Combinatorial Effects of miRNAs in HSV-2 Infection of Macrophages: An In Silico and In Vitro Integration Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell and Virus Culture
2.2. miRNA Microarray
2.3. TaqMan-Based Quantitative PCR for miRNA Expression Validation
2.4. SYBR Green-Based Quantitative PCR for Viral Gene Expression
2.5. miRNA Target Prediction
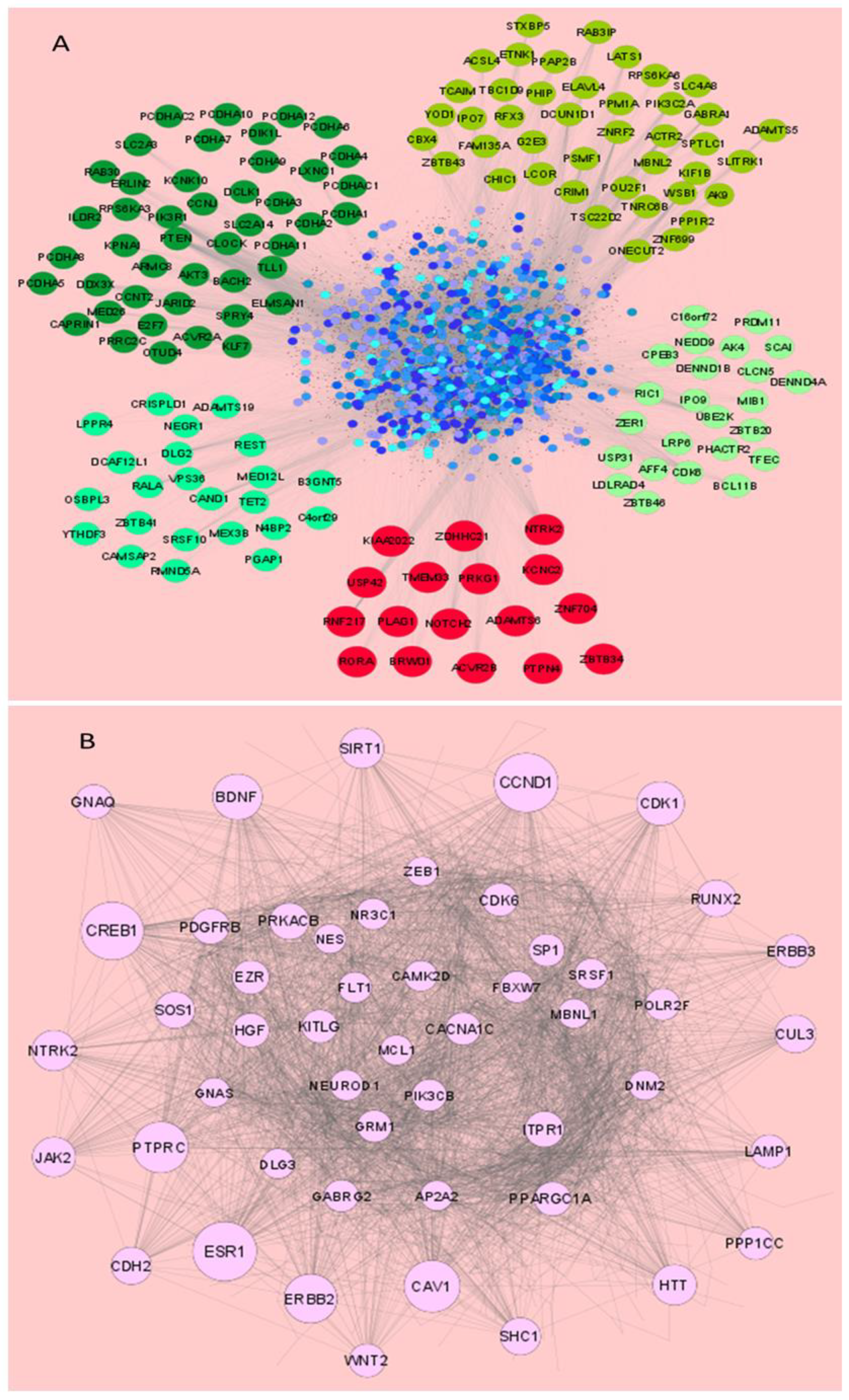
2.6. Construction of miRNA Target Interaction Network
2.7. Screening of Common Interacting Targets and Network Biological Parameters
2.8. Functional Annotation of miRNA Targets
2.9. Cloning and Transfection
2.10. Luciferase Reporter Assay
2.11. Immunoblotting
2.12. Plaque Reduction Assay
2.13. Box–Behnken Design (BBD) Based Design of Experimental Model
2.14. Optimization and Validation of the Applied Model
2.15. Cell Viability Assay
2.16. Statistical Analyses of In Vitro Analyses
3. Results
3.1. miRNAs of the Human Inflammatory Response Pathway Are Differentially Expressed in the HSV-2 Infected Macrophages
3.2. miR-195, miR-374a, miR-29b and miR-181a Are Validated to Be Reciprocally Expressed to miR-211 upon HSV-2 Infection in Macrophages
3.3. miRNAs Dysregulated in HSV-2 Infection Target a Vast Array of Signaling Intermediates That Participate in Several Crucial Cellular Pathways
3.4. miRNAs Modulated by HSV-2 Infection Target the Components of the PI3K/AKT/mTOR Signaling Cascade
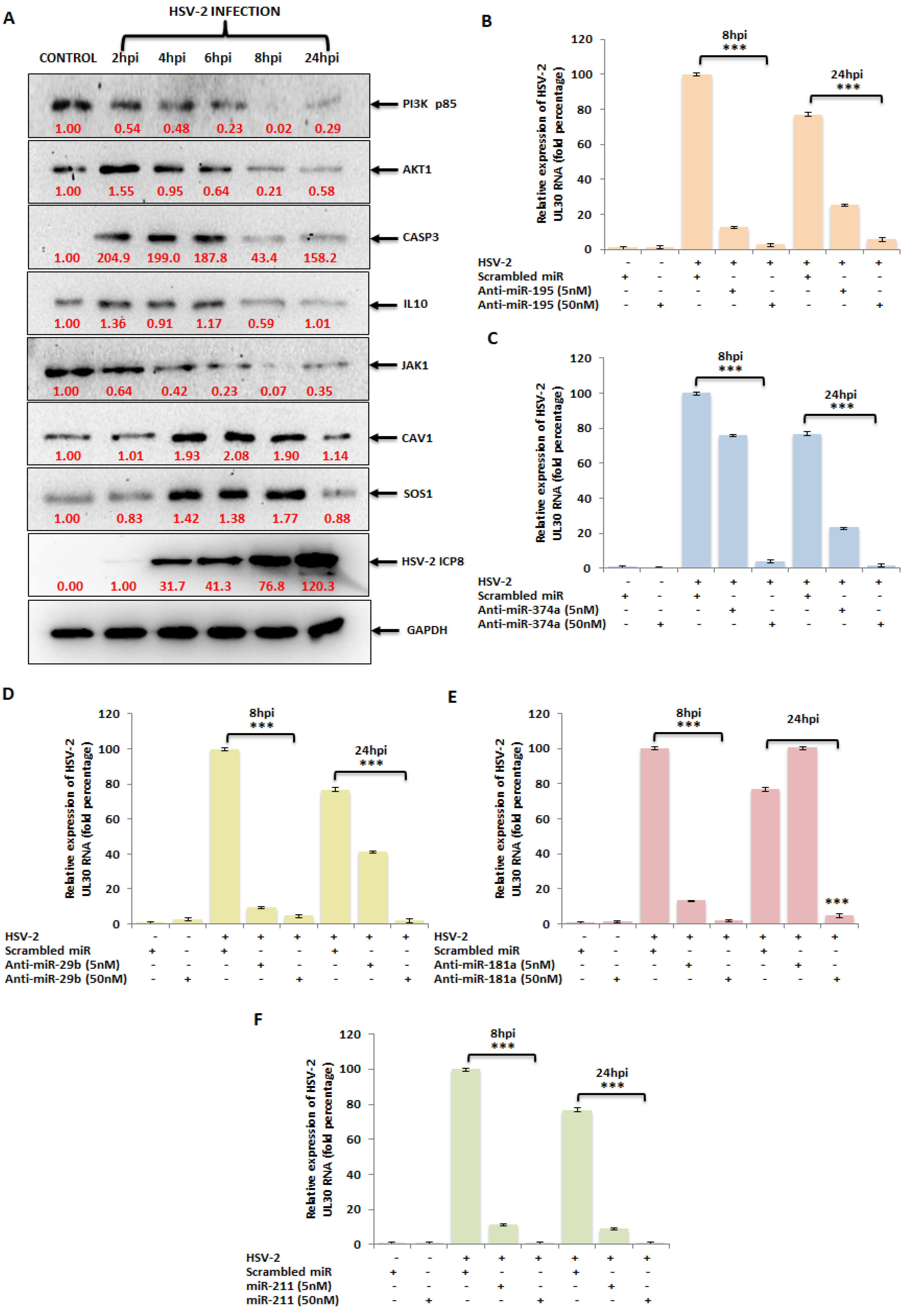
3.5. The miRNA Targets in the PI3K/AKT/mTOR Pathway Are Dysregulated upon HSV-2 Infection of the Macrophages
3.6. Ectopic Expression of HSV-2-Dysregulated miRNAs Suppress the Expression of the HSV-2 DNA Polymerase Catalytic Subunit Gene
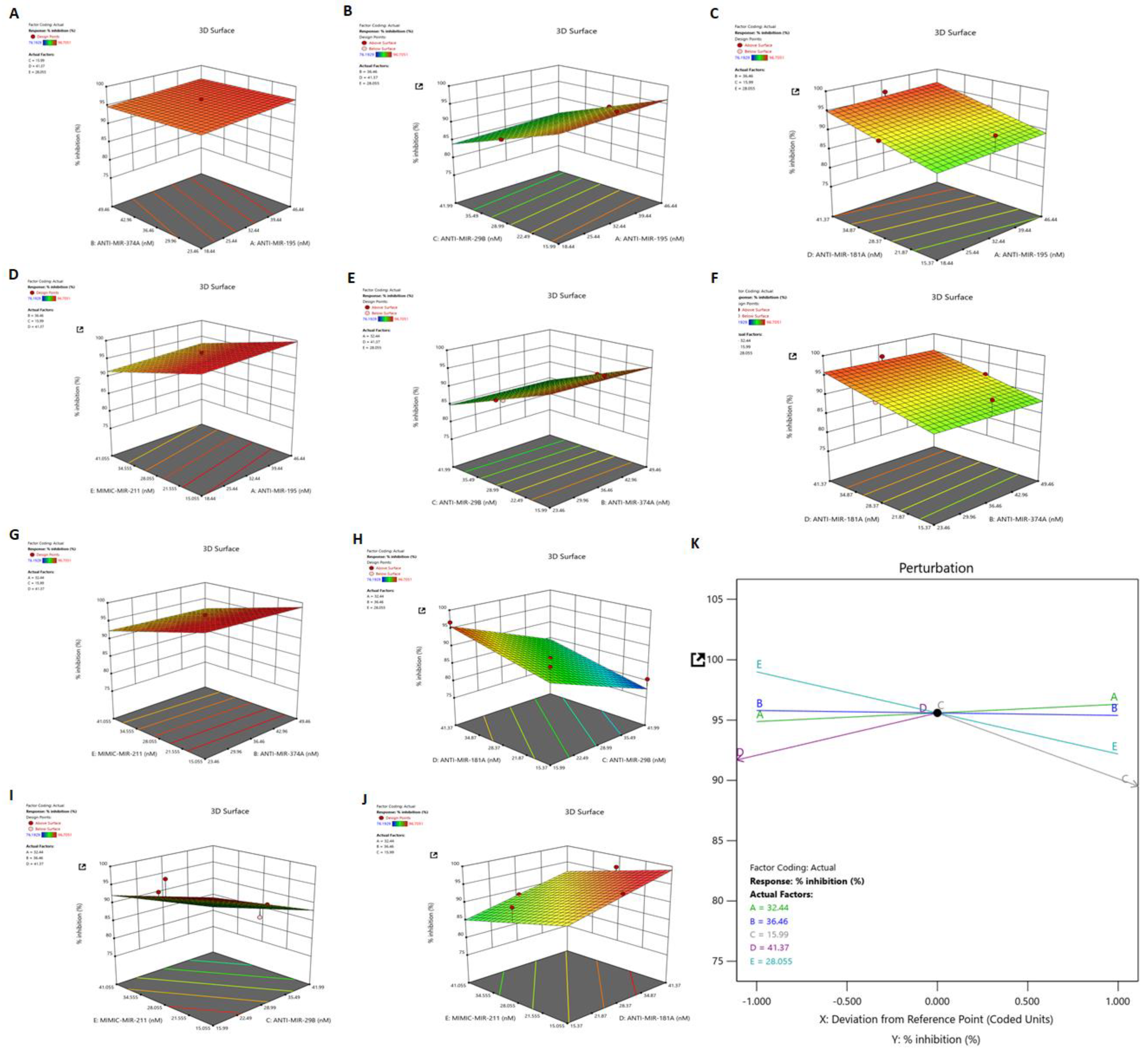
3.7. Identification of Synergistic Combination of Five Dysregulated miRNAs on HSV-2 Infection: Box–Behnken Method
3.8. Dependant Variable: R1 (Response 1)—Percent Inhibition
3.9. Validation of the Applied Model and Identification of Synergistic Combination of Five Dysregulated miRNAs on HSV-2 Infection
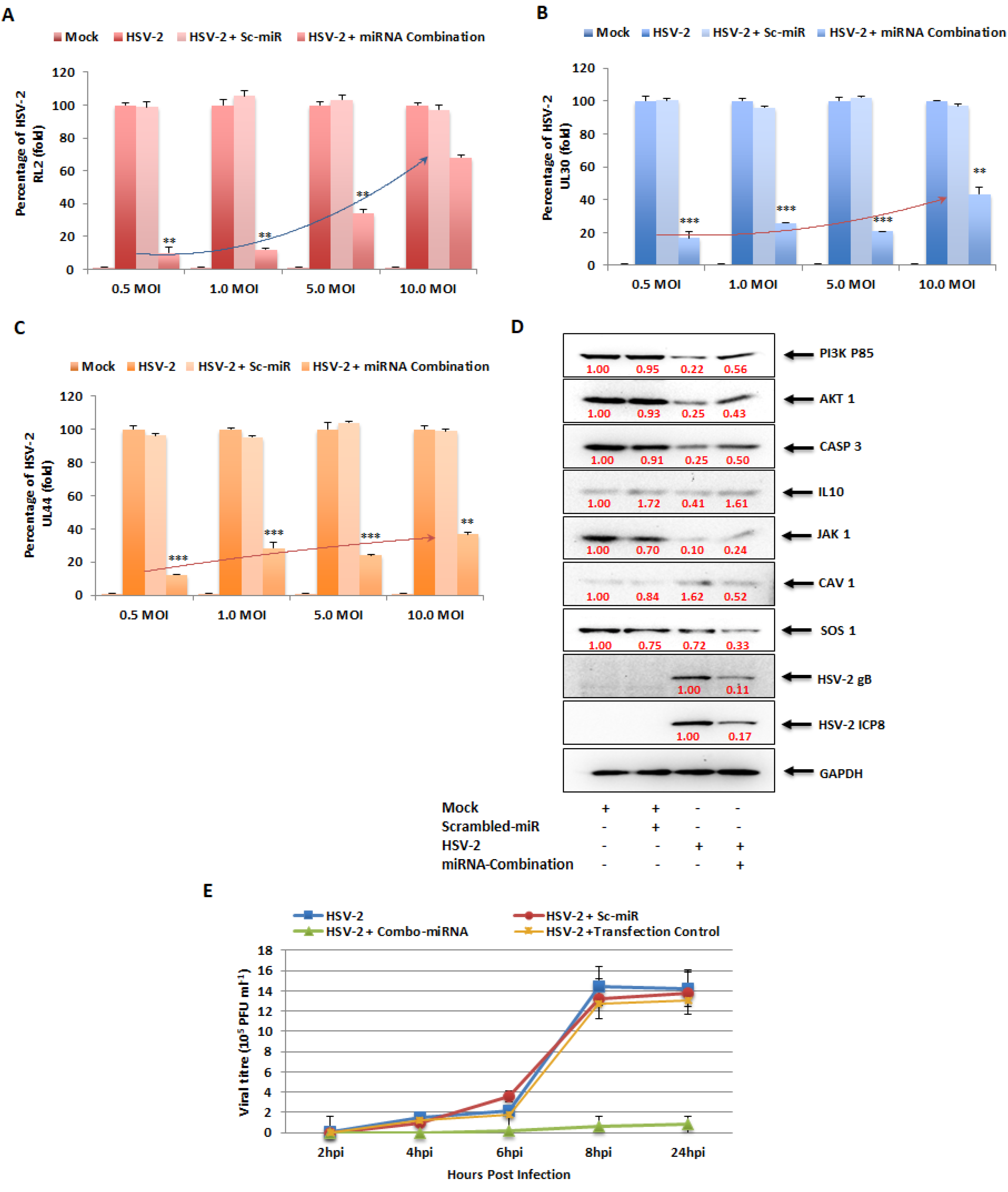
3.10. In Vitro Combinatorial Effect of miRNAs on Attenuation of HSV-2 Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhang, M.; Liu, Y.; Wang, P.; Guan, X.; He, S.; Luo, S.; Li, C.; Hu, K.; Jin, W.; Du, T.; et al. HSV-2 Immediate-Early Protein US1 Inhibits IFN-β Production by Suppressing Association of IRF-3 with IFN-β Promoter. J. Immunol. 2015, 194, 3102–3115. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, E.; Tharinger, H.; Frank, I.; Arthos, J.; Piatak, M.; Lifson, J.D.; Blanchard, J.; Gettie, A.; Robbiani, M. HSV-2 Infection of Dendritic Cells Amplifies a Highly Susceptible HIV-1 Cell Target. PLoS Pathog. 2011, 7, e1002109. [Google Scholar] [CrossRef] [PubMed]
- Le Sage, V.; Jung, M.; Alter, J.D.; Wills, E.G.; Johnston, S.M.; Kawaguchi, Y.; Baines, J.D.; Banfield, B.W. The Herpes Simplex Virus 2 UL21 Protein Is Essential for Virus Propagation. J. Virol. 2013, 87, 5904–5915. [Google Scholar] [CrossRef] [PubMed]
- James, C.; Harfouche, M.; Welton, N.J.; Turner, K.M.; Abu-Raddad, L.J.; Gottlieb, S.L.; Looker, K.J. Herpes Simplex Virus: Global Infection Prevalence and Incidence Estimates, 2016. Bull. World Health Organ. 2020, 98, 315–329. [Google Scholar] [CrossRef]
- Banerjee, A.; Mukherjee, A. Herpesviridae and MicroRNAs. In Current Perspectives on Viral Disease Outbreaks—Epidemiology, Detection and Control; Claborn, D., Ed.; IntechOpen: London, UK, 2022; ISBN 978-1-83881-910-1. [Google Scholar]
- Schneider, J.A.; Lakshmi, V.; Dandona, R.; Kumar, G.A.; Sudha, T.; Dandona, L. Population-Based Seroprevalence of HSV-2 and Syphilis in Andhra Pradesh State of India. BMC Infect. Dis. 2010, 10, 59. [Google Scholar] [CrossRef]
- Whitley, R.; Baines, J. Clinical Management of Herpes Simplex Virus Infections: Past, Present, and Future. F1000Res 2018, 7, F1000. [Google Scholar] [CrossRef]
- Hanna, J.; Hossain, G.S.; Kocerha, J. The Potential for MicroRNA Therapeutics and Clinical Research. Front. Genet. 2019, 10, 478. [Google Scholar] [CrossRef]
- Huleihel, M.; Ishanu, V.; Tal, J.; Arad, S. (Malis) Antiviral Effect of Red Microalgal Polysaccharides on Herpes Simplex and Varicella Zoster Viruses. J. Appl. Phycol. 2001, 13, 127–134. [Google Scholar] [CrossRef]
- Wang, X. MiRDB: A MicroRNA Target Prediction and Functional Annotation Database with a Wiki Interface. RNA 2008, 14, 1012–1017. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Ferreira, S.L.C.; Bruns, R.E.; Ferreira, H.S.; Matos, G.D.; David, J.M.; Brandão, G.C.; da Silva, E.G.P.; Portugal, L.A.; dos Reis, P.S.; Souza, A.S.; et al. Box-Behnken Design: An Alternative for the Optimization of Analytical Methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, J.; He, W.; Zhao, P.; Ye, C. MicroRNA-433 Targets AKT3 and Inhibits Cell Proliferation and Viability in Breast Cancer. Oncol. Lett. 2018, 15, 3998. [Google Scholar] [CrossRef] [PubMed]
- Elkady, M.A.; Doghish, A.S.; Elshafei, A.; Elshafey, M.M. MicroRNA-567 Inhibits Cell Proliferation and Induces Cell Apoptosis in A549 NSCLC Cells by Regulating Cyclin-Dependent Kinase 8. Saudi J. Biol. Sci. 2021, 28, 2581–2590. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gao, S.; Gong, Y.; Lin, T.; Tong, S.; Xiong, W.; Shi, C.; Wang, W.; Fang, J. Anti-HSV-1 Effect of Dihydromyricetin from Ampelopsis Grossedentata via the TLR9-Dependent Anti-Inflammatory Pathway. J. Glob. Antimicrob. Resist. 2020, 23, 370–376. [Google Scholar] [CrossRef]
- Krzyzowska, M.; Baska, P.; Grochowska, A.; Orlowski, P.; Nowak, Z.; Winnicka, A. Fas/FasL Pathway Participates in Resolution of Mucosal Inflammatory Response Early during HSV-2 Infection. Immunobiology 2014, 219, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, P.; Tomaszewska, E.; Gniadek, M.; Baska, P.; Nowakowska, J.; Sokolowska, J.; Nowak, Z.; Donten, M.; Celichowski, G.; Grobelny, J.; et al. Tannic Acid Modified Silver Nanoparticles Show Antiviral Activity in Herpes Simplex Virus Type 2 Infection. PLoS ONE 2014, 9, e104113. [Google Scholar] [CrossRef]
- Ferreira, V.H.; Nazli, A.; Dizzell, S.E.; Mueller, K.; Kaushic, C. The Anti-Inflammatory Activity of Curcumin Protects the Genital Mucosal Epithelial Barrier from Disruption and Blocks Replication of HIV-1 and HSV-2. PLoS ONE 2015, 10, e0124903. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 Cell Line: An in Vitro Cell Model for Immune Modulation Approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef]
- Wu, Z.; Chen, J.; Zhao, W.; Zhuo, C.-H.; Chen, Q. Inhibition of MiR-181a Attenuates Sepsis-Induced Inflammation and Apoptosis by Activating Nrf2 and Inhibiting NF-ΚB Pathways via Targeting SIRT1. Kaohsiung J. Med. Sci. 2021, 37, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yi, N.; Chen, R.; Meng, Y.; Wang, Y.; Liu, H.; Cao, W.; Hu, Y.; Gu, Y.; Tong, C.; et al. MiR-29b-3p Protects Cardiomyocytes against Endotoxin-Induced Apoptosis and Inflammatory Response through Targeting FOXO3A. Cell. Signal. 2020, 74, 109716. [Google Scholar] [CrossRef] [PubMed]
- Vitali, D.; Bagri, P.; Wessels, J.M.; Arora, M.; Ganugula, R.; Parikh, A.; Mandur, T.; Felker, A.; Garg, S.; Kumar, M.N.V.R.; et al. Curcumin Can Decrease Tissue Inflammation and the Severity of HSV-2 Infection in the Female Reproductive Mucosa. Int. J. Mol. Sci. 2020, 21, 337. [Google Scholar] [CrossRef] [PubMed]
- Cokarić Brdovčak, M.; Zubković, A.; Jurak, I. Herpes Simplex Virus 1 Deregulation of Host MicroRNAs. Non-Coding RNA 2018, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Li, H.; Wang, H. MicroRNA-203 Inhibits Epithelial-Mesenchymal Transition, Migration, and Invasion of Renal Cell Carcinoma Cells via the Inactivation of the PI3K/AKT Signaling Pathway by Inhibiting CAV1. Cell Adhes. Migr. 2020, 14, 227–241. [Google Scholar] [CrossRef]
- Xie, Y.; He, S.; Wang, J. MicroRNA-373 Facilitates HSV-1 Replication through Suppression of Type I IFN Response by Targeting IRF1. Biomed. Pharmacother. 2018, 97, 1409–1416. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, J.; Tang, J.; Zhou, L.; Zhou, M. MicroRNA-649 Promotes HSV-1 Replication by Directly Targeting MALT1. J. Med. Virol. 2017, 89, 1069–1079. [Google Scholar] [CrossRef]
- Zheng, S.; Li, Y.; Zhang, Y.; Li, X.; Tang, H. MiR-101 Regulates HSV-1 Replication by Targeting ATP5B. Antivir. Res. 2011, 89, 219–226. [Google Scholar] [CrossRef]
- Dass, D.; Dhotre, K.; Chakraborty, M.; Nath, A.; Banerjee, A.; Bagchi, P.; Mukherjee, A. MiRNAs in Herpesvirus Infection: Powerful Regulators in Small Packages. Viruses 2023, 15, 429. [Google Scholar] [CrossRef]
- Frappier, L. Regulation of Herpesvirus Reactivation by Host MicroRNAs. J. Virol. 2015, 89, 2456–2458. [Google Scholar] [CrossRef]
- Piedade, D.; Azevedo-Pereira, J.M. The Role of MicroRNAs in the Pathogenesis of Herpesvirus Infection. Viruses 2016, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Deng, Y.; Chen, H.; Lin, Y.; Yang, X.; Sun, B.; Pan, D. Neuronal MiR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1. J. Virol. 2022, 96, e00349-22. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. MiRDB: An Online Database for Prediction of Functional MicroRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Vara, J.Á.F.; Casado, E.; de Castro, J.; Cejas, P.; Belda-Iniesta, C.; González-Barón, M. PI3K/Akt Signalling Pathway and Cancer. Cancer Treat. Rev. 2004, 30, 193–204. [Google Scholar] [CrossRef]
- Kirsch, J.M.; Mlera, L.; Offerdahl, D.K.; VanSickle, M.; Bloom, M.E. Tick-Borne Flaviviruses Depress AKT Activity during Acute Infection by Modulating AKT1/2. Viruses 2020, 12, 1059. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.-D.; Xun, Y.; Lu, J.-L.; Lu, Y.-C.; Yang, Y.-Y.; Zhou, P.; Hu, J.; Li, C.; Wang, S.-G. Network Pharmacology and Molecular Docking Analyses on Lianhua Qingwen Capsule Indicate Akt1 Is a Potential Target to Treat and Prevent COVID-19. Cell Prolif. 2020, 53, e12949. [Google Scholar] [CrossRef]
- Kuroda, M.; Halfmann, P.; Kawaoka, Y. HER2-Mediated Enhancement of Ebola Virus Entry. PLoS Pathog. 2020, 16, e1008900. [Google Scholar] [CrossRef]
- Icyuz, M.; Bryant, S.M.J.; Fortinberry, H.K.; Molakandov, K.; Siegal, G.P.; Contreras, J.L.; Wu, H. Adenovirus Infection Activates Akt1 and Induces Cell Proliferation in Pancreatic Islets. Transplantation 2009, 87, 821–824. [Google Scholar] [CrossRef]
- Vergadi, E.; Ieronymaki, E.; Lyroni, K.; Vaporidi, K.; Tsatsanis, C. Akt Signaling Pathway in Macrophage Activation and M1/M2 Polarization. J. Immunol. 2017, 198, 1006–1014. [Google Scholar] [CrossRef]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt Pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef]
- Linton, M.F.; Moslehi, J.J.; Babaev, V.R. Akt Signaling in Macrophage Polarization, Survival, and Atherosclerosis. Int. J. Mol. Sci. 2019, 20, 2703. [Google Scholar] [CrossRef]
- Zhou, J.-H.; Broussard, S.R.; Strle, K.; Freund, G.G.; Johnson, R.W.; Dantzer, R.; Kelley, K.W. IL-10 Inhibits Apoptosis of Promyeloid Cells by Activating Insulin Receptor Substrate-2 and Phosphatidylinositol 3′-Kinase1. J. Immunol. 2001, 167, 4436–4442. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.P.; Brown, J.R.; Sag, D.; Zhang, L.; Suttles, J. Adenosine 5′-Monophosphate-Activated Protein Kinase Regulates IL-10-Mediated Anti-Inflammatory Signaling Pathways in Macrophages. J. Immunol. 2015, 194, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Franke, T.F.; Hornik, C.P.; Segev, L.; Shostak, G.A.; Sugimoto, C. PI3K/Akt and Apoptosis: Size Matters. Oncogene 2003, 22, 8983–8998. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-Q.; Chong, Z.Z.; Maiese, K. Critical Role for Akt1 in the Modulation of Apoptotic Phosphatidylserine Exposure and Microglial Activation. Mol. Pharmacol. 2003, 64, 557–569. [Google Scholar] [CrossRef]
- Chong, Z.Z.; Kang, J.-Q.; Maiese, K. AKT1 Drives Endothelial Cell Membrane Asymmetry and Microglial Activation through Bcl-XL and Caspase 1, 3, and 9. Exp. Cell Res. 2004, 296, 196–207. [Google Scholar] [CrossRef]
- Wu, Y.; Ma, J.; Sun, Y.; Tang, M.; Kong, L. Effect and Mechanism of PI3K/AKT/MTOR Signaling Pathway in the Apoptosis of GC-1 Cells Induced by Nickel Nanoparticles. Chemosphere 2020, 255, 126913. [Google Scholar] [CrossRef]
- Xue, W.; Wang, J.; Jiang, W.; Shi, C.; Wang, X.; Huang, Y.; Hu, C. Caveolin-1 Alleviates Lipid Accumulation in NAFLD Associated with Promoting Autophagy by Inhibiting the Akt/MTOR Pathway. Eur. J. Pharmacol. 2020, 871, 172910. [Google Scholar] [CrossRef]
- Rodriguez-Viciana, P.; Warne, P.H.; Dhand, R.; Vanhaesebroeck, B.; Gout, I.; Fry, M.J.; Waterfield, M.D.; Downward, J. Phosphatidylinositol-3-OH Kinase Direct Target of Ras. Nature 1994, 370, 527–532. [Google Scholar] [CrossRef]
- Mattox, T.E.; Chen, X.; Maxuitenko, Y.Y.; Keeton, A.B.; Piazza, G.A. Exploiting RAS Nucleotide Cycling as a Strategy for Drugging RAS-Driven Cancers. Int. J. Mol. Sci. 2019, 21, 141. [Google Scholar] [CrossRef]
- Su, A.-R.; Qiu, M.; Li, Y.-L.; Xu, W.-T.; Song, S.-W.; Wang, X.-H.; Song, H.-Y.; Zheng, N.; Wu, Z.-W. BX-795 Inhibits HSV-1 and HSV-2 Replication by Blocking the JNK/P38 Pathways without Interfering with PDK1 Activity in Host Cells. Acta Pharmacol. Sin. 2017, 38, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, C.; Hao, C.; Zhang, Y.; Wang, Z.; Wang, S.; Wang, W. Inhibition of Herpes Simplex Virus by Myricetin through Targeting Viral GD Protein and Cellular EGFR/PI3K/Akt Pathway. Antivir. Res. 2020, 177, 104714. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Liu, T.; Wang, W.; Duan, Q.; Liu, T.; Xu, L.; Huang, G.; Chen, Z. The Chinese Herbal Prescription JZ-1 Induces Autophagy to Protect against Herpes Simplex Virus-2 in Human Vaginal Epithelial Cells by Inhibiting the PI3K/Akt/MTOR Pathway. J. Ethnopharmacol. 2020, 254, 112611. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cohen, J.I. The Role of PI3K/Akt in Human Herpesvirus Infection: From the Bench to the Bedside. Virology 2015, 479–480, 568–577. [Google Scholar] [CrossRef]
- Siracusano, G.; Venuti, A.; Lombardo, D.; Mastino, A.; Esclatine, A.; Sciortino, M.T. Early Activation of MyD88-Mediated Autophagy Sustains HSV-1 Replication in Human Monocytic THP-1 Cells. Sci. Rep. 2016, 6, 31302. [Google Scholar] [CrossRef]
- Smith, C.C.; Luo, J.H.; Hunter, J.C.R.; Ordonez, J.V.; Aurelian, L. The Transmembrane Domain of the Large Subunit of HSV-2 Ribonucleotide Reductase (ICP10) Is Required for Protein Kinase Activity and Transformation-Related Signaling Pathways That Result in Ras Activation. Virology 1994, 200, 598–612. [Google Scholar] [CrossRef]
- Hussein, H.A.M.; Akula, S.M. MiRNA-36 Inhibits KSHV, EBV, HSV-2 Infection of Cells via Stifling Expression of Interferon Induced Transmembrane Protein 1 (IFITM1). Sci. Rep. 2017, 7, 17972. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banerjee, A.; Dass, D.; Dhotre, K.; Wakchoure, P.; More, A.; Rana, S.; Khan, A.A.; Mukherjee, A. Combinatorial Effects of miRNAs in HSV-2 Infection of Macrophages: An In Silico and In Vitro Integration Approach. Vaccines 2023, 11, 1488. https://doi.org/10.3390/vaccines11091488
Banerjee A, Dass D, Dhotre K, Wakchoure P, More A, Rana S, Khan AA, Mukherjee A. Combinatorial Effects of miRNAs in HSV-2 Infection of Macrophages: An In Silico and In Vitro Integration Approach. Vaccines. 2023; 11(9):1488. https://doi.org/10.3390/vaccines11091488
Chicago/Turabian StyleBanerjee, Anwesha, Debashree Dass, Kishore Dhotre, Pooja Wakchoure, Ashwini More, Santanu Rana, Abdul A. Khan, and Anupam Mukherjee. 2023. "Combinatorial Effects of miRNAs in HSV-2 Infection of Macrophages: An In Silico and In Vitro Integration Approach" Vaccines 11, no. 9: 1488. https://doi.org/10.3390/vaccines11091488





