Fetal Nasal Bone Hypoplasia in the Second Trimester as a Marker of Multiple Genetic Syndromes
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cicero, S.; Longo, D.; Rembouskos, G.; Sacchini, C.; Nicolaides, K.H. Absent nasal bone at 11–14 weeks of gestation and chromosomal defects. Ultrasound Obstet. Gynecol. 2003, 22, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Cicero, S.; Sonek, J.D.; McKenna, D.S.; Croom, C.S.; Johnson, L.; Nicolaides, K.H. Nasal bone hypoplasia in trisomy 21 at 15–22 weeks’ gestation. Ultrasound Obstet. Gynecol. 2003, 21, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Cai, M.; Ma, W.; Lin, N.; Xu, L. Chromosomal Microarray Analysis for the Prenatal Diagnosis in Fetuses with Nasal Bone Hypoplasia: A Retrospective Cohort Study. Risk Manag. Healthc. Policy 2021, 14, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 2015, 519, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Petrovski, S.; Aggarwal, V.; Giordano, J.L.; Stosic, M.; Wou, K.; Bier, L.; Spiegel, E.; Brennan, K.; Stong, N.; Jobanputra, V.; et al. Whole-exome sequencing in the evaluation of fetal structural anomalies: A prospective cohort study. Lancet 2019, 393, 758–767. [Google Scholar] [CrossRef]
- Sonek, J.D.; McKenna, D.; Webb, D.; Croom, C.; Nicolaides, K. Nasal bone length throughout gestation: Normal ranges based on 3537 fetal ultrasound measurements. Ultrasound Obstet. Gynecol. 2003, 21, 152–155. [Google Scholar] [CrossRef]
- Ting, Y.H.; Lao, T.T.; Lau, T.K.; Chung, M.K.; Leung, T.Y. Isolated absent or hypoplastic nasal bone in the second trimester fetus: Is amniocentesis necessary? J. Matern.-Fetal Neonatal Med. 2011, 24, 555–558. [Google Scholar] [CrossRef]
- Society for Maternal-Fetal Medicine (SMFM); Prabhu, M.; Kuller, J.; Biggio, J.R. Society for Maternal-Fetal Medicine Consult Series #57: Evaluation and management of isolated soft ultrasound markers for aneuploidy in the second trimester. Am. J. Obstet. Gynecol. 2021, 225, B2–B15. [Google Scholar]
- Gu, Y.Z.; Nisbet, D.L.; Reidy, K.L.; Palma-Dias, R. Hypoplastic nasal bone: A potential marker for facial dysmorphism associated with pathogenic copy number variants on microarray. Prenat. Diagn. 2019, 39, 116–123. [Google Scholar] [CrossRef]
- Zhang, F.; Long, W.; Zhou, Q.; Wang, J.; Shi, Y.; Liu, J.; Wang, Q. Is Prenatal Diagnosis Necessary for Fetal Isolated Nasal Bone Absence or Hypoplasia? Int. J. Gen. Med. 2021, 14, 4435–4441. [Google Scholar] [CrossRef] [PubMed]
- Dukhovny, S.; Wilkins-Haug, L.; Shipp, T.D.; Benson, C.B.; Kaimal, A.J.; Reiss, R. Absent fetal nasal bone: What does it mean for the euploid fetus? J. Ultrasound Med. 2013, 32, 2131–2134. [Google Scholar] [CrossRef] [PubMed]
- Mak, A.S.L.; Ma, T.W.L.; Chan, K.Y.K.; Kan, A.S.Y.; Tang, M.H.Y.; Leung, K.Y. Prenatal diagnosis of 5p deletion syndrome: Report of five cases. J. Obstet. Gynaecol. Res. 2019, 45, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Teoh, X.H.; Tan, T.Y.; Chow, K.K.; Lee, I.W. Prenatal diagnosis of cri-du-chat syndrome: Importance of ultrasonographical markers. Singap. Med. J. 2009, 50, e181–e184. [Google Scholar]
- Liberati, M.; Melchiorre, K.; D’Emilio, I.; Guanciali-Franchi, P.E.; Iezzi, I.; Rotmensch, S.; Celentano, C. Fetal facial profile in Pallister-Killian syndrome. Fetal Diagn. Ther. 2008, 23, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Grati, F.R.; Lalatta, F.; Turolla, L.; Cavallari, U.; Gentilin, B.; Rossella, F.; Cetin, I.; Antonazzo, P.; Bellotti, M.; Dulcetti, F.; et al. Three cases with de novo 6q imbalance and variable prenatal phenotype. Am. J. Med. Genet. A 2005, 136, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rodriguez, R.; Rodriguez-Rodriguez, R.; Garcia-Delgado, R.; Romero-Requejo, A.; Medina-Castellano, M.; Cruz, L.G.; Rodriguez, A.S.; Garcia-Hernandez, J.A. Prenatal diagnosis of Greig Cephalopolysyndactyly Syndrome. When to suspect it. J. Matern.-Fetal Neonatal Med. 2020, 4, 1–4. [Google Scholar] [CrossRef]
- Allderdice, P.W.; Davis, J.G.; Miller, O.J.; Klinger, H.P.; Warburton, D.; Miller, D.A.; Allen, F.H., Jr.; Abrams, C.A.; McGilvray, E. The 13q-deletion syndrome. Am. J. Hum. Genet. 1969, 21, 499–512. [Google Scholar] [PubMed]
- Dhandha, S.; Hogge, W.A.; Surti, U.; McPherson, E. Three cases of tetrasomy 9p. Am. J. Med. Genet. 2002, 113, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Podolsky, R.; Saltzman, D.; Auerbach, M.; Roman, A.S. Absent nasal bone as a marker of tetrasomy 9p. Prenat. Diagn. 2011, 31, 1313. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; McCalman, M.T.; Bossuyt, T.P.; Barakat, T.S. Case Report: Two New Cases of Chromosome 12q14 Deletions and Review of the Literature. Front. Genet. 2021, 12, 716874. [Google Scholar] [CrossRef] [PubMed]
- Mary, L.; Scheidecker, S.; Kohler, M.; Lombardi, M.P.; Delezoide, A.L.; Auberger Triau, S.; Colin, E.; Gerard, M.; Grzeschik, K.H.; Dollfus, H.; et al. Prenatal diagnosis of focal dermal hypoplasia: Report of three fetuses and review of the literature. Am. J. Med. Genet. A 2017, 173, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Sankar, V.H.; Phadke, S.R. Asphyxiating thoracic dystrophy with facial dysmorphism. Indian J. Pediatr. 2006, 73, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Mistry, K.A.; Suthar, P.P.; Bhesania, S.R.; Patel, A. Antenatal Diagnosis of Jeune Syndrome (Asphyxiating Thoracic Dysplasia) with Micromelia and Facial Dysmorphism on Second-Trimester Ultrasound. Pol. J. Radiol. 2015, 80, 296–299. [Google Scholar] [PubMed] [Green Version]
- Chen, M.; Lin, S.M.; Li, N.; Li, Y.; Li, Y.; Zhang, L. An induced pluripotent stem cell line (GZHMCi003-A) derived from a fetus with exon 3 heterozygous deletion in RUNX2 gene causing cleidocranial dysplasia. Stem Cell Res. 2021, 51, 102166. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.P.; Hung, H.Y.; Chang, T.Y.; Lin, S.P.; Wang, W. Second-trimester nasal bone hypoplasia/aplasia associated with cleidocranial dysplasia. Prenat. Diagn. 2004, 24, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Singh, C.; Thakur, S.; Arora, N.; Khurana, D. Revisiting absent nasal bone in the second trimester. J. Clin. Ultrasound 2021, 49, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Lostchuck, E.; Hui, L. Should second-trimester hypoplastic nasal bone be sole indication for diagnostic testing with chromosomal microarray analysis? Ultrasound Obstet. Gynecol. 2019, 53, 848–850. [Google Scholar] [CrossRef]
- Du, Y.; Ren, Y.; Yan, Y.; Cao, L. Absent fetal nasal bone in the second trimester and risk of abnormal karyotype in a prescreened population of Chinese women. Acta Obstet. Gynecol. Scand. 2018, 97, 180–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]

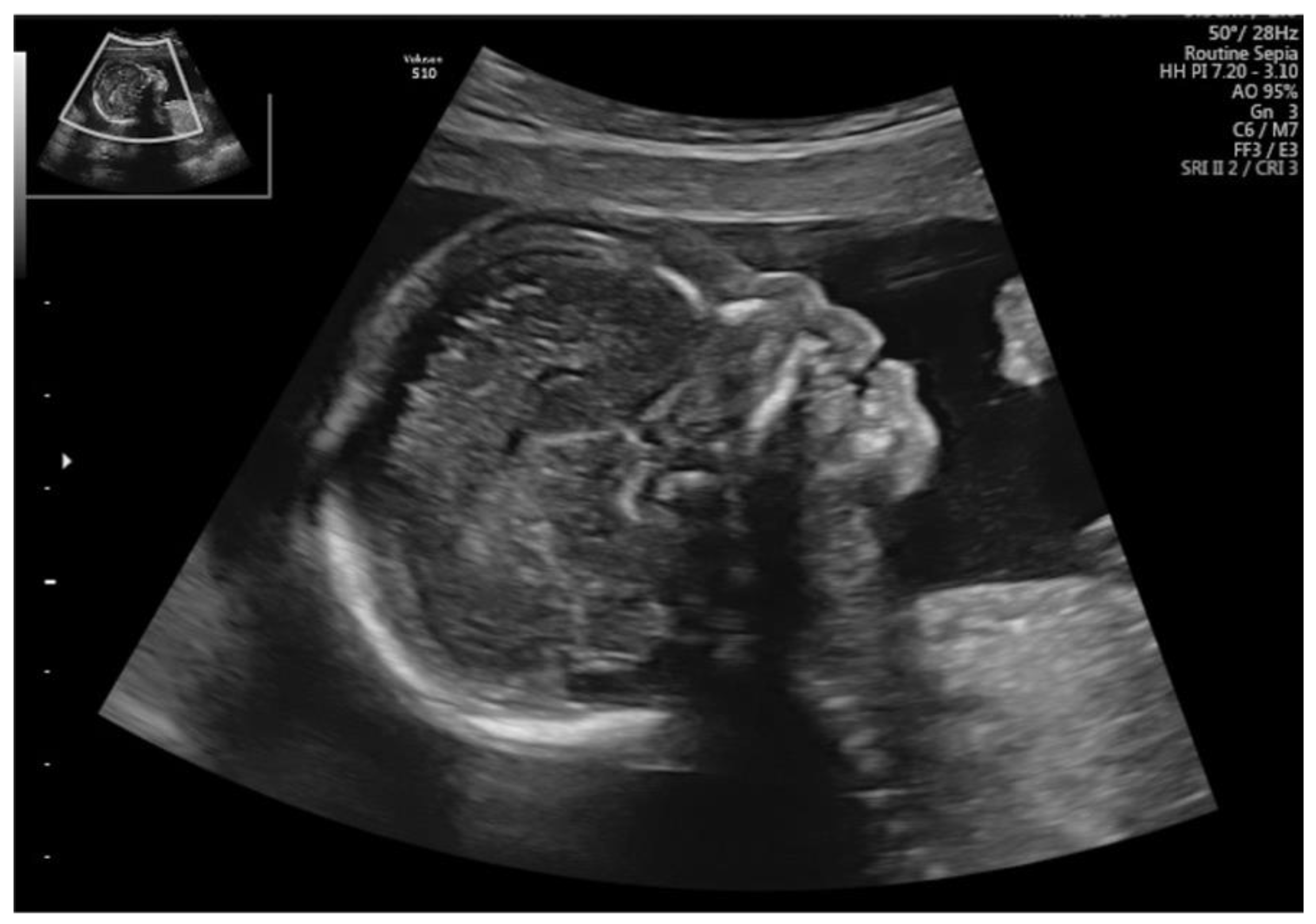
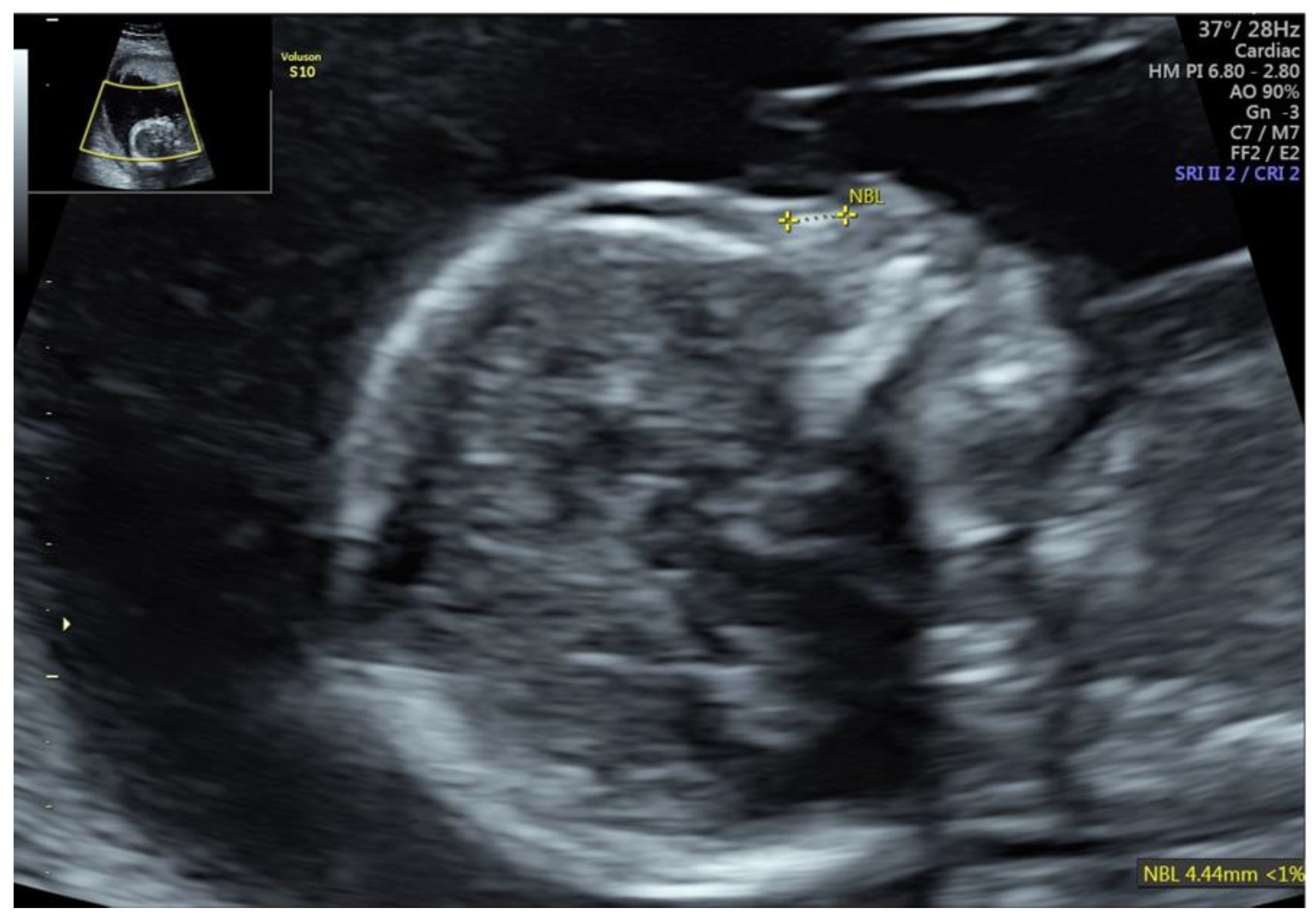
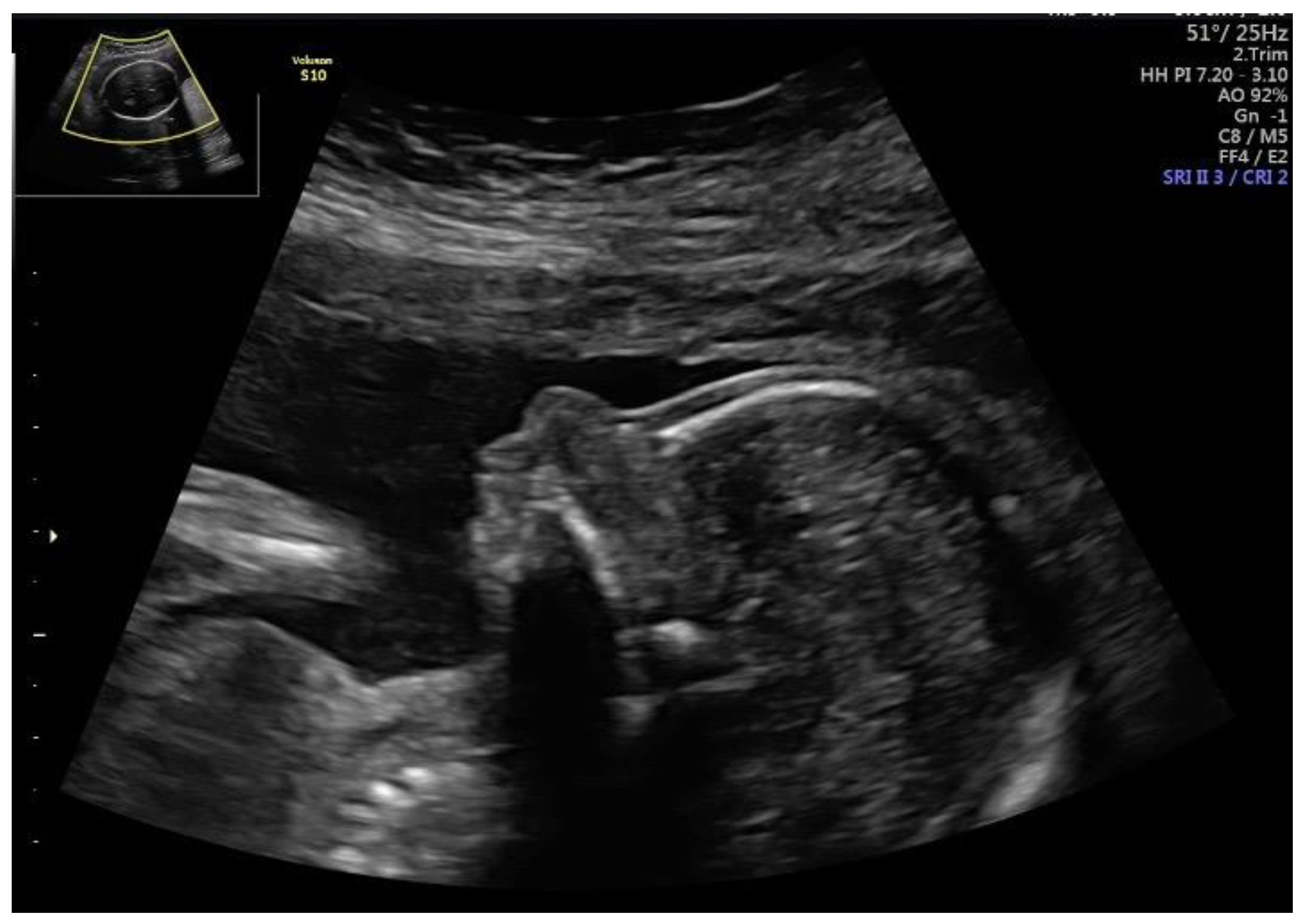
| Fetal Ultrasound in the First Trimester | GA at AC | Ultrasound Findings in the Second Trimester | Chromosome Region | Size | Variant Type | Classification | Pregnancy Outcome | |
|---|---|---|---|---|---|---|---|---|
| 1 | CIII PIII 37 y/o NT 1.4 mm; CRL 70 mm; NB (+) cFTS: low risk | 20 | isolated NB hypoplasia | Xp22.31 * | 1.67 Mb | deletion | pathogenic | LB, 39 weeks GA, normal newborn examination |
| 2 | CV PIII 35 y/o NT 2.1 mm; CRL 68 mm; NB (+) cFTS: low risk | 19 | isolated NB hypoplasia | 8p23.3p23.1 ** 8p23.1p12 *** | 6.68 Mb 22.26 Mb | deletion amplifiaction | potentially pathogenic potentially pathogenic | LB, 39 weeks GA, cerebellar hypoplasia, hypotonia, facial dysmorphia, strabismus |
| Ultrasound Findings in the Second Trimester | Number of Cases | Chromosome Region |
|---|---|---|
| NB hypoplasia, CPC and pyelectasis | 1 | normal CMA |
| NB hypoplasia, ICEF, hyperechogenic bowels | 1 | normal CMA |
| NB hypoplasia and ICEF | 1 | normal CMA |
| NB hypoplasia and CPC | 2 | normal CMA |
| NB hypoplasia and NF | 2 | normal CMA |
| NB hypoplasia and pyelectasis | 3 | normal CMA |
| NB hypoplasia and pyelectasis | 1 | 18q22.1 * |
| Fetal Ultrasound in the First Trimester | GA at AC | Ultrasound Findings in the Second Trimester | Chromosome Region | Size | Variant Type | Classification | Pregnancy Outcome | |
|---|---|---|---|---|---|---|---|---|
| 1a * | CIII PI 37 y/o NT 3.7 mm; CRL 65 mm cFTS: high risk | 16 | NB hypoplasia, ventriculomegaly, CCA, NF, hypotelorism, RAA, hypospadias with bifid scrotum | 13q32.3q34 | 30.4 Mb | deletion | pathogenic | TOP |
| 1b * | CIII PI 37 y/o NT 3.6 mm; CRL 66 mm cFTS: high risk | 16 | NB hypoplasia, ventriculomegaly, CCA, NF, hypotelorism, PE, hypospadias with bifid scrotum | 13q32.3q34 | 30.4 Mb | deletion | pathogenic | TOP |
| 2 | CI PI 23 y/o cFTS: NA | 20 | NB hypoplasia, abnormal profile, ventricular septal defect, diaphragmatic hernia | 12p13.33p11.21 47,XY,+i(12)(p10) | 33 Mb | amplification | pathogenic (Pallister–Killian syndrome) | TOP |
| 3 | CIII PI 33 y/o NT 3.0 mm; CRL 75 mm; NB (-) cFTS: high risk | 16 | NB hypoplasia, cleft lip, abnormal posterior fossa, ventricular septal defect | 9p24.3p13.1 47,XY,+i(9) | 38.8 Mb | amplification | pathogenic (tetrasomy 9p) | TOP |
| 4 | CII PI 33 y/o NT 2.6 mm; CRL 79 mm; NB (-), TR cFTS: high risk | 19 | ventriculomegaly, NB hypoplasia, aberrant right subclavian artery, pulmonary stenosis, TR | 9p24.3p13.2 9q12q21.11 22q11.1q11.22 | 37.3 Mb 5.33 Mb 7.1 Mb | amplification amplification amplification | pathogenic potentially pathogenic | NA |
| 5 | CIV PI 32 y/o cFTS: NA | 20 | NB hypoplasia, ventriculomegaly, micrognathia, abnormal posterior fossa, empty stomach | 5p15.33p15.32 11q22.3q25 | 5.6 Mb 29.5 Mb | deletion amplification | pathogenic (Cri-du-chat syndrome) pathogenic | TOP |
| 6 | CI PI 33 y/o cFTS: NA | 16 | NB hypoplasia, ventriculomegaly, hyperechogenic kidneys, SUA | 12q14.3q21.1 | 6.13 Mb | deletion | pathogenic | TOP |
| 7 | CVI PV 40 y/o NT 2.0 mm; CRL 78mm; NB (-) cFTS: high risk | 17 | NB hypoplasia, micrognathia | 6q14.1q16.3 | 20.4 Mb | deletion | pathogenic | TOP |
| 8 | CII PI 28 y/o NT 1.4 mm, CRL 53 mm cFTS: low risk | 22 | NB hypoplasia, DORV, PLSVC CCA, club foot | normal CMA | NA | |||
| 9 | CI PI 21 y/o NT 2.1mm; CRL 50mm, NB(-); TR cFTS: high risk | 15 | NB hypoplasia; tetralogy of Fallot | normal CMA | NA | |||
| 10 | CIIPII 36 y/o NT 1.0 mm, CRL 71 mm cFTS: low risk | 19 | NB hypoplasia, anomaly of the sacral spine—“human tail” | normal CMA | NA | |||
| 11 | CII PII 29 y/o NT 2.0 mm, CRL 62 mm cFTS: NA | 19 | NB hypoplasia, severe shortening and bowing of the long bones, trigonocephaly | normal CMA | NA | |||
| 12 | CVI PII 33 y/o NT 5.0 mm; CRL 65 mm NB(-), foetal oedema, TR cFTS: high risk | 13 (CVS) 16 | NB hypoplasia, body stalk anomaly | CVS: arr(7)x3 AC: normal CMA | trisomy 7 | pathogenic | stillbirth 27 weeks GA | |
| 13 | CIV PII 32 y/o NT 2.0 mm; CRL 58 mm cFTS: NA | 19 | NB hypoplasia, hypotrophy | triploidy 69,XXY | pathogenic | TOP | ||
| 14 | CV PII 37 y/o NT 1.2 mm; CRL 55 mm cFTS: low risk | 20 | NB hypoplasia, SUA, skeletal defects, shortening of long bones, cerebellar hypoplasia, preaxial polydactyly in the feet, ventricular septal defect, hypospadias with bifid scrotum | 7p12.3p14.1 | 7 Mb | deletion | pathogenic (Greig cephalopolysyndactyly syndrome) | TOP |
| 15 | CI PI 32 y/o cFTS: NA | 17 | NB hypoplasia, omphalocele, diaphragmatic hernia | 4q28.2q28.3 | 1.8 Mb | deletion | potentially pathogenic | TOP |
| 16 | CII PII 32 y/o cFTS: NA | 18 | NB hypoplasia, NF, polydactyly, kidney hypoplasia, cerebellum abnormality | normal CMA | NA | |||
| 17 | CII PII 37 y/o cFTS: NA | 18 | NB hypoplasia, SUA, shortening of long bones, hypospadias, hyperechogenic kidneys, cerebellar vermis hypoplasia | normal CMA | NA | |||
| 18 | CI PI 24 y/o cFTS: NA maternal cleidocranial dysplasia | 19 | NB hypoplasia, hypertelorism, shortening of long bones, significantly shortened clavicles, small shoulder blades | normal CMA cleidocranial dysplasia | LB, 39 weeks GA, 2690 g, cleidocranial dysplasia | |||
| 19 | CII PII 34 y/o cFTS: NA maternal focal dermal hypoplasia | 16 | NB hypoplasia, retrognathia, hand cleft, syndactyly 2 and 3 fingers of both hands, syndactyly 4 and 5 left hand, splits of the left foot, pulmonary stenosis | normal CMA focal dermal hypoplasia | LB, 36 weeks GA, focal dermal hypoplasia | |||
| 20 | CI PI 30 y/o NT 2.1 mm; CRL 61 mm cFTS: intermediate risk maternal craniofacial-deafness-hand syndrome | 18 | NB hypoplasia, hypertelorism, clinodactyly, ulnar deviations of the fingers, brachydactyly | normal CMA craniofacial-deafness-hand syndrome | NA | |||
| 21 | CVPII 35 y/o NT 5.4 mm CRL 64 mm cFTS: high risk | 15 | NB hypoplasia, NF, hypotelorism, short ribs, narrow chest, shortening of long bones | normal CMA Jeune syndrome | TOP |
| Number of Cases | Normal CMA Results | Abnormal CMA Results (% of Cases) | |
|---|---|---|---|
| isolated NB hypoplasia | 28 | 26 | 2 (7.1%) |
| soft marker + NB hypoplasia | 11 | 10 | 1 (9.1%) |
| multiple abnormalities + NB hypoplasia | 21 | 9 | 12 (57%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moczulska, H.; Serafin, M.; Wojda, K.; Borowiec, M.; Sieroszewski, P. Fetal Nasal Bone Hypoplasia in the Second Trimester as a Marker of Multiple Genetic Syndromes. J. Clin. Med. 2022, 11, 1513. https://doi.org/10.3390/jcm11061513
Moczulska H, Serafin M, Wojda K, Borowiec M, Sieroszewski P. Fetal Nasal Bone Hypoplasia in the Second Trimester as a Marker of Multiple Genetic Syndromes. Journal of Clinical Medicine. 2022; 11(6):1513. https://doi.org/10.3390/jcm11061513
Chicago/Turabian StyleMoczulska, Hanna, Marcin Serafin, Katarzyna Wojda, Maciej Borowiec, and Piotr Sieroszewski. 2022. "Fetal Nasal Bone Hypoplasia in the Second Trimester as a Marker of Multiple Genetic Syndromes" Journal of Clinical Medicine 11, no. 6: 1513. https://doi.org/10.3390/jcm11061513
APA StyleMoczulska, H., Serafin, M., Wojda, K., Borowiec, M., & Sieroszewski, P. (2022). Fetal Nasal Bone Hypoplasia in the Second Trimester as a Marker of Multiple Genetic Syndromes. Journal of Clinical Medicine, 11(6), 1513. https://doi.org/10.3390/jcm11061513






