Prognostic Values of METTL3 and Its Roles in Tumor Immune Microenvironment in Pan-Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression Analysis of METTL3 and Genomic Alterations of METTL3 in Pan-Cancer and Cancer Cell Lines
2.2. Prognostic Value of METTL3 in Pan-Cancer
2.3. Relationships between Expression Levels of METTL3 and Immune Infiltrating Levels, Immune Checkpoint Gene Levels, Immune Neoantigens, Tumor Mutational Burden, Microsatellite Instability, and DNA Mismatch Repair Genes in Pan-Cancer
2.4. Protein–Protein Interaction Network, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis and Gene Set Enrichment Analysis of METTL3
2.5. Validation of Expression Profiles of METTL3 in Gene Expression Omnibus and Clinical Tissues
2.6. Statistical Analysis
3. Results
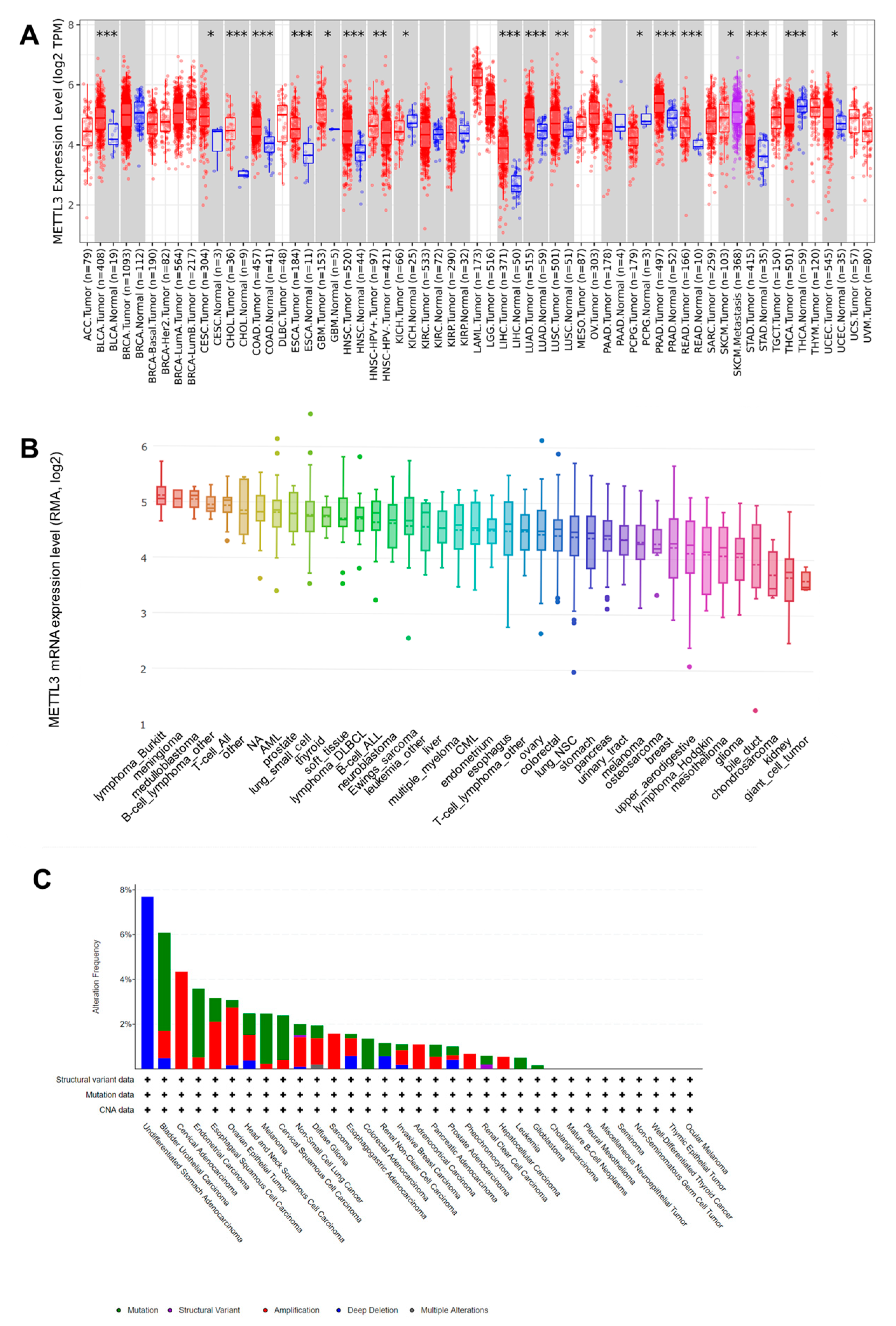
3.1. Upregualted Expression of METTL3 in Pan-Cancer Tissues and Cell Lines and Its Different Genetic Alterations in Cancers
3.2. METTL3 Expression Is Associated with Prognosis in Pan-Cancer
3.3. METTL3 Expression Is Correlated with Immune Infiltrating Levels in Pan-Cancer
3.4. METTL3 Expression Is Correlated with Immune Checkpoint Gene Levels and Immune Neoantigen Loads in Pan-Cancer
3.5. METTL3 Expression Is Correlated with TMB, MSI, and DNA MMR Genes in Pan-Cancer
3.6. METTL3 Is Enriched in Pathways Related to RNA Modification and Metabolism as Well as EMT
3.7. Upregulation of METTL3 Validated Using the GEO Database and Clinical Tissues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, H.; Weng, H.; Chen, J. m6A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019, 12, 121. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Wang, H.; Zhu, L.; Jin, H.; Wang, X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018, 9, 124. [Google Scholar] [CrossRef]
- Xu, X.; Huang, J.; Ocansey, D.K.W.; Xia, Y.; Zhao, Z.; Xu, Z.; Yan, Y.; Zhang, X.; Mao, F. The Emerging Clinical Application of m6A RNA Modification in Inflammatory Bowel Disease and Its Associated Colorectal Cancer. J. Inflamm. Res. 2021, 14, 3289–3306. [Google Scholar] [CrossRef]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wu, Q.; Li, B.; Wang, D.; Wang, L.; Zhou, Y.L. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol. Cancer 2020, 19, 53. [Google Scholar] [CrossRef]
- Zhu, J.; Xiao, J.; Wang, M.; Hu, D. Pan-Cancer Molecular Characterization of m6A Regulators and Immunogenomic Perspective on the Tumor Microenvironment. Front. Oncol. 2021, 10, 618374. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Chen, Z.; Liu, B.; Liu, J.; Wang, H. N6-methyladenine-related genes affect biological behavior and the prognosis of glioma. Cancer Med. 2020, 10, 98–108. [Google Scholar] [CrossRef]
- Lin, G.; Zhang, J.; Wu, Y.; Zhu, S.; Li, G. Signatures and Prognostic Values of N6-methyladenosine (m6A)—Related Immune Genes in Bladder Cancer. Bioengineered 2021, 12, 2649–2663. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96.e11. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hu, X.; Huang, M.; Liu, J.; Gu, Y.; Ma, L.; Zhou, Q.; Cao, X. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019, 10, 1898. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zha, X.; Wang, S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1875, 188522. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhang, Y.; Li, D.; Cai, H.; Cai, L.; Xu, Q. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell. Signal. 2020, 69, 109553. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Huang, J.; Tang, C.; Zou, T.; et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef]
- Maibach, F.; Sadozai, H.; Jafari, S.M.S.; Hunger, R.E.; Schenk, M. Tumor-Infiltrating Lymphocytes and Their Prognostic Value in Cutaneous Melanoma. Front. Immunol. 2020, 11, 2105. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Wang, B.; Zhong, J.-L.; Li, H.-Z.; Wu, B.; Sun, D.-F.; Jiang, N.; Shang, J.; Chen, Y.-F.; Xu, X.-H.; Lu, H.-D. Diagnostic and therapeutic values of PMEPA1 and its correlation with tumor immunity in pan-cancer. Life Sci. 2021, 277, 119452. [Google Scholar] [CrossRef]
- Xu, C.; Zang, Y.; Zhao, Y.; Cui, W.; Zhang, H.; Zhu, Y.; Xu, M. Comprehensive Pan-Cancer Analysis Confirmed That ATG5 Promoted the Maintenance of Tumor Metabolism and the Occurrence of Tumor Immune Escape. Front. Oncol. 2021, 11, 652211. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, Q.; Cheng, C.; Yi, J.; Sun, H.; Wang, Q.; Quan, W.; Xue, Y.; Sun, L.; Cong, X.; et al. Analysis of Bulk RNA Sequencing Data Reveals Novel Transcription Factors Associated With Immune Infiltration Among Multiple Cancers. Front. Immunol. 2021, 12, 3292. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, Q.; Zheng, J.; Hsueh, C.-Y.; Yuan, X.; Heng, Y.; Zhou, L. Diagnostic Role of Dysregulated Circular RNA hsa_circ_0036722 in Laryngeal Squamous Cell Carcinoma. OncoTargets Ther. 2020, 13, 5709–5719. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Hsueh, C.; Guo, Y.; Wu, X.; Li, J.; Zhou, L. Lack of miR-1246 in small extracellular vesicle blunts tumorigenesis of laryngeal carcinoma cells by regulating Cyclin G2. IUBMB Life 2020, 72, 1491–1503. [Google Scholar] [CrossRef]
- Tong, J.; Cao, G.; Zhang, T.; Sefik, E.; Vesely, M.C.A.; Broughton, J.P.; Zhu, S.; Li, H.-B.; Li, B.; Chen, L.; et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018, 28, 253–256. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Wang, C.-X.; Cui, G.-S.; Liu, X.; Xu, K.; Wang, M.; Zhang, X.-X.; Jiang, L.-Y.; Li, A.; Yang, Y.; Lai, W.-Y.; et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018, 16, e2004880. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef]
- Wang, Q.; Geng, W.; Guo, H.; Wang, Z.; Xu, K.; Chen, C.; Wang, S. Emerging role of RNA methyltransferase METTL3 in gastrointestinal cancer. J. Hematol. Oncol. 2020, 13, 57. [Google Scholar] [CrossRef]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 2017, 37, 522–533. [Google Scholar] [CrossRef]
- Chen, H.; Gao, S.; Liu, W.; Wong, C.-C.; Wu, J.; Wu, J.; Liu, D.; Gou, H.; Kang, W.; Zhai, J.; et al. RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 2021, 160, 1284–1300.e16. [Google Scholar] [CrossRef]
- Badalamenti, G.; Fanale, D.; Incorvaia, L.; Barraco, N.; Listi, A.; Maragliano, R.; Vincenzi, B.; Calò, V.; Iovanna, J.L.; Bazan, V.; et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: Can a drop dig a stone? Cell. Immunol. 2019, 343, 103753. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Renz, P.; Wegner, R.E.; Finley, G.G.; Raj, M.S.; Monga, D.K.; McCormick, J.; Kirichenko, A.V. Microsatellite instability (MSI) as an independent predictor of pathologic complete response (PCR) in locally advanced rectal cancer: A National Cancer Database (NCDB) analysis. Ann. Surg. 2020, 271, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Mann, S.A.; Cheng, L. Microsatellite instability and mismatch repair deficiency in the era of precision immuno-oncology. Expert Rev. Anticancer Ther. 2019, 20, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2018, 30, 44–56. [Google Scholar] [CrossRef]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Ritterhouse, L.L. Tumor mutational burden. Cancer Cytopathol. 2019, 127, 735–736. [Google Scholar] [CrossRef]
- Sieviläinen, M.; Almahmoudi, R.; Al-Samadi, A.; Salo, T.; Pirinen, M.; Almangush, A. The prognostic value of immune checkpoints in oral squamous cell carcinoma. Oral Dis. 2018, 25, 1435–1445. [Google Scholar] [CrossRef]
- Toor, S.M.; Nair, V.S.; Decock, J.; Elkord, E. Immune checkpoints in the tumor microenvironment. Semin. Cancer Biol. 2019, 65, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yin, K.; Zhang, Y.; Tian, J.; Wang, S. The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2021, 1876, 188609. [Google Scholar] [CrossRef] [PubMed]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 2019, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhuo, L.; Wang, J.; Zhang, Q.; Li, Q.; Li, G.; Yan, L.; Jin, T.; Pan, T.; Sui, X.; et al. METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 2020, 10, 1631–1646. [Google Scholar] [CrossRef]
- Zeng, C.; Huang, W.; Li, Y.; Weng, H. Roles of METTL3 in cancer: Mechanisms and therapeutic targeting. J. Hematol. Oncol. 2020, 13, 117. [Google Scholar] [CrossRef]
- Zheng, W.; Dong, X.; Zhao, Y.; Wang, S.; Jiang, H.; Zhang, M.; Zheng, X.; Gu, M. Multiple Functions and Mechanisms Underlying the Role of METTL3 in Human Cancers. Front. Oncol. 2019, 9, 1403. [Google Scholar] [CrossRef]
- Zhou, Y.; Kong, Y.; Fan, W.; Tao, T.; Xiao, Q.; Li, N.; Zhu, X. Principles of RNA methylation and their implications for biology and medicine. Biomed. Pharmacother. 2020, 131, 110731. [Google Scholar] [CrossRef]
- Lin, X.; Chai, G.; Wu, Y.; Li, J.; Chen, F.; Liu, J.; Luo, G.; Tauler, J.; Du, J.; Lin, S.; et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019, 10, 2065. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, R.; Yuan, L. Crosstalk of intracellular post-translational modifications in cancer. Arch. Biochem. Biophys. 2019, 676, 108138. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Heng, Y.; Chen, H.; Huang, Q.; Wu, C.; Tao, L.; Zhou, L. Prognostic Values of METTL3 and Its Roles in Tumor Immune Microenvironment in Pan-Cancer. J. Clin. Med. 2023, 12, 155. https://doi.org/10.3390/jcm12010155
Guo Y, Heng Y, Chen H, Huang Q, Wu C, Tao L, Zhou L. Prognostic Values of METTL3 and Its Roles in Tumor Immune Microenvironment in Pan-Cancer. Journal of Clinical Medicine. 2023; 12(1):155. https://doi.org/10.3390/jcm12010155
Chicago/Turabian StyleGuo, Yang, Yu Heng, Hui Chen, Qiang Huang, Chunping Wu, Lei Tao, and Liang Zhou. 2023. "Prognostic Values of METTL3 and Its Roles in Tumor Immune Microenvironment in Pan-Cancer" Journal of Clinical Medicine 12, no. 1: 155. https://doi.org/10.3390/jcm12010155
APA StyleGuo, Y., Heng, Y., Chen, H., Huang, Q., Wu, C., Tao, L., & Zhou, L. (2023). Prognostic Values of METTL3 and Its Roles in Tumor Immune Microenvironment in Pan-Cancer. Journal of Clinical Medicine, 12(1), 155. https://doi.org/10.3390/jcm12010155




