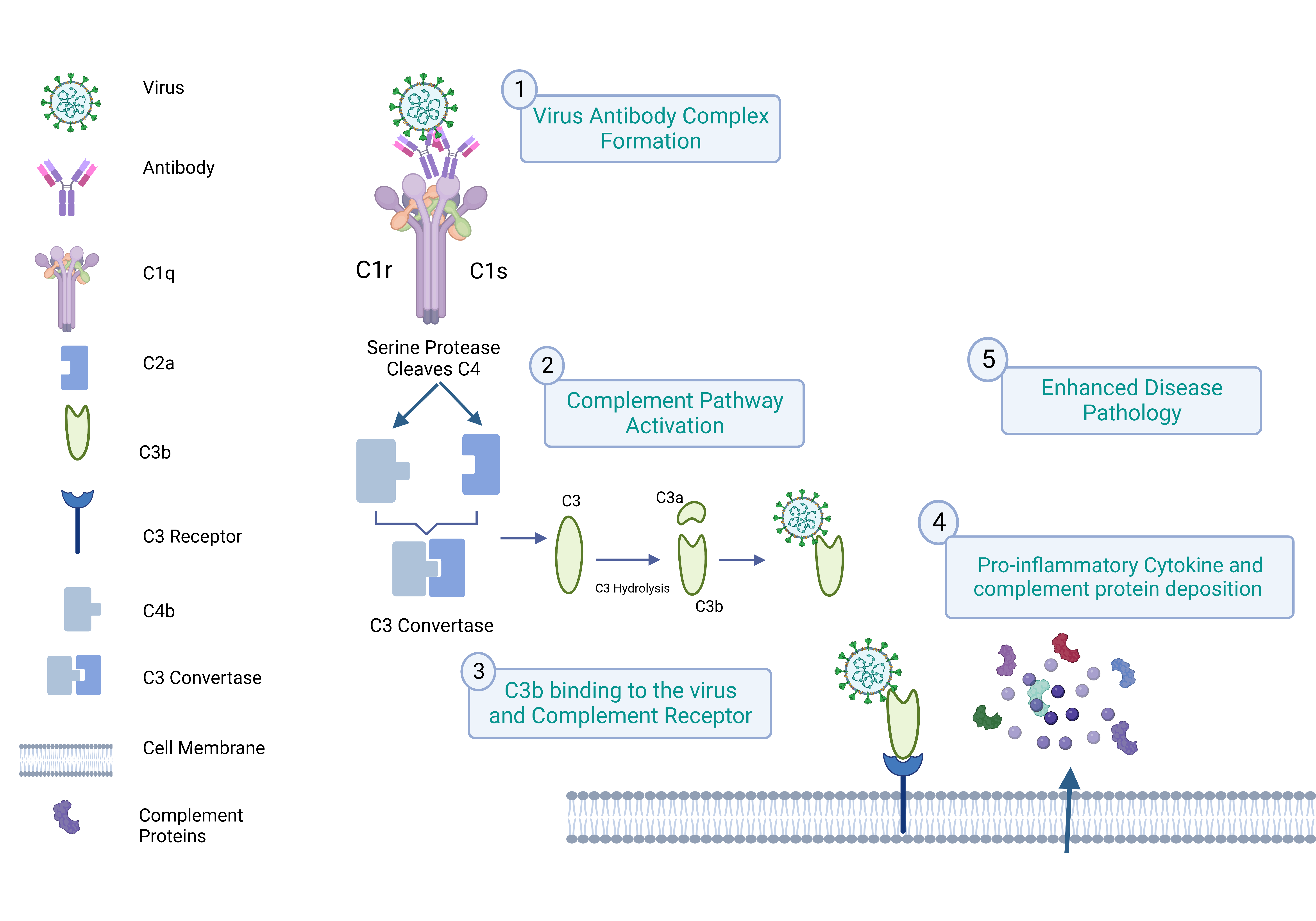
3. Complement and ADE
3.1. Virus Binding to the Complement Receptor
3.2. Virus Capsule and Cell Membrane Fusion Mechanism
C1q-Mediated ADE in SARS-CoV-2
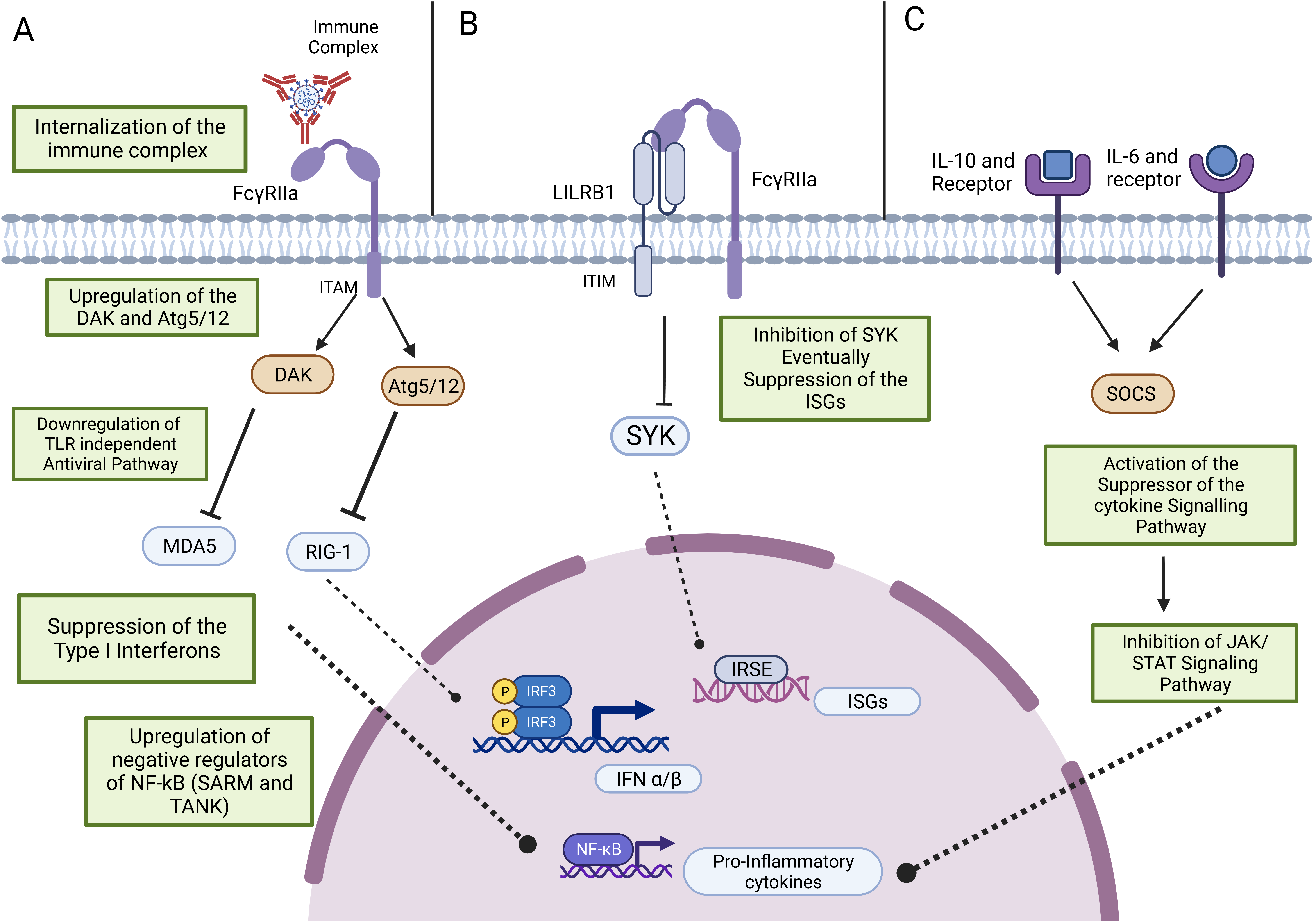
4. Antiviral Activity and ADE
4.1. Antiviral Response upon Virus Entry through Its Receptor
4.2. Antiviral Response and ADE
5. ADE Assays
| Virus Studied | Cell Line | Pseudovirus | Reporter System | Readout | References |
|---|---|---|---|---|---|
| DENV | K562 and Mylc cells. | Single-round infection particles (SRIPs). | Luciferase. | Luminometer. | [60] |
| Ebola | Human embryonated kidney (293) cells (HEK293) or Vero E6 | Pseudotyped vesicular stomatitis virus (VSV) and VSVΔG-EBOV GP. | GFP. | Fluorescent microscopy. | [11] |
| HIV | CD21-expressing cell line and T cell line naturally expressing complement receptor 2 (CR2; CD21). | HIV isolates and simian-human immunodeficiency viruses (SHIVs). | Intracellular staining for P24 expression. | Flow cytometry. | [62] |
| SARS-CoV-2 | Raji cells, K562 cells, primary B cells, and Vero E6. | Spike protein expressing pseudovirus and VSV pseudotyped with SARS CoV-2 S (VSV-SARS2). | Luciferase and GFP. | Luminometer, flow cytometry, and fluorescent microscopy. | [20] |
6. SARS-CoV-2 Vaccines and ADE Mechanism
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahangarzadeh, S.; Payandeh, Z.; Arezumand, R.; Shahzamani, K.; Yarian, F.; Alibakhshi, A. An update on antiviral antibody-based biopharmaceuticals. Int. Immunopharmacol. 2020, 86, 106760. [Google Scholar] [CrossRef] [PubMed]
- Sedova, E.S.; Scherbinin, D.N.; Lysenko, A.A.; Alekseeva, S.V.; Artemova, E.A.; Shmarov, M.M. Non-neutralizing Antibodies Directed at Conservative Influenza Antigens. Acta Nat. 2019, 11, 22–32. [Google Scholar] [CrossRef]
- Hawkes, R.A.; Lafferty, K.J. The enhancement of virus infectivity by antibody. Virology 1967, 33, 250–261. [Google Scholar] [CrossRef]
- Dejnirattisai, W.; Jumnainsong, A.; Onsirisakul, N.; Fitton, P.; Vasanawathana, S.; Limpitikul, W.; Puttikhunt, C.; Edwards, C.; Duangchinda, T.; Supasa, S.; et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010, 328, 745–748. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Bhattacharjee, S. Dengue virus: Epidemiology, biology, and disease aetiology. Can. J. Microbiol. 2021, 67, 687–702. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Pillat, M.M.; Tarnok, A. Dengue Fever, COVID-19 (SARS-CoV-2), and Antibody-Dependent Enhancement (ADE): A Perspective. Cytom. A 2020, 97, 662–667. [Google Scholar] [CrossRef]
- Wen, J.; Cheng, Y.; Ling, R.; Dai, Y.; Huang, B.; Huang, W.; Zhang, S.; Jiang, Y. Antibody-dependent enhancement of coronavirus. Int. J. Infect. Dis. 2020, 100, 483–489. [Google Scholar] [CrossRef]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef]
- Cancel-Tirado, S.M.; Evans, R.B.; Yoon, K.J. Monoclonal antibody analysis of porcine reproductive and respiratory syndrome virus epitopes associated with antibody-dependent enhancement and neutralization of virus infection. Vet. Immunol. Immunopathol. 2004, 102, 249–262. [Google Scholar] [CrossRef]
- Lidbury, B.A.; Mahalingam, S. Specific ablation of antiviral gene expression in macrophages by antibody-dependent enhancement of Ross River virus infection. J. Virol. 2000, 74, 8376–8381. [Google Scholar] [CrossRef]
- Takada, A.; Feldmann, H.; Ksiazek, T.G.; Kawaoka, Y. Antibody-dependent enhancement of Ebola virus infection. J. Virol. 2003, 77, 7539–7544. [Google Scholar] [CrossRef] [PubMed]
- Karthik, K.; Senthilkumar, T.M.A.; Udhayavel, S.; Raj, G.D. Role of antibody-dependent enhancement (ADE) in the virulence of SARS-CoV-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter COVID-19. Hum. Vaccines Immunother. 2020, 16, 3055–3060. [Google Scholar] [CrossRef]
- Garcia-Nicolas, O.; Ricklin, M.E.; Liniger, M.; Vielle, N.J.; Python, S.; Souque, P.; Charneau, P.; Summerfield, A. A Japanese Encephalitis Virus Vaccine Inducing Antibodies Strongly Enhancing In Vitro Infection Is Protective in Pigs. Viruses 2017, 9, 124. [Google Scholar] [CrossRef]
- Elfessi, Z.; Doyle, R.; Young, L.; Knaub, M.; Yamanaka, T. Antibody dependent enhancement-induced hypoxic respiratory failure: A case report. Vis. J. Emerg. Med. 2023, 30, 101602. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Smatti, M.K.; Ouhtit, A.; Cyprian, F.S.; Almaslamani, M.A.; Thani, A.A.; Yassine, H.M. Antibody-Dependent Enhancement (ADE) and the role of complement system in disease pathogenesis. Mol. Immunol. 2022, 152, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R. Antibody-Dependent Enhancement of Viral Infections. In Dynamics of Immune Activation in Viral Diseases; Springer Nature: Singapore, 2019; pp. 9–41. [Google Scholar]
- Furuyama, W.; Nanbo, A.; Maruyama, J.; Marzi, A.; Takada, A. A complement component C1q-mediated mechanism of antibody-dependent enhancement of Ebola virus infection. PLoS Negl. Trop. Dis. 2020, 14, e0008602. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.G.; Vazquez, S. The complexity of antibody-dependent enhancement of dengue virus infection. Viruses 2010, 2, 2649–2662. [Google Scholar] [CrossRef]
- Okuya, K.; Hattori, T.; Saito, T.; Takadate, Y.; Sasaki, M.; Furuyama, W.; Marzi, A.; Ohiro, Y.; Konno, S.; Hattori, T.; et al. Multiple Routes of Antibody-Dependent Enhancement of SARS-CoV-2 Infection. Microbiol. Spectr. 2022, 10, e0155321. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Yu, X.; Jiang, W.; Chen, S.; Wang, R.; Wang, M.; Jiao, S.; Yang, Y.; Wang, W.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 pseudoviral infection requires FcgammaRIIB and virus-antibody complex with bivalent interaction. Commun. Biol. 2022, 5, 262. [Google Scholar] [CrossRef]
- Fust, G. Enhancing antibodies in HIV infection. Parasitology 1997, 115, S127–S140. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Sun, S.; Tai, W.; Chen, J.; Geng, Q.; He, L.; Chen, Y.; Wu, J.; Shi, Z.; et al. Molecular Mechanism for Antibody-Dependent Enhancement of Coronavirus Entry. J. Virol. 2020, 94, e02015. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Foo, S.S.; Bruzzone, R.; Dinh, L.V.; King, N.J.; Mahalingam, S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015, 268, 340–364. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef]
- Mancardi, D.; Daëron, M. Fc Receptors in Immune Responses. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar] [CrossRef]
- Junker, F.; Gordon, J.; Qureshi, O. Fc Gamma Receptors and Their Role in Antigen Uptake, Presentation, and T Cell Activation. Front. Immunol. 2020, 11, 1393. [Google Scholar] [CrossRef] [PubMed]
- Bruhns, P.; Iannascoli, B.; England, P.; Mancardi, D.A.; Fernandez, N.; Jorieux, S.; Daeron, M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009, 113, 3716–3725. [Google Scholar] [CrossRef]
- Ben Mkaddem, S.; Benhamou, M.; Monteiro, R.C. Understanding Fc Receptor Involvement in Inflammatory Diseases: From Mechanisms to New Therapeutic Tools. Front. Immunol. 2019, 10, 811. [Google Scholar] [CrossRef]
- Getahun, A.; Cambier, J.C. Of ITIMs, ITAMs, and ITAMis: Revisiting immunoglobulin Fc receptor signaling. Immunol. Rev. 2015, 268, 66–73. [Google Scholar] [CrossRef]
- Halstead, S.B.; O’Rourke, E.J. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977, 146, 201–217. [Google Scholar] [CrossRef]
- Moi, M.L.; Lim, C.K.; Takasaki, T.; Kurane, I. Involvement of the Fc gamma receptor IIA cytoplasmic domain in antibody-dependent enhancement of dengue virus infection. J. Gen. Virol. 2010, 91, 103–111. [Google Scholar] [CrossRef]
- Ayala-Nunez, N.V.; Hoornweg, T.E.; van de Pol, D.P.; Sjollema, K.A.; Flipse, J.; van der Schaar, H.M.; Smit, J.M. How antibodies alter the cell entry pathway of dengue virus particles in macrophages. Sci. Rep. 2016, 6, 28768. [Google Scholar] [CrossRef]
- Kuzmina, N.A.; Younan, P.; Gilchuk, P.; Santos, R.I.; Flyak, A.I.; Ilinykh, P.A.; Huang, K.; Lubaki, N.M.; Ramanathan, P.; Crowe, J.E., Jr.; et al. Antibody-Dependent Enhancement of Ebola Virus Infection by Human Antibodies Isolated from Survivors. Cell Rep. 2018, 24, 1802–1815.e5. [Google Scholar] [CrossRef] [PubMed]
- Janoff, E.N.; Wahl, S.M.; Thomas, K.; Smith, P.D. Modulation of human immunodeficiency virus type 1 infection of human monocytes by IgA. J. Infect. Dis. 1995, 172, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Kozlowski, P.A.; Black, K.P.; Shen, L.; Jackson, S. High prevalence of serum IgA HIV-1 infection-enhancing antibodies in HIV-infected persons. Masking by IgG. J. Immunol. 1995, 154, 6163–6173. [Google Scholar] [CrossRef]
- Shi, P.; Zhang, L.; Wang, J.; Lu, D.; Li, Y.; Ren, J.; Shen, M.; Zhang, L.; Huang, J. Porcine FcεRI Mediates Porcine Reproductive and Respiratory Syndrome Virus Multiplication and Regulates the Inflammatory Reaction. Virol. Sin. 2018, 33, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The Dark Side of Antibody Dependent Enhancement During Dengue Infection. Front. Cell Infect. Microbiol. 2020, 10, 580096. [Google Scholar] [CrossRef]
- Ubol, S.; Phuklia, W.; Kalayanarooj, S.; Modhiran, N. Mechanisms of immune evasion induced by a complex of dengue virus and preexisting enhancing antibodies. J. Infect. Dis. 2010, 201, 923–935. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Cheung, C.Y.; Poon, L.L.; Ng, I.H.; Luk, W.; Sia, S.F.; Wu, M.H.; Chan, K.H.; Yuen, K.Y.; Gordon, S.; Guan, Y.; et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: Possible relevance to pathogenesis. J. Virol. 2005, 79, 7819–7826. [Google Scholar] [CrossRef]
- Li, L.; Wo, J.; Shao, J.; Zhu, H.; Wu, N.; Li, M.; Yao, H.; Hu, M.; Dennin, R.H. SARS-coronavirus replicates in mononuclear cells of peripheral blood (PBMCs) from SARS patients. J. Clin. Virol. 2003, 28, 239–244. [Google Scholar] [CrossRef]
- Maemura, T.; Kuroda, M.; Armbrust, T.; Yamayoshi, S.; Halfmann, P.J.; Kawaoka, Y. Antibody-Dependent Enhancement of SARS-CoV-2 Infection Is Mediated by the IgG Receptors FcgammaRIIA and FcgammaRIIIA but Does Not Contribute to Aberrant Cytokine Production by Macrophages. mBio 2021, 12, e0198721. [Google Scholar] [CrossRef]
- Hegazy, A.N.; Krönke, J.; Angermair, S.; Schwartz, S.; Weidinger, C.; Keller, U.; Treskatsch, S.; Siegmund, B.; Schneider, T. Anti-SARS-CoV2 antibody-mediated cytokine release syndrome in a patient with acute promyelocytic leukemia. BMC Infect. Dis. 2022, 22, 537. [Google Scholar] [CrossRef] [PubMed]
- Dustin, M.L. Complement Receptors in Myeloid Cell Adhesion and Phagocytosis. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.M.; Cabezas-Falcon, S.; Dubowsky, J.G.; Hulme-Jones, J.; Gordon, D.L. Dengue virus and the complement alternative pathway. FEBS Lett. 2020, 594, 2543–2555. [Google Scholar] [CrossRef]
- Smatti, M.K.; Al Thani, A.A.; Yassine, H.M. Viral-Induced Enhanced Disease Illness. Front. Microbiol. 2018, 9, 2991. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.E., Jr.; Montefiori, D.C.; Gillespie, D.H.; Mitchell, W.M. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J. Acquir. Immune Defic. Syndr. 1989, 2, 33–42. [Google Scholar]
- Von Kietzell, K.; Pozzuto, T.; Heilbronn, R.; Grossl, T.; Fechner, H.; Weger, S. Antibody-mediated enhancement of parvovirus B19 uptake into endothelial cells mediated by a receptor for complement factor C1q. J. Virol. 2014, 88, 8102–8115. [Google Scholar] [CrossRef]
- Naito, A.T.; Sumida, T.; Nomura, S.; Liu, M.L.; Higo, T.; Nakagawa, A.; Okada, K.; Sakai, T.; Hashimoto, A.; Hara, Y.; et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell 2012, 149, 1298–1313. [Google Scholar] [CrossRef]
- Henry, B.M.; Szergyuk, I.; de Oliveira, M.H.S.; Lippi, G.; Benoit, J.L.; Vikse, J.; Benoit, S.W. Complement levels at admission as a reflection of coronavirus disease 2019 (COVID-19) severity state. J. Med. Virol. 2021, 93, 5515–5522. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef]
- Cui, J.; Chen, Y.; Wang, H.Y.; Wang, R.F. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum. Vaccin. Immunother. 2014, 10, 3270–3285. [Google Scholar] [CrossRef]
- Flipse, J.; Diosa-Toro, M.A.; Hoornweg, T.E.; van de Pol, D.P.; Urcuqui-Inchima, S.; Smit, J.M. Antibody-Dependent Enhancement of Dengue Virus Infection in Primary Human Macrophages; Balancing Higher Fusion against Antiviral Responses. Sci. Rep. 2016, 6, 29201. [Google Scholar] [CrossRef]
- Chareonsirisuthigul, T.; Kalayanarooj, S.; Ubol, S. Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 2007, 88, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Khandia, R.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Malik, Y.S.; Singh, R.K.; Chaicumpa, W. Modulation of Dengue/Zika Virus Pathogenicity by Antibody-Dependent Enhancement and Strategies to Protect Against Enhancement in Zika Virus Infection. Front. Immunol. 2018, 9, 597. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.R.; Ong, E.Z.; Tan, H.C.; Zhang, S.L.; Zhang, Q.; Tang, K.F.; Kaliaperumal, N.; Lim, A.P.; Hibberd, M.L.; Chan, S.H.; et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc. Natl. Acad. Sci. USA 2014, 111, 2722–2727. [Google Scholar] [CrossRef] [PubMed]
- Modhiran, N.; Kalayanarooj, S.; Ubol, S. Subversion of innate defenses by the interplay between DENV and pre-existing enhancing antibodies: TLRs signaling collapse. PLoS Negl. Trop. Dis. 2010, 4, e924. [Google Scholar] [CrossRef] [PubMed]
- Hueston, L.; Ramirez, R.; Mahalingam, S. Enhancement of zika infection by dengue virus–specific antibody is associated with low levels of antiviral factors. J. Infect. Dis. 2017, 216, 612–614. [Google Scholar] [CrossRef]
- Mahalingam, S.; Lidbury, B.A. Suppression of lipopolysaccharide-induced antiviral transcription factor (STAT-1 and NF-kappa B) complexes by antibody-dependent enhancement of macrophage infection by Ross River virus. Proc. Natl. Acad. Sci. USA 2002, 99, 13819–13824. [Google Scholar] [CrossRef]
- Yamanaka, A.; Rattanaamnuaychai, P.; Matsuda, M.; Suzuki, R.; Shimizu, J.; Shioda, T.; Miyazaki, K. Development of a rapid assay system for detecting antibody-dependent enhancement of dengue virus infection. J. Virol. Methods 2023, 311, 114641. [Google Scholar] [CrossRef]
- Li, D.; Edwards, R.J.; Manne, K.; Martinez, D.R.; Schäfer, A.; Alam, S.M.; Wiehe, K.; Lu, X.; Parks, R.; Sutherland, L.L.; et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell 2021, 184, 4203–4219.e32. [Google Scholar] [CrossRef]
- Marasini, B.; Vyas, H.K.; Lakhashe, S.K.; Hariraju, D.; Akhtar, A.; Ratcliffe, S.J.; Ruprecht, R.M. Mucosal AIDS virus transmission is enhanced by antiviral IgG isolated early in infection. AIDS 2021, 35, 2423–2432. [Google Scholar] [CrossRef]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, F.; Li, Y.; Yan, L.; Liu, L.; Zhu, W.; Ma, P.; Shi, X.; Yang, G. Potent NTD-Targeting Neutralizing Antibodies against SARS-CoV-2 Selected from a Synthetic Immune System. Vaccines 2023, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin-Bussières, G.; Tauzin, A.; Dionne, K.; Gendron-Lepage, G.; Medjahed, H.; Perreault, J.; Levade, I.; Alfadhli, L.; Bo, Y.; Bazin, R.; et al. A Recent SARS-CoV-2 Infection Enhances Antibody-Dependent Cellular Cytotoxicity against Several Omicron Subvariants following a Fourth mRNA Vaccine Dose. Viruses 2023, 15, 1274. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, J.; Sasaki, T.; Koketsu, R.; Morita, R.; Yoshimura, Y.; Murakami, A.; Saito, Y.; Kusunoki, T.; Samune, Y.; Nakayama, E.E.; et al. Reevaluation of antibody-dependent enhancement of infection in anti-SARS-CoV-2 therapeutic antibodies and mRNA-vaccine antisera using FcR- and ACE2-positive cells. Sci. Rep. 2022, 12, 15612. [Google Scholar] [CrossRef]
- Wu, F.; Yan, R.; Liu, M.; Liu, Z.; Wang, Y.; Luan, D.; Wu, K.; Song, Z.; Sun, T.; Ma, Y.; et al. Antibody-dependent enhancement (ADE) of SARS-CoV-2 infection in recovered COVID-19 patients: Studies based on cellular and structural biology analysis. MedRxiv 2020. [Google Scholar] [CrossRef]
- Kan, A.K.C.; Li, P.H. Inactivated COVID-19 vaccines: Potential concerns of antibody-dependent enhancement and original antigenic sin. Immunol. Lett. 2023, 259, 21–23. [Google Scholar] [CrossRef]
- Sun, C.; Kong, D.; Guo, E.; Zhao, J.; Jia, J.; Wang, R.; Ma, J.; Chen, M.; Lu, J.; Yu, C.; et al. A Polysaccharide-RBD-Fc-Conjugated COVID-19 Vaccine, SCTV01A, Showed High Immunogenicity and Low Toxicity in Animal Models. Vaccines 2023, 11, 526. [Google Scholar] [CrossRef]
- Ikewaki, N.; Kurosawa, G.; Levy, G.A.; Preethy, S.; Abraham, S.J.K. Antibody dependent disease enhancement (ADE) after COVID-19 vaccination and beta glucans as a safer strategy in management. Vaccine 2023, 41, 2427–2429. [Google Scholar] [CrossRef]
- House, R.V.; Broge, T.A.; Suscovich, T.J.; Snow, D.M.; Tomic, M.T.; Nonet, G.; Bajwa, K.; Zhu, G.; Martinez, Z.; Hackett, K.; et al. Evaluation of strategies to modify Anti-SARS-CoV-2 monoclonal antibodies for optimal functionality as therapeutics. PLoS ONE 2022, 17, e0267796. [Google Scholar] [CrossRef] [PubMed]
- Dippel, A.; Gallegos, A.; Aleti, V.; Barnes, A.; Chen, X.; Christian, E.; Delmar, J.; Du, Q.; Esfandiary, R.; Farmer, E.; et al. Developability profiling of a panel of Fc engineered SARS-CoV-2 neutralizing antibodies. mAbs 2023, 15, 2152526. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, O.; Nechipurenko, Y.; Lagutkin, D.; Yegorov, Y.E.; Kzhyshkowska, J. SARS-CoV-2 infection of phagocytic immune cells and COVID-19 pathology: Antibody-dependent as well as independent cell entry. Front. Immunol. 2022, 13, 1050478. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Yang, M.; Lai, H.; Neupane, B.; Teh, A.Y.; Jugler, C.; Ma, J.K.; Steinkellner, H.; Bai, F.; Chen, Q. A Dual-Approach Strategy to Optimize the Safety and Efficacy of Anti-Zika Virus Monoclonal Antibody Therapeutics. Viruses 2023, 15, 1156. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Li, R.; Liu, S. Immunoregulation with mTOR inhibitors to prevent COVID-19 severity: A novel intervention strategy beyond vaccines and specific antiviral medicines. J. Med. Virol. 2020, 92, 1495–1500. [Google Scholar] [CrossRef]
- Karam, B.S.; Morris, R.S.; Bramante, C.T.; Puskarich, M.; Zolfaghari, E.J.; Lotfi-Emran, S.; Ingraham, N.E.; Charles, A.; Odde, D.J.; Tignanelli, C.J. mTOR inhibition in COVID-19: A commentary and review of efficacy in RNA viruses. J. Med. Virol. 2021, 93, 1843–1846. [Google Scholar] [CrossRef]
| FcγRI | FcγRIIa | FcγRIIb | FcγRIIc | FcγRllla | FcRIIIb | |
|---|---|---|---|---|---|---|
| (CD64) | (CD32a) | (CD32b) | (CD32c) | (CD16a) | (CD16b) | |
| Expressed by human immune cells | Macrophages, Eosinophils, Neutrophils, and Dendritic cells. | Platelets, Macrophages, Neutrophils, and Eosinophils. | Platelets, Dendritic cells, B cells, Mast cells, Neutrophils, and Eosinophils. | Natural Killer cells depend on allele status. | Natural Killer cells, Macrophages, Neutrophils, and Eosinophils. | Macrophages, Neutrophils, and Follicular dendritic cells. |
| Binding specificity | IgG1, IgG3, and IgG4. | IgG. | IgG. | IgG. | IgG1 and IgG3. | IgG1 and IgG3. |
| Functions | Phagocytosis, respiratory burst activation, and cell activation. | Degranulation, Phagocytosis, and ROI production. | Inhibits the phagocytosis and release of the pro-inflammatory cytokines, Degranulation, platelet activation, and B cell activation. | Phagocytosis and clearing of immune complexes. | Antibody-dependent cell-mediated cytotoxicity (ADCC) initiation, and cytokine release. | Phagocytosis, cytokine, and chemokine release. |
| ADE participation of viruses | DENV, Ebola virus, PRRSV, and JEV. | DENV, FIPV, MERS-CoV, SARS-CoV, and WNV SARS-CoV-2. | SARS-CoV-2, DENV, WNV, and PRRSV. | Unknown. | DENV, Ebola virus, HIV, PRRSV, and JEV. | Unknown. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawant, J.; Patil, A.; Kurle, S. A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections. Vaccines 2023, 11, 1240. https://doi.org/10.3390/vaccines11071240
Sawant J, Patil A, Kurle S. A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections. Vaccines. 2023; 11(7):1240. https://doi.org/10.3390/vaccines11071240
Chicago/Turabian StyleSawant, Jyoti, Ajit Patil, and Swarali Kurle. 2023. "A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections" Vaccines 11, no. 7: 1240. https://doi.org/10.3390/vaccines11071240
APA StyleSawant, J., Patil, A., & Kurle, S. (2023). A Review: Understanding Molecular Mechanisms of Antibody-Dependent Enhancement in Viral Infections. Vaccines, 11(7), 1240. https://doi.org/10.3390/vaccines11071240







