A 69 kb Deletion in chr19q13.42 including PRPF31 Gene in a Chinese Family Affected with Autosomal Dominant Retinitis Pigmentosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Ocular Examinations
2.3. Whole-Genome Sequencing
2.4. Mutation and Breakpoint Analysis
2.5. Multiplex Ligation-Dependent Probe Amplification (MLPA)
2.6. Parametric Linkage Analysis and Haplotype Construction
2.7. RNA-Sequencing and Data Analysis
3. Results
3.1. Ocular Examinations
3.2. Mutation Analysis
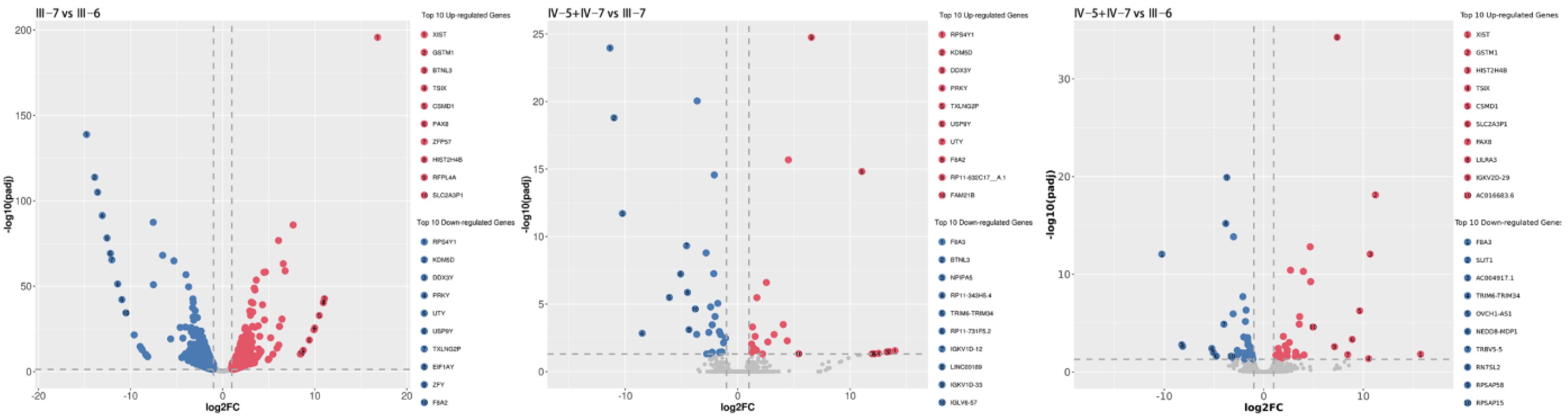
3.3. RNA-Sequencing and Data Analysis
4. Discussion
4.1. Discovery of Pathogenic Mutation in the Pedigree
4.2. Gene Expression of PRPF31 and Potential Mechanisms of RP
4.3. Incomplete Penetrance of RP
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Daiger, S.P.; Sullivan, L.S.; Bowne, S.J. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013, 84, 132–141. [Google Scholar] [CrossRef]
- Mansergh, F.C.; Millington-Ward, S.; Kennan, A.; Kiang, A.S.; Humphries, M.; Farrar, G.J.; Humphries, P.; Kenna, P.F. Retinitis pigmentosa and progressive sensorineural hearing loss caused by a C12258A mutation in the mitochondrial MTTS2 gene. Am. J. Hum. Genet. 1999, 64, 971–985. [Google Scholar] [CrossRef] [Green Version]
- Kajiwara, K.; Berson, E.L.; Dryja, T.P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 1994, 264, 1604–1608. [Google Scholar] [CrossRef] [Green Version]
- Katsanis, N.; Ansley, S.J.; Badano, J.L.; Eichers, E.R.; Lewis, R.A.; Hoskins, B.E.; Scambler, P.J.; Davidson, W.S.; Beales, P.L.; Lupski, J.R. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 2001, 293, 2256–2259. [Google Scholar] [CrossRef] [Green Version]
- Koyanagi, Y.; Akiyama, M.; Nishiguchi, K.M.; Momozawa, Y.; Kamatani, Y.; Takata, S.; Inai, C.; Iwasaki, Y.; Kumano, M.; Murakami, Y.; et al. Regional differences in genes and variants causing retinitis pigmentosa in Japan. Jpn. J. Ophthalmol. 2021, 65, 338–343. [Google Scholar] [CrossRef]
- Trapani, I.; Auricchio, A. Has retinal gene therapy come of age? From bench to bedside and back to bench. Hum. Mol. Genet. 2019, 28, R108–R118. [Google Scholar] [CrossRef] [Green Version]
- Bennett, J.; Wellman, J.; Marshall, K.A.; McCague, S.; Ashtari, M.; DiStefano-Pappas, J.; Elci, O.U.; Chung, D.C.; Sun, J.; Wright, J.F.; et al. Safety and durability of effect of contralateral-eye administration of AAV2 gene therapy in patients with childhood-onset blindness caused by RPE65 mutations: A follow-on phase 1 trial. Lancet 2016, 388, 661–672. [Google Scholar] [CrossRef] [Green Version]
- FDA approves hereditary blindness gene therapy. Nat. Biotechnol. 2018, 36, 6. [CrossRef]
- Verbakel, S.K.; van Huet, R.A.C.; Boon, C.J.F.; den Hollander, A.I.; Collin, R.W.J.; Klaver, C.C.W.; Hoyng, C.B.; Roepman, R.; Klevering, B.J. Non-syndromic retinitis pigmentosa. Prog. Retin. Eye Res. 2018, 66, 157–186. [Google Scholar] [CrossRef]
- Wheway, G.; Douglas, A.; Baralle, D.; Guillot, E. Mutation spectrum of PRPF31, genotype-phenotype correlation in retinitis pigmentosa, and opportunities for therapy. Exp. Eye Res. 2020, 192, 107950. [Google Scholar] [CrossRef]
- Weidenhammer, E.M.; Ruiz-Noriega, M.; Woolford, J.L., Jr. Prp31p promotes the association of the U4/U6 x U5 tri-snRNP with prespliceosomes to form spliceosomes in Saccharomyces cerevisiae. Mol. Cell Biol. 1997, 17, 3580–3588. [Google Scholar] [CrossRef] [Green Version]
- Farkas, M.H.; Lew, D.S.; Sousa, M.E.; Bujakowska, K.; Chatagnon, J.; Bhattacharya, S.S.; Pierce, E.A.; Nandrot, E.F. Mutations in pre-mRNA processing factors 3, 8, and 31 cause dysfunction of the retinal pigment epithelium. Am. J. Pathol. 2014, 184, 2641–2652. [Google Scholar] [CrossRef] [Green Version]
- Tanackovic, G.; Ransijn, A.; Thibault, P.; Abou Elela, S.; Klinck, R.; Berson, E.L.; Chabot, B.; Rivolta, C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011, 20, 2116–2130. [Google Scholar] [CrossRef] [Green Version]
- Farkas, M.H.; Grant, G.R.; White, J.A.; Sousa, M.E.; Consugar, M.B.; Pierce, E.A. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genom. 2013, 14, 486. [Google Scholar] [CrossRef] [Green Version]
- Rio Frio, T.; Civic, N.; Ransijn, A.; Beckmann, J.S.; Rivolta, C. Two trans-acting eQTLs modulate the penetrance of PRPF31 mutations. Hum. Mol. Genet. 2008, 17, 3154–3165. [Google Scholar] [CrossRef] [Green Version]
- Rivolta, C.; McGee, T.L.; Rio Frio, T.; Jensen, R.V.; Berson, E.L.; Dryja, T.P. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum. Mutat. 2006, 27, 644–653. [Google Scholar] [CrossRef]
- Vithana, E.N.; Abu-Safieh, L.; Pelosini, L.; Winchester, E.; Hornan, D.; Bird, A.C.; Hunt, D.M.; Bustin, S.A.; Bhattacharya, S.S. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: A molecular clue for incomplete penetrance? Investig. Ophthalmol. Vis. Sci. 2003, 44, 4204–4209. [Google Scholar] [CrossRef] [Green Version]
- Vithana, E.N.; Abu-Safieh, L.; Allen, M.J.; Carey, A.; Papaioannou, M.; Chakarova, C.; Al-Maghtheh, M.; Ebenezer, N.D.; Willis, C.; Moore, A.T.; et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell 2001, 8, 375–381. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Hamel, C.P. Gene discovery and prevalence in inherited retinal dystrophies. Comptes Rendus Biol. 2014, 337, 160–166. [Google Scholar] [CrossRef]
- Grover, D.; Mukerji, M.; Bhatnagar, P.; Kannan, K.; Brahmachari, S.K. Alu repeat analysis in the complete human genome: Trends and variations with respect to genomic composition. Bioinformatics 2004, 20, 813–817. [Google Scholar] [CrossRef] [Green Version]
- Abu-Safieh, L.; Vithana, E.N.; Mantel, I.; Holder, G.E.; Pelosini, L.; Bird, A.C.; Bhattacharya, S.S. A large deletion in the adRP gene PRPF31: Evidence that haploinsufficiency is the cause of disease. Mol. Vis. 2006, 12, 384–388. [Google Scholar]
- Deininger, P. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef] [Green Version]
- Köhn, L.; Bowne, S.J.; SSullivan, L.; Daiger, S.P.; Burstedt, M.S.; Kadzhaev, K.; Sandgren, O.; Golovleva, I. Breakpoint characterization of a novel approximately 59 kb genomic deletion on 19q13.42 in autosomal-dominant retinitis pigmentosa with incomplete penetrance. Eur. J. Hum. Genet. 2009, 17, 651–655. [Google Scholar] [CrossRef]
- Rose, A.M.; Mukhopadhyay, R.; Webster, A.R.; Bhattacharya, S.S.; Waseem, N.H. A 112 kb deletion in chromosome 19q13.42 leads to retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6597–6603. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L.; Hellmann-Blumberg, U.; Jurka, J.; Labuda, D.; Rubin, C.M.; Schmid, C.W.; Zietkiewicz, E.; Zuckerkandl, E. Standardized nomenclature for Alu repeats. J. Mol. Evol. 1996, 42, 3–6. [Google Scholar] [CrossRef]
- McGee, T.L.; Devoto, M.; Ott, J.; Berson, E.L.; Dryja, T.P. Evidence that the penetrance of mutations at the RP11 locus causing dominant retinitis pigmentosa is influenced by a gene linked to the homologous RP11 allele. Am. J. Hum. Genet. 1997, 61, 1059–1066. [Google Scholar] [CrossRef] [Green Version]
- Venturini, G.; Rose, A.M.; Shah, A.Z.; Bhattacharya, S.S.; Rivolta, C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 2012, 8, e1003040. [Google Scholar] [CrossRef] [Green Version]
- Rose, A.M.; Shah, A.Z.; Venturini, G.; Krishna, A.; Chakravarti, A.; Rivolta, C.; Bhattacharya, S.S. Transcriptional regulation of PRPF31 gene expression by MSR1 repeat elements causes incomplete penetrance in retinitis pigmentosa. Sci. Rep. 2016, 6, 19450. [Google Scholar] [CrossRef] [Green Version]
- Buskin, A.; Zhu, L.; Chichagova, V.; Basu, B.; Mozaffari-Jovin, S.; Dolan, D.; Droop, A.; Collin, J.; Bronstein, R.; Mehrotra, S.; et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat. Commun. 2018, 9, 4234. [Google Scholar] [CrossRef] [PubMed]
- McLenachan, S.; Zhang, D.; Grainok, J.; Zhang, X.; Huang, Z.; Chen, S.C.; Zaw, K.; Lima, A.; Jennings, L.; Roshandel, D.; et al. Determinants of Disease Penetrance in PRPF31-Associated Retinopathy. Genes 2021, 12, 1542. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.M.; Bhattacharya, S.S. Variant haploinsufficiency and phenotypic non-penetrance in PRPF31-associated retinitis pigmentosa. Clin. Genet. 2016, 90, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Ruberto, F.P.; Balzano, S.; Namburi, P.; Kimchi, A.; Pescini-Gobert, R.; Obolensky, A.; Banin, E.; Ben-Yosef, T.; Sharon, D.; Rivolta, C. Heterozygous deletions of noncoding parts of the PRPF31 gene cause retinitis pigmentosa via reduced gene expression. Mol. Vis. 2021, 27, 107–116. [Google Scholar]
- Brambillasca, F.; Mosna, G.; Colombo, M.; Rivolta, A.; Caslini, C.; Minuzzo, M.; Giudici, G.; Mizzi, L.; Biondi, A.; Privitera, E. Identification of a novel molecular partner of the E2A gene in childhood leukemia. Leukemia 1999, 13, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Brambillasca, F.; Mosna, G.; Ballabio, E.; Biondi, A.; Boulukos, K.E.; Privitera, E. Promoter analysis of TFPT (FB1), a molecular partner of TCF3 (E2A) in childhood acute lymphoblastic leukemia. Biochem. Biophys. Res. Commun. 2001, 288, 1250–1257. [Google Scholar] [CrossRef]
- Gan, Y.; Taira, E.; Irie, Y.; Fujimoto, T.; Miki, N. Arrest of cell cycle by amida which is phosphorylated by Cdc2 kinase. Mol. Cell Biochem. 2003, 246, 179–185. [Google Scholar] [CrossRef]
- Franchini, C.; Fontana, F.; Minuzzo, M.; Babbio, F.; Privitera, E. Apoptosis promoted by up-regulation of TFPT (TCF3 fusion partner) appears p53 independent, cell type restricted and cell density influenced. Apoptosis 2006, 11, 2217–2224. [Google Scholar] [CrossRef]
- Irie, Y.; Yamagata, K.; Gan, Y.; Miyamoto, K.; Do, E.; Kuo, C.H.; Taira, E.; Miki, N. Molecular cloning and characterization of Amida, a novel protein which interacts with a neuron-specific immediate early gene product arc, contains novel nuclear localization signals, and causes cell death in cultured cells. J. Biol. Chem. 2000, 275, 2647–2653. [Google Scholar] [CrossRef] [Green Version]
- Andrysik, Z.; Kim, J.; Tan, A.C.; Espinosa, J.M. A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep. 2013, 3, 1346–1354. [Google Scholar] [CrossRef] [Green Version]
- Olivares-González, L.; Velasco, S.; Campillo, I.; Rodrigo, R. Retinal Inflammation, Cell Death and Inherited Retinal Dystrophies. Int. J. Mol. Sci. 2021, 22, 2096. [Google Scholar] [CrossRef] [PubMed]
- Da Pozzo, P.; Cardaioli, E.; Malfatti, E.; Gallus, G.N.; Malandrini, A.; Gaudiano, C.; Berti, G.; Invernizzi, F.; Zeviani, M.; Federico, A. A novel mutation in the mitochondrial tRNA(Pro) gene associated with late-onset ataxia, retinitis pigmentosa, deafness, leukoencephalopathy and complex I deficiency. Eur. J. Hum. Genet. 2009, 17, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Loeffen, J.; van den Heuvel, L.; Smeets, R.; Triepels, R.; Sengers, R.; Trijbels, F.; Smeitink, J. cDNA sequence and chromosomal localization of the remaining three human nuclear encoded iron sulphur protein (IP) subunits of complex I: The human IP fraction is completed. Biochem. Biophys. Res. Commun. 1998, 247, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Starr, J.M.; Shiels, P.G.; Harris, S.E.; Pattie, A.; Pearce, M.S.; Relton, C.L.; Deary, I.J. Oxidative stress, telomere length and biomarkers of physical aging in a cohort aged 79 years from the 1932 Scottish Mental Survey. Mech. Ageing Dev. 2008, 129, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Rak, M.; Rustin, P. Supernumerary subunits NDUFA3, NDUFA5 and NDUFA12 are required for the formation of the extramembrane arm of human mitochondrial complex I. FEBS Lett. 2014, 588, 1832–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, S.; Thorburn, D. Nuclear Gene-Encoded Leigh Syndrome Spectrum Overview. In GeneReviews(®); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Mirzaa, G.M., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2020. [Google Scholar]
- Ma, Q.; Wang, C.; Wang, M.; Li, Y.; Li, P.; Wang, J.; Cheng, L.; An, Y.; Dai, H.; Duan, Y.; et al. Investigation of brain damage mechanism in middle cerebral artery occlusion/reperfusion rats based on i-TRAQ quantitative proteomics. Exp. Brain Res. 2021, 239, 1247–1260. [Google Scholar] [CrossRef]
- Pagano, G.; Pallardó, F.V.; Lyakhovich, A.; Tiano, L.; Trifuoggi, M. Mitigating the pro-oxidant state and melanogenesis of Retinitis pigmentosa: By counteracting mitochondrial dysfunction. Cell Mol. Life Sci. 2021, 78, 7491–7503. [Google Scholar] [CrossRef]
- Campochiaro, P.A.; Mir, T.A. The mechanism of cone cell death in Retinitis Pigmentosa. Prog. Retin. Eye Res. 2018, 62, 24–37. [Google Scholar] [CrossRef]
- Kohno, H.; Koso, H.; Okano, K.; Sundermeier, T.R.; Saito, S.; Watanabe, S.; Tsuneoka, H.; Sakai, T. Expression pattern of Ccr2 and Cx3cr1 in inherited retinal degeneration. J. Neuroinflamm. 2015, 12, 188. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zabel, M.K.; Wang, X.; Ma, W.; Shah, P.; Fariss, R.N.; Qian, H.; Parkhurst, C.N.; Gan, W.B.; Wong, W.T. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol. Med. 2015, 7, 1179–1197. [Google Scholar] [CrossRef]
- Funatsu, J.; Murakami, Y.; Shimokawa, S.; Nakatake, S.; Fujiwara, K.; Okita, A.; Fukushima, M.; Shibata, K.; Yoshida, N.; Koyanagi, Y.; et al. Circulating inflammatory monocytes oppose microglia and contribute to cone cell death in retinitis pigmentosa. PNAS Nexus 2022, 1, pgac003. [Google Scholar] [CrossRef]
- Steevels, T.A.; Lebbink, R.J.; Westerlaken, G.H.; Coffer, P.J.; Meyaard, L. Signal inhibitory receptor on leukocytes-1 is a novel functional inhibitory immune receptor expressed on human phagocytes. J. Immunol. 2010, 184, 4741–4748. [Google Scholar] [CrossRef]
- Steevels, T.A.; van Avondt, K.; Westerlaken, G.H.; Stalpers, F.; Walk, J.; Bont, L.; Coffer, P.J.; Meyaard, L. Signal inhibitory receptor on leukocytes-1 (SIRL-1) negatively regulates the oxidative burst in human phagocytes. Eur. J. Immunol. 2013, 43, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, Y.; Liu, Y.; He, Y.; Shi, G. Expression of VSTM1-v2 Is Increased in Peripheral Blood Mononuclear Cells from Patients with Rheumatoid Arthritis and Is Correlated with Disease Activity. PLoS ONE 2016, 11, e0146805. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; En, Z.; Li, D.J.; Mao, C.Y.; He, Q.; Zhang, J.F.; Fan, Y.Q.; Wang, C.Q. VSTM1 regulates monocyte/macrophage function via the NF-κB signaling pathway. Open Med. 2021, 16, 1513–1524. [Google Scholar] [CrossRef] [PubMed]
- Radjabova, V.; Mastroeni, P.; Skjødt, K.; Zaccone, P.; de Bono, B.; Goodall, J.C.; Chilvers, E.R.; Juss, J.K.; Jones, D.C.; Trowsdale, J.; et al. TARM1 Is a Novel Leukocyte Receptor Complex-Encoded ITAM Receptor That Costimulates Proinflammatory Cytokine Secretion by Macrophages and Neutrophils. J. Immunol. 2015, 195, 3149–3159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merck, E.; Gaillard, C.; Scuiller, M.; Scapini, P.; Cassatella, M.A.; Trinchieri, G.; Bates, E.E. Ligation of the FcR gamma chain-associated human osteoclast-associated receptor enhances the proinflammatory responses of human monocytes and neutrophils. J. Immunol. 2006, 176, 3149–3156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrow, A.D.; Palarasah, Y.; Bugatti, M.; Holehouse, A.S.; Byers, D.E.; Holtzman, M.J.; Vermi, W.; Skjødt, K.; Crouch, E.; Colonna, M. OSCAR is a receptor for surfactant protein D that activates TNF-α release from human CCR2+ inflammatory monocytes. J. Immunol. 2015, 194, 3317–3326. [Google Scholar] [CrossRef] [Green Version]
- Azizzadeh Pormehr, L.; Ahmadian, S.; Daftarian, N.; Mousavi, S.A.; Shafiezadeh, M. PRPF31 reduction causes mis-splicing of the phototransduction genes in human organotypic retinal culture. Eur. J. Hum. Genet. 2020, 28, 491–498. [Google Scholar] [CrossRef]
- Kuroda, T.; Yasuda, S.; Nakashima, H.; Takada, N.; Matsuyama, S.; Kusakawa, S.; Umezawa, A.; Matsuyama, A.; Kawamata, S.; Sato, Y. Identification of a Gene Encoding Slow Skeletal Muscle Troponin T as a Novel Marker for Immortalization of Retinal Pigment Epithelial Cells. Sci. Rep. 2017, 7, 8163. [Google Scholar] [CrossRef] [Green Version]
- Uppugunduri, C.R.S.; Muthukumaran, J.; Robin, S.; Santos-Silva, T.; Ansari, M. In silico and in vitro investigations on the protein-protein interactions of glutathione S-transferases with mitogen-activated protein kinase 8 and apoptosis signal-regulating kinase 1. J. Biomol. Struct. Dyn. 2022, 40, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Bui, D.S.; Lodge, C. Glutathione S-Transferase Gene Associations and Gene-Environment Interactions for Asthma. Curr. Allergy Asthma Rep. 2021, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Osiecka, O.; Skrzeczynska-Moncznik, J.; Morytko, A.; Mazur, A.; Majewski, P.; Bilska, B.; Kapinska-Mrowiecka, M.; Kosalka-Wegiel, J.; Pastuszczak, M.; Pyza, E.; et al. Secretory Leukocyte Protease Inhibitor Is Present in Circulating and Tissue-Recruited Human Eosinophils and Regulates Their Migratory Function. Front. Immunol. 2021, 12, 737231. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tang, Y.D.; Zheng, C. Revisiting IRF1-mediated antiviral innate immunity. Cytokine Growth Factor Rev. 2022, 64, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Venema, W.J.; Hiddingh, S.; de Boer, J.H.; Claas, F.H.J.; Mulder, A.; den Hollander, A.I.; Stratikos, E.; Sarkizova, S.; van der Veken, L.T.; Janssen, G.M.C.; et al. ERAP2 Increases the Abundance of a Peptide Submotif Highly Selective for the Birdshot Uveitis-Associated HLA-A29. Front. Immunol. 2021, 12, 634441. [Google Scholar] [CrossRef]
- Zhao, J.; Izumi, T.; Nunomura, K.; Satoh, S.; Watanabe, S. MARCKS-like protein, a membrane protein identified for its expression in developing neural retina, plays a role in regulating retinal cell proliferation. Biochem. J. 2007, 408, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Prieto, D.; Zolessi, F.R. Functional Diversification of the Four MARCKS Family Members in Zebrafish Neural Development. J. Exp. Zool. B Mol. Dev. Evol. 2017, 328, 119–138. [Google Scholar] [CrossRef]
- Huang, W.; Wang, W.; Zhou, M.; Chen, S.; Zhang, X. Association of glutathione S-transferase polymorphisms (GSTM1 and GSTT1) with primary open-angle glaucoma: An evidence-based meta-analysis. Gene 2013, 526, 80–86. [Google Scholar] [CrossRef]
- Micheal, S.; Hogewind, B.F.; Khan, M.I.; Siddiqui, S.N.; Zafar, S.N.; Akhtar, F.; Qamar, R.; Hoyng, C.B.; den Hollander, A.I. Variants in the PRPF8 Gene are Associated with Glaucoma. Mol. Neurobiol. 2018, 55, 4504–4510. [Google Scholar] [CrossRef]
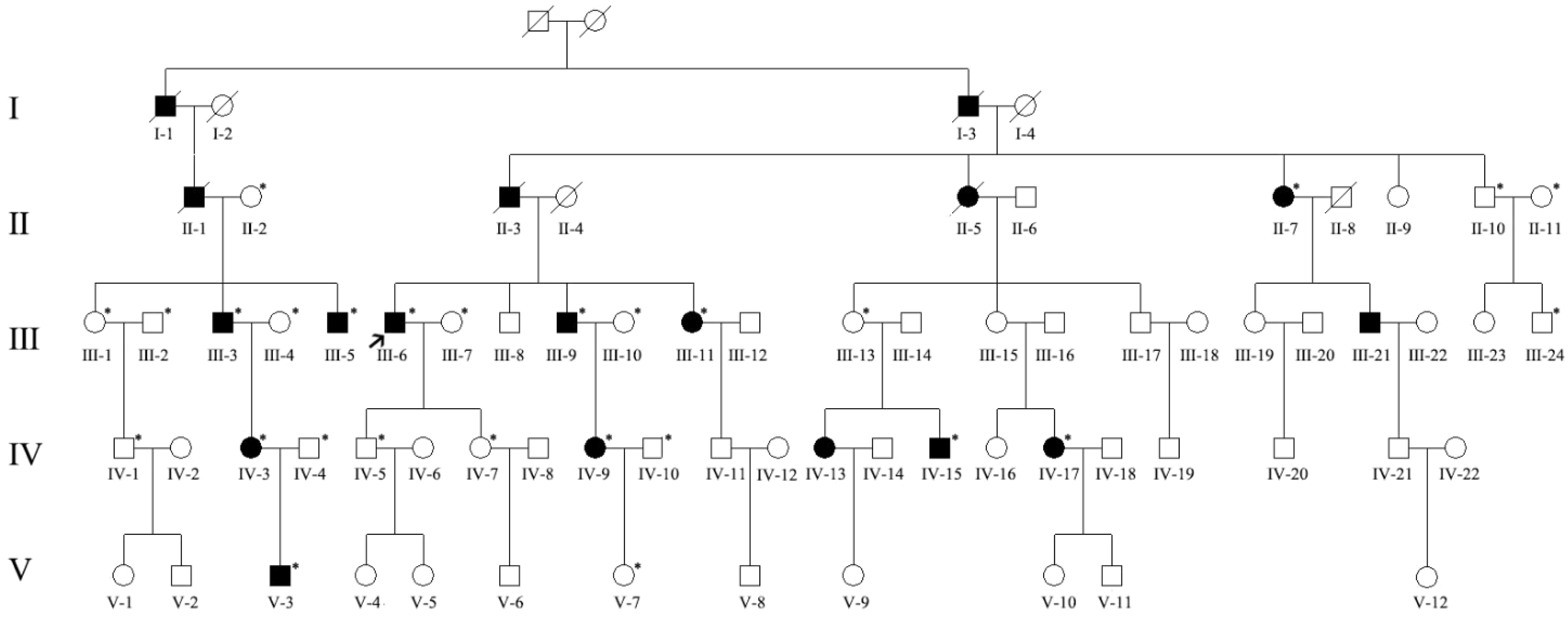

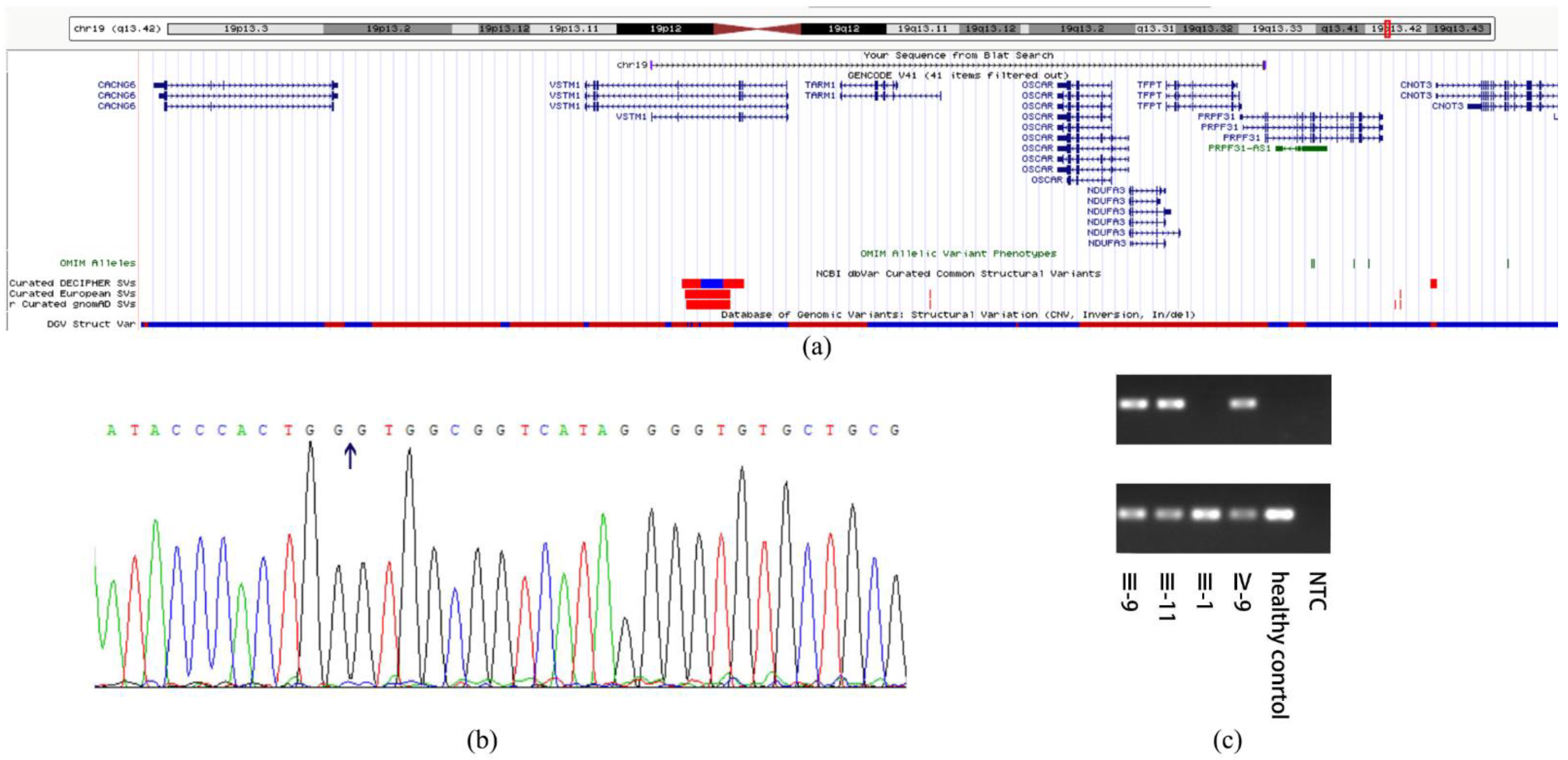
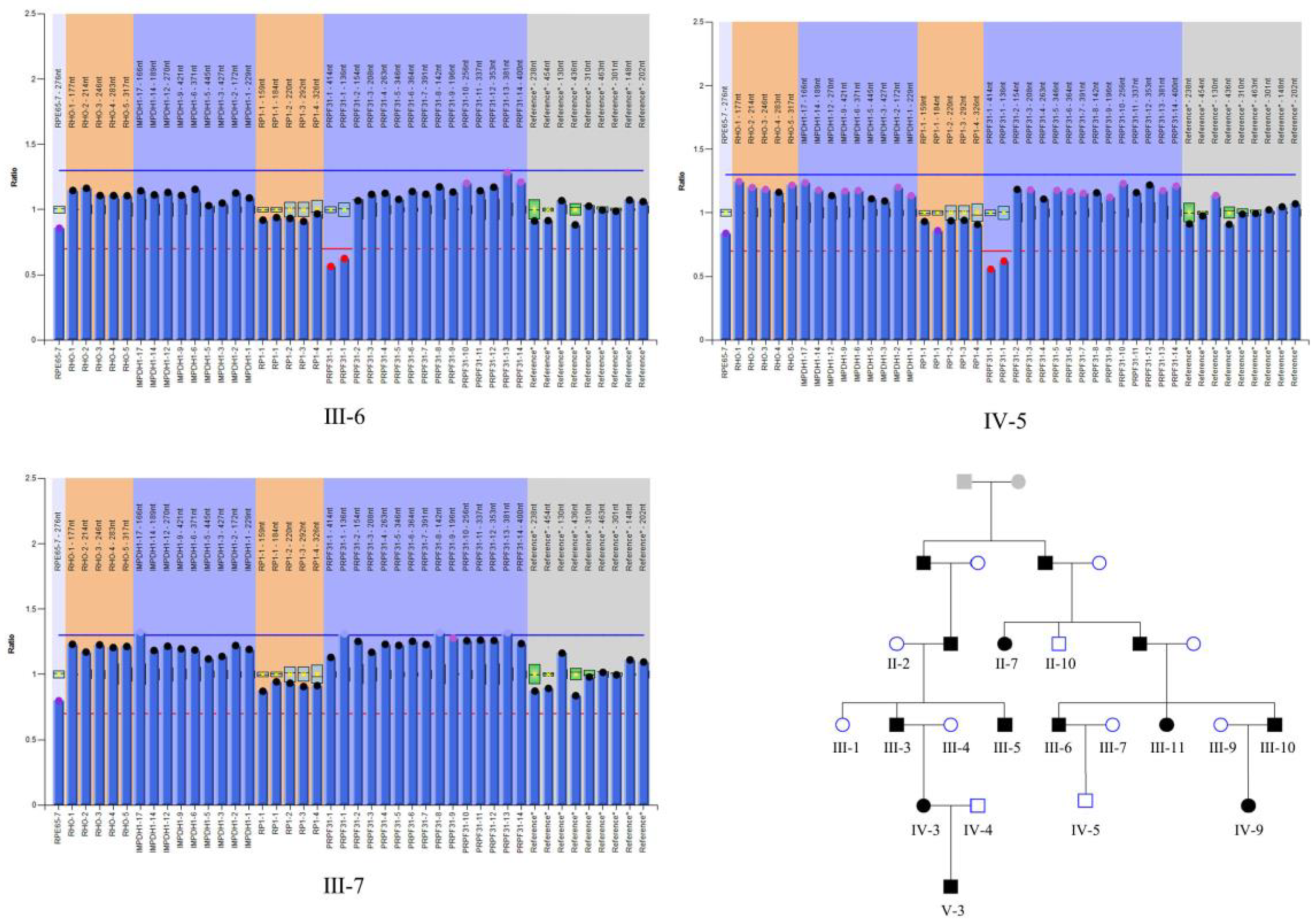
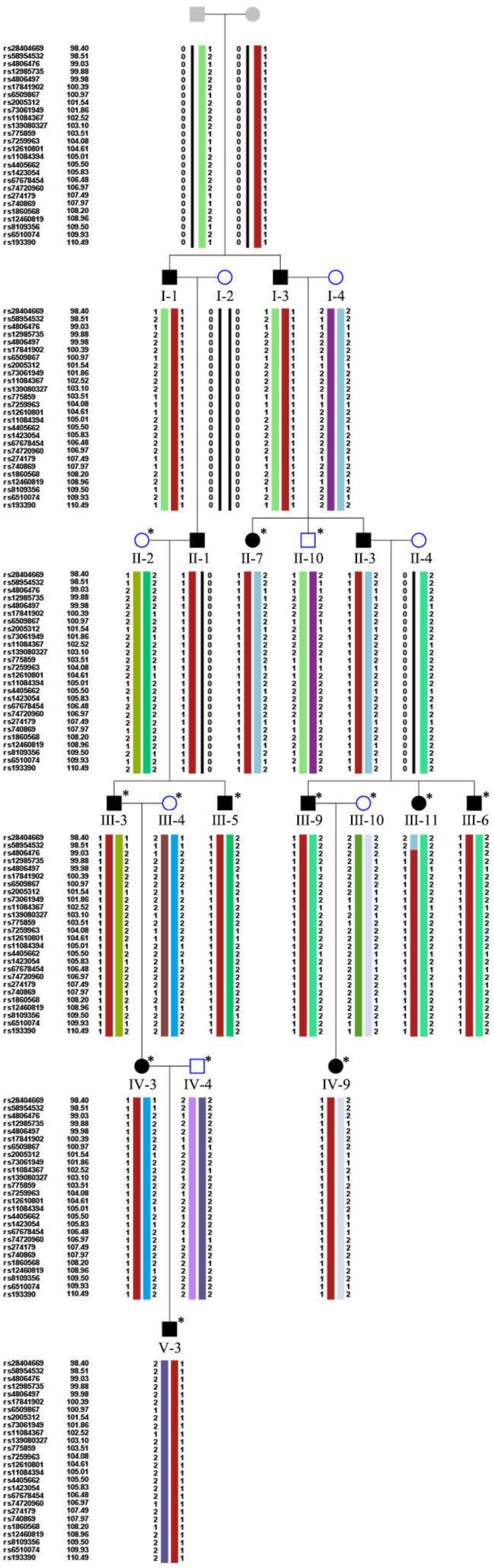
| Pedigree ID | Age at Examination | Sex | Phenotype | BCVA | IOP (mmHg) | Cup-Disc Ratio | Avg_Axial (mm) | Avg_ACD (mm) | Avg_lens (mm) |
|---|---|---|---|---|---|---|---|---|---|
| II-2 | 83 | Female | Normal | 4.7/4.5 | 16/14 | 0.3/0.3 | 21.86/21.75 | 2.92/2.86 | 4.69/4.57 |
| II-7 | 78 | Female | RP | HM/LP | 13/14 | 0.4/0.4 | 21.58/21.43 | 3.64/2.74 | 4.94/4.65 |
| II-10 | 72 | Male | Normal | 5.3/5.2 | 15/16 | 0.4/0.4 | 22.54/22.44 | 2.88/2.76 | 4.74/4.77 |
| II-11 | 68 | Female | Normal | 4.7/4.8 | 18/20 | 0.3/0.3 | 21.54/21.64 | 2.64/2.72 | 4.72/4.75 |
| III-1 | 58 | Female | Normal | 5.2/5.3 | 15/18 | 0.4/0.4 | 22.98/23.08 | 3.08/3.22 | 4.57/4.22 |
| III-2 | 63 | Male | Normal | 4.9/4.9 | 19/20 | 0.6/0.4 | 22.61/22.47 | 2.92/2.77 | 4.76/4.91 |
| III-3 | 56 | Male | RP | - | 7/10 | 0.3/0.3 | 23.46/23.37 | 2.24/2.14 | 5.30/5.33 |
| III-5 | 54 | Male | RP | - | 9/10 | 0.3/0.3 | 25.77/25.70 | 2.83/2.72 | 4.61/4.69 |
| III-6 | 72 | Male | RP | Fc20/HM | 18/17 | 0.9/1.0 | 22.95/23.51 | 4.10/4.15 | IOL/IOL |
| III-7 | 70 | Female | Normal | 4.7/4.8 | 14/13 | 0.4/0.4 | 22.55/22.66 | 2.59/2.49 | 4.68/4.71 |
| III-9 | 62 | Male | RP | 4.1/Fc150 | 22/13 | 0.6/0.5 | 24.23/24.07 | 4.83/3.22 | 5.38/4.11 |
| III-10 | 61 | Female | Normal | 4.8/4.7 | 11/14 | 0.3/0.3 | 21.98/22.12 | 2.32/2.50 | 4.48/5.15 |
| III-11 | 56 | Female | RP | 4.8/4.7 | 19/14 | 0.3/0.3 | 21.50/22.08 | 2.71/2.87 | 3.80/4.23 |
| III-13 | 61 | Female | Normal | 5.0/5.0 | 15/15 | 0.4/0.4 | 22.18/22.23 | 2.68/2.47 | 4.31/4.43 |
| III-24 | 45 | Male | Normal | 5.2/5.1 | 21/18 | 0.3/0.3 | 22.83/22.76 | 3.02/3.03 | 4.1/4.05 |
| IV-1 | 35 | Male | Normal | 5.2/5.2 | 19/19 | 0.3/0.3 | 24.75/24.66 | 3.46/3.22 | 4.15/4.20 |
| IV-7 | 46 | Female | Normal | 5.2/5.2 | 17/18 | 0.4/0.4 | 22.83/22.95 | 3.13/3.36 | 4.02/3.94 |
| IV-10 | 47 | Male | Normal | 5.1/5.1 | 19/17 | 0.3/0.3 | 21.46/21.54 | 2.86/2.84 | 4.66/4.65 |
| IV-15 | 28 | Male | RP | 4.7/4.7 | 15/16 | 0.3/0.3 | 26.23/24.03 | 2.57/2.43 | 4.46/4.55 |
| IV-17 | 30 | Female | RP | 5.0/5.0 | 19/15 | 0.3/0.3 | 21.76/21.35 | 2.83/2.69 | 4.09/4.10 |
| V-3 | 4 | Female | RP | - | - | 0.3/0.3 | 21.63/21.58 | 2.65/2.63 | 3.60/3.58 |
| V-7 | 16 | Female | Normal | 5.2/5.3 | 18/21 | 0.3/0.3 | 23.18/23.92 | 3.18/3.63 | 3.67/3.61 |
| Pedigree ID | Str | End | Chr | SVTYPE | Variant Type |
|---|---|---|---|---|---|
| II-2 | - | - | - | - | - |
| II-7 | 54048499 | 54118055 | 19 | DEL | Het |
| II-10 | - | - | - | - | - |
| II-11 | - | - | - | - | - |
| III-1 | - | - | - | - | - |
| III-2 | - | - | - | - | - |
| III-3 | 54048499 | 54118055 | 19 | DEL | Het |
| III-4 | - | - | - | - | - |
| III-5 | 54048499 | 54118055 | 19 | DEL | Het |
| III-6 | 54048499 | 54118055 | 19 | DEL | Het |
| III-7 | - | - | - | - | - |
| III-9 | 54048499 | 54118055 | 19 | DEL | Het |
| III-10 | - | - | - | - | - |
| III-11 | 54048499 | 54118055 | 19 | DEL | Het |
| III-13 | 54048499 | 54118055 | 19 | DEL | Het |
| III-24 | - | - | - | - | - |
| IV-1 | - | - | - | - | - |
| IV-3 | 54048499 | 54118055 | 19 | DEL | Het |
| IV-4 | - | - | - | - | - |
| IV-5 | 54048499 | 54118055 | 19 | DEL | Het |
| IV-7 | 54048499 | 54118055 | 19 | DEL | Het |
| IV-9 | 54048499 | 54118055 | 19 | DEL | Het |
| IV-10 | - | - | - | - | - |
| IV-15 | 54048499 | 54118055 | 19 | DEL | Het |
| IV-17 | 54048499 | 54118055 | 19 | DEL | Het |
| V-3 | 54048499 | 54118055 | 19 | DEL | Het |
| V-7 | - | - | - | - | - |
| CHR | POS | LABEL | MODEL | LOD | HLOD |
|---|---|---|---|---|---|
| 19 | 0.9903 | rs4806476 | dominant_autosomal | 3.1885 | 3.1885 |
| 19 | 0.9988 | rs12985735 | dominant_autosomal | 3.6074 | 3.6074 |
| 19 | 0.9998 | rs4806497 | dominant_autosomal | 3.6096 | 3.6096 |
| 19 | 1.0039 | rs17841902 | dominant_autosomal | 3.6095 | 3.6095 |
| 19 | 1.0097 | rs6509867 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0154 | rs2005312 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0186 | rs73061949 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0252 | rs11084367 | dominant_autosomal | 3.6092 | 3.6092 |
| 19 | 1.031 | rs139080327 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0351 | rs775859 | dominant_autosomal | 3.609 | 3.609 |
| 19 | 1.0408 | rs7259963 | dominant_autosomal | 3.609 | 3.609 |
| 19 | 1.0461 | rs12610801 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0501 | rs11084394 | dominant_autosomal | 3.6094 | 3.6094 |
| 19 | 1.055 | rs4405662 | dominant_autosomal | 3.6094 | 3.6094 |
| 19 | 1.0583 | rs1423054 | dominant_autosomal | 3.6094 | 3.6094 |
| 19 | 1.0648 | rs67678454 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0697 | rs74720960 | dominant_autosomal | 3.6093 | 3.6093 |
| 19 | 1.0749 | rs274179 | dominant_autosomal | 3.6091 | 3.6091 |
| 19 | 1.0797 | rs740869 | dominant_autosomal | 3.6091 | 3.6091 |
| 19 | 1.082 | rs1860568 | dominant_autosomal | 3.609 | 3.609 |
| 19 | 1.0896 | rs12460819 | dominant_autosomal | 3.6078 | 3.6078 |
| 19 | 1.095 | rs8109356 | dominant_autosomal | 3.6066 | 3.6066 |
| 19 | 1.0993 | rs6510074 | dominant_autosomal | 3.6043 | 3.6043 |
| 19 | 1.1049 | rs193390 | dominant_autosomal | 3.5849 | 3.5849 |
| Group | Up-Regulated | Down-Regulated |
|---|---|---|
| Patient vs. healthy control | 588 | 532 |
| NPCs vs. healthy control | 24 | 29 |
| NPCs vs. patient | 42 | 40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lan, Y.; Chen, Y.; Qiao, Y.; Xu, Q.; Zhai, R.; Sun, X.; Wu, J.; Chen, X. A 69 kb Deletion in chr19q13.42 including PRPF31 Gene in a Chinese Family Affected with Autosomal Dominant Retinitis Pigmentosa. J. Clin. Med. 2022, 11, 6682. https://doi.org/10.3390/jcm11226682
Lan Y, Chen Y, Qiao Y, Xu Q, Zhai R, Sun X, Wu J, Chen X. A 69 kb Deletion in chr19q13.42 including PRPF31 Gene in a Chinese Family Affected with Autosomal Dominant Retinitis Pigmentosa. Journal of Clinical Medicine. 2022; 11(22):6682. https://doi.org/10.3390/jcm11226682
Chicago/Turabian StyleLan, Yuanzheng, Yuhong Chen, Yunsheng Qiao, Qingdan Xu, Ruyi Zhai, Xinghuai Sun, Jihong Wu, and Xueli Chen. 2022. "A 69 kb Deletion in chr19q13.42 including PRPF31 Gene in a Chinese Family Affected with Autosomal Dominant Retinitis Pigmentosa" Journal of Clinical Medicine 11, no. 22: 6682. https://doi.org/10.3390/jcm11226682
APA StyleLan, Y., Chen, Y., Qiao, Y., Xu, Q., Zhai, R., Sun, X., Wu, J., & Chen, X. (2022). A 69 kb Deletion in chr19q13.42 including PRPF31 Gene in a Chinese Family Affected with Autosomal Dominant Retinitis Pigmentosa. Journal of Clinical Medicine, 11(22), 6682. https://doi.org/10.3390/jcm11226682






