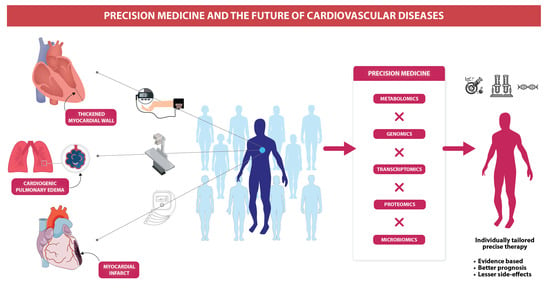
Precision Medicine and the future of Cardiovascular Diseases: A Clinically Oriented Comprehensive Review
Abstract
1. Introduction
2. Precision Medicine and Cardiology
2.1. Myocardial Infarction
| S.N. | Authors | Year | Study Design | Clinically Relevant Findings | Comments | References |
|---|---|---|---|---|---|---|
| PROTEOMICS | ||||||
| 1 | Felicita Andreotti et al. | 2021 | Literature Review | Biological sex (sex chromosomes, hormones, body size, social behavior, etc) affects the incidence of Chronic Coronary Syndrome (CCS). | Gender affects the incidence of CCS. | [31] |
| 2 | Zhanpeng Wen., et al. | 2021 | Scientific Report | Telmisartan-doped co-assembly nanofibers (TDCNfs). | Telmisartan-doped co-assembly nanofibers (TDCNfs) synergistically counter-regulate RAS and can be used in treatment post-MI. | [32] |
| 3 | Gaozan Tong., et al. | 2020 | Scientific Report | Basic fibroblast growth factor (bFGF). | The activation of NrF2 by bFGF lowers oxidative stress. This can significantly lessen the apoptosis of cardiomyocytes and the size of the infarcts brought on by MI, thereby lessening the damage to the heart. | [33] |
| 4 | Apurva Shrivastava., et al. | 2020 | Scientific Report | High-sensitive troponin, miRNAs, natriuretic peptides, CRP, and metabolites. | Deep phenotyping data, blood biobank, cardiac stress-MRI test, and identified candidate biomarkers (such as miRNAs, troponin, CRP, etc.) will be used to derive a biomarker specifically for ischemia. | [34] |
| 5. | Filippo Crea., et al. | 2019 | Review | Myeloperoxidase and hyaluronidase-2. | Potential biomarkers that can non-invasively detect plaque erosion in ACS patients include myeloperoxidase and hyaluronidase-2. | [35] |
| 6. | Rocco A Montone., et al. | 2018 | Review | - Radiographic biomarker: Unstable plaques; - Interruption of treatment with Ca antagonist during follow-up leads to a worse outcome. | A patient-specific treatment plan for MINOCA requires determining the cause. Some diagnostic tools for this purpose include the electrocardiogram (ECG), cardiac enzymes, echocardiography, coronary angiography, left ventricular angiography, coronary vasomotion, and intravascular imaging techniques. | [36] |
| 7. | Kenneth Mangion., et al. | 2017 | Literature Review | Biomechanical parameters of LV function. | The biomechanical aspects of LV function, such as contractility, stiffness, strain, and stress, which are related to LV pump performance and, consequently, prognosis, can be learned through computational modeling. | [37] |
| 8 | Laura Pasea., et al. | 2017 | Scientific Report | BMI, WBC, haemoglobin, creatinine, and cholesterol ratio. | When deciding whether to continue DAPT therapy (in patients 1 year after acute MI), prognostic factors such as demographics, behavior, cardiovascular history, non-cardiovascular history, biomarkers, and medications should be taken into account. | [38] |
| 9 | Emeline Goretti., et al. | 2014 | Review | miRNAs. | miRNAs can be used to diagnose acute MI, stratify MI risk, and other medical conditions. After an MI, circulating miRNA levels have some prognostic value, and BNP’s diagnostic value is enhanced. | [39] |
| 10 | Elena Kaschina., et al. | 2014 | Review | AT2-receptor can be used post-MI for cardiac protection. | AT2R has a protective role in the heart after MI, leading to improved cardiovascular health. The protective function includes improvement of LV contractility, protection from early LV dilatation, and an antifibrotic effect under certain conditions. | [40] |
| 11 | Akinyemi Oni-Orisan., et al. | 2014 | Review | Epoxyeicosatrienoic acids (EETs). | Epoxyeicosatrienoic acids (EETs) possess cardioprotective effects as they elicit anti-inflammatory, vasodilatory, fibrinolytic, and anti-apoptotic effects. They can be used as a treatment for acute MI. | [41] |
| 12 | Nisha Arenja., et al. | 2013 | Scientific Report | Infarct size, LVEF Independent predictors of silent MI are: history of diabetes and CAD. | Better diagnostic capability is provided by imaging methods such as myocardial perfusion single-photon emission computed tomography or cardiac magnetic resonance. Infarct size, left ventricular ejection fraction, a history of diabetes, CAD, and other factors are independent predictors of silent MI. | [42] |
| GENOMICS | ||||||
| 13 | Naveen Pereira., et al. | 2020 | RCT | CYP2C19 LOF mutation (or CYP2C19 LOF alleles carriers). | A genotype (CYP2C19 LOF mutation)-selected P2Y12 inhibitor instead of traditional clopidogrel shows no difference in ischemic events, thereby suggesting the incorporation of precision medicine earlier in the course of PCI. | [43] |
| 14 | Daniel Claassens., et al. | 2019 | RCT | CYP2C19 mutation (or CYP2C19 LOF alleles carriers). | After PCI, genotype (CYP2C19)-guided P2Y12 inhibitor selection was superior to standard treatment in terms of thrombotic events and had less bleeding events. | [44] |
| 15 | Robert A Scott., et al. | 2016 | RCT | Six genes (CNR2, DPP4, GLP1R, SLC5A1, HTR2C, MCHR1) that encode therapeutic targets in development of type 2 diabetes or obesity. | Six genes (CNR2, DPP4, GLP1R, SLC5A1, HTR2C, MCHR1) encode the therapeutic targets in development of type 2 diabetes or obesity. Therefore, these genes can be used to develop drugs with more efficacy and minimal side effects. | [45] |
| METABOLOMICS | ||||||
| 16 | Gaetano Tanzilli., et al. | 2021 | RCT | Glutathione. | Institution of early and prolonged infusion of glutathione in a ST-segment elevation MI patients having undergone PCI can reduce the inflammatory response, thereby protecting the myocardial cells and improving cardiac remodeling. | [46] |
| 17 | Bicciré F., et al. | 2021 | Observational study | Serum albumin level. | Less than normal serum albumin levels were predictive of complications in STEMI patients. | [47] |
| 18 | Sayanti Bhattacharya., et al. | 2014 | Case-Control Study | Branched-chain amino acid metabolites. | BCAA and related metabolites are associated with atherosclerosis. | [48] |
| 19 | Svati Shah., et al. | 2012 | Case-Control Study | Medium-chain acylcarnitines, short-chain dicarboxylacylcarnitines, long-chain dicarboxylacylcarnitines, long-chain acylcarnitines, short-chain acylcarnitines, medium-chain acylcarnitines, ketone-related, cholesterol, lipids, fatty acids, glucose, branched-chain amino acids and related catabolites, amino acids. | Branch-chain amino acids, medium-chain acylcarnitines, short-chain and long-chain dicarboxylacylcarnitines, and fatty acids were all independently linked to mortality. Fatty acids, on the other hand, were predictors of MI/death, as were short-chain, long-chain, and dicarboxylacylcarnitines. | [49] |
2.1.1. Metabolomics
2.1.2. Genomics
2.1.3. Biomarkers
2.2. Hypertension
| S.N. | Authors | Year | Study Design | Clinically Relevant Findings | Comments | References |
|---|---|---|---|---|---|---|
| METABOLOMICS | ||||||
| 1 | Phil Chowienczyk et al. | 2014–2023 | Ongoing RCT | Pharmaco-metabolomics of antihypertensive drug response. | AIM HY study is underway to investigate biochemical pathways, pharmaco-metabolomics, and pharmacogenomics in antihypertensive drug response which can further be used for precision therapy in hypertension. | [55] |
| 2 | Diana M Rydberg et al. | 2018 | Cross-sectional study | Sex in antihypertensive drug response. | More adverse events (ADE) were seen in women compared to most antihypertensive drugs, however, aldosterone antagonists showed an increased incidence of ADE in men. | [56] |
| 3 | Yan Gong et al. | 2016 | Meta-Analysis | Obesity and ancestry in response to beta blockers. | In terms of ethnic diversity, diuretics and CCBs work better than ACEIs in African Americans due to the genetic variations in the RAAS. Obesity also influences antihypertensive drug response. Obese patients report better response to beta blockers due to their enhanced sympathetic activity, causing increased cardiac output. | [57] |
| 4 | Yidan Zhao et al. | 2014 | Experimental Study | Dicarboxylic acid (hexadecanedioate) as a causal pathway for hypertension. | Metabolomic profiling study on pulmonary hypertension supported a novel mechanism for blood pressure regulation that involved dicarboxylic acid (hexadecanedioate). It disrupted beta-oxidation and increased omega-oxidation pertaining to its novel putative role in both pulmonary and systemic hypertension. Identification of this metabolomic pathway potentiates further research in precision medicine for hypertension. | [58] |
| 5 | Lizzy M Brewster et al. | 2013 | Systematic Review | African ancestry in antihypertensive drug response. | African Americans are prone to severe hypertension due to higher vascular contractility and salt-holding capacity. Thus, ethnicity and ancestry provide a valuable marker, and a predictor of blood pressure lowering response to antihypertensives for devising individual treatment options is imperative. | [59] |
| 6 | William R Wikoff et al. | 2013 | RCT | Atenolol-induced changes in metabolome help understand the differential response to beta-blockers. | Metabolomic analysis of hypertension helps understand the disease as it is closely knit with the alteration of metabolic pathways. Atenolol-induced changes in the metabolome have been shown to depend on race and genotype, and this may explain the differential metabolomics of response to β-blockers in terms of blood pressure control. | [60] |
| GENOMICS | ||||||
| 7 | Lakshmanan Loganathan et al. | 2020 | Literature Review | Identification of deleterious SNPs and target proteins in rational drug design through computational screening of mutagenesis. | The identification of deleterious SNPs involved in RAAS signaling genes that play an associative role with hypertension could contribute to antihypertensive drug discovery and diagnosis. Moreover, Cytochrome P (CYP) polymorphisms vouch for individual and population differences in drug tolerance. | [61] |
| 8 | Ian M. Kronish et al. | 2019 | Pilot Study | - | Personalized trial with antihypertensive treatment carried out with seven patients. The study found that patients were able to choose a particular antihypertensive drug at the end of the study period based on personal preference through daily blood pressure monitoring and were able to decide which treatment was personally more beneficial to them in lowering their blood pressure. | [62] |
| 9 | Boon-Peng Hoh et al. | 2019 | Literature Review | Ancestral alleles in salt homeostasis and identification of “Intermediate Phenotypes” for hypertension through GWAS. | Natural selection and evolution have a role in blood pressure variability due to varying frequencies of ancestral alleles that regulated salt homeostasis in humans. In a search carried out in the GWAS catalog in 2018, seven candidate genes with an established pathophysiological mechanism pertaining to hypertension were replicated in GWAS, namely ACE1, ACE2, ADRB1, ADRB2, MME, CACNA2D2, and UMOD. This implies that genetic variants concerning hypertension are comparatively more common (>1% in a population), with negligible natural selection in the past populations. | [63] |
| 10 | Yoshihiro Kokubo et al. | 2019 | Literature Review | Gene–environment and gene–lifestyle interactions in hypertension treatment. | Hypertension management must incorporate gene–environment interactions in addition to lifestyle components to prevent and treat hypertension. Gene–obesity, gene–physical activity, gene–sodium, gene–alcohol, gene–smoking, and gene–healthy-diet interactions contribute to the choice of treatment for hypertension as it is a multifactorial disease. | [64] |
| 11 | Evangelos Evangelou et al. | 2017 | Meta-Analysis | Polygenic risk score and genetic risk score, drug target genetic loci identified for drug development. | Application of polygenic risk score and genetic risk score in estimation of hypertension manifestation and drug response. It has been reported that five loci (PKD2L1, SLC12A2, CACNA1C, CACNB4, and CA7) are drug targets for hypertension medications. More than 1000 BP-determining loci have now been identified and drug-target genes are expected to expand in the future. | [65] |
| 12 | David L. Mattson et al. | 2017 | Editorial | Functional genomics in disease-relevant tissues. | Taking into account only DNA sequence for precision medicine in hypertension is an incomplete approach because hypertension is also influenced by environmental factors, which pose a challenge to quantify and identify. Functional genomics should be carried out in the disease-relevant tissues to contribute to precision medicine for hypertension. | [66] |
| 13 | Surendran P. et al. | 2016 | Meta-Analysis | Allelic variations in hypertension. | Identified 31 novel hypertension-associated genetic regions that include three unidentified missense variants in RBM47, COL21A1, and RRAS. Various associations were found in A2ML1 and ENPEP. These allelic variations lay the foundation for new drug targets for clinical interventions and management in precision hypertension. | [67] |
| 14 | Wenzheng Zhang | 2015 | Editorial | Modified sodium channel epithelial 1α subunit (SCNN1A). | Epigenomics has helped to define groups of hypertensive patients who respond superiorly to aldosterone-based therapies. It targets the epigenetically modified sodium channel epithelial 1α subunit (SCNN1A) in a specific group of hypertensive individuals. | [68] |
| 15 | Timo P Hiltunen et al. | 2015 | RCT | TET2, CSMD1 response to thiazide diuretic. | Identified TET2 and CSMD1 as loci linked to systolic blood pressure lowering response to hydrochlorothiazides. | [69] |
| 16 | Matteo Trudu et al. | 2014 | Cohort | UMOD gene polymorphisms in differential response to loop diuretics in hypertensive patients. | UMOD polymorphisms have been linked to a better BP response to loop diuretics in essential hypertension patients. “High” UMOD group has better hypertension control to loop diuretics while “low” UMOD group has a poorer response to loop diuretics. | [70] |
| 17 | Anne M Muskalla et al. | 2014 | Observational Study | G-protein receptor kinase 4 (GRK4) polymorphism response to antihypertensives. | GRK4 polymorphisms R65L, A142V, and A486V were analyzed, and it was found that homozygote double variants of 65 L and 142 V required more antihypertensive therapy than did homozygous single variants or heterozygous carriers to reach the optimal mean arterial BP reading. | [71] |
| 18 | Hye Duck Choi et al. | 2013 | Meta-Analysis | ADD1 and ACE I/D polymorphisms’ role in HCT response. | ADD1 polymorphism Gly460Trp: The carriers of the Gly/Gly genotype had an enhanced response to hydrochlorothiazide compared to carriers of the Trp/Trp genotype. | [72] |
| 19 | J D Duarte et al. | 2012 | Original Study | PRKCA and GNAS-EDN3 in response to thiazide. | On administration of thiazide diuretics, PRKCA (rs16960228) allele carriers had better hypertension control than GG homozygote patients. Additionally, GNAS–EDN3 rs2273359 G allele carriers showed a better blood pressure response than CC homozygote patients. | [73] |
| TRANSCRIPTOMICS | ||||||
| 20. | Brian G Bazzell et al. | 2018 | Experimental Study | The RNA-sequencing of urinary vesicles (extracellular) on molecular modification in the mineralocorticoid receptor activation showed changes in the mRNA found in urine supernatant. This may contribute to better-tailored pharmacological treatment in mineralocorticoid signaling disorders such as resistant hypertension. Furthermore, it may help in the noninvasive identification of new putative biomarkers for cardiovascular and renal diseases and also predict drug responses. | Measurement of mRNA transcripts in the urine as a marker for mineralocorticoid receptor activation can help predict the response to mineralocorticoid receptor antagonists in hypertension. | [74] |
2.2.1. Hypertension Pharmacogenomics
2.2.2. Hypertension Metabolomics
2.2.3. Resistant Hypertension
2.3. Heart Failure
| S.N. | Authors | Year | Study Design | Clinically Relevant Findings | Comments | References |
|---|---|---|---|---|---|---|
| PROTEOMICS | ||||||
| 1 | Leanne Dumeny et al. | 2021 | RCT | NR3C2 rs5522 G allele. | NR3C2, which codes for the spironolactone target protein, and CYP11B2, which is implicated in aldosterone synthesis, were linked to improved spironolactone responsiveness in diastolic HF patients. | [84] |
| 2 | Shah S et al. | 2020 | Meta-Analysis | KLHL3 and SYNPOL2–AGAP5. | Causative of HF. | [49] |
| 3 | Maurer, M. S et al. | 2018 | RCT | Gene-encoding autosomal dominant Val122Ile variant. | Tafamidis reduced death and hospitalizations associated with cardiovascular events in patients with transthyretin-associated cardiomyopathy. | [85] |
| 4 | Dominguez, F et al. | 2018 | Review | BLC2-associated athanogene 3 (BAG3). | Loci KLHL3 and SYNPOL2–AGAP5 are implicated in of HF, and also BAG3 and CDKN1A are associated with LV systolic dysfunction. | [86] |
| GENOMICS | ||||||
| 5 | Julio Núñez et al. | 2021 | Original Study | Plasma carbohydrate antigen 125 (CA125). | CA125 is a surrogate of fluid overload, hence potentially valuable for guiding decongestion therapy, and a CA125-guided diuretic strategy improved eGFR in patients with acute heart failure with renal dysfunction. | [87] |
| 6 | Pierpaolo Pellicori et al. | 2020 | RCT | Collagen type I C-terminal telopeptide (CITP) and galectin-3. | Spironolactone affects various pathways that contribute to HF progression. | [88] |
| 7 | Feng SD et al. | 2017 | Observational Study | Beta-endorphin (β-EP) and brain natriuretic peptide (BNP) plasma concentrations. | Patients with acute left heart failure or atrial fibrillation can be identified early with excellent specificity and sensitivity using β-endorphin and brain natriuretic peptide. | [89] |
| 8 | Chester L. Drum et al. | 2017 | Comparative Study | Plasma Thymosin Beta-4 (TB4). | Heart failure with preserved ejection fraction (HFpEF) in females is associated with an increase in plasma TB4, which independently predicts mortality | [90,91] |
| 9 | G Michael Felker et al. | 2015 | RCT | High-sensitivity cardiac troponin T (hs-cTnT). | High levels of hs-cTnT were found in those with AHF. An increase in hs-cTnT, whether at baseline or peak, is associated with poor outcomes, most notably cardiovascular mortality at 180 days. | |
| 10 | Zoltán Pozsonyi, et al. | 2014 | Clinical Trial | Copeptin. | Copeptin can be used to predict a 5-year all-cause mortality in patients with heart failure. | [92] |
| 11 | Glick D et al. | 2013 | Observational Study | cTnT and NT-proBNP. | Systolic dysfunction, incident HF, and CV death can all be predicted from the long-term trajectory of cardiac troponin T(cTnT) and N-terminal pro-brain natriuretic peptide(NT-proBNP) in older adults without HF. | [93] |
| 12 | Hanna K Gaggin et al. | 2013 | Clinical Trial | sST2. | Soluble suppression of tumorigenesis (sST2) measurement identifies patients with chronic heart failure in whom higher beta-blocker doses may be beneficial | [94] |
| 13 | Shah RV et al. | 2012 | Original Study | MR-proANP and MR-proANP and MR-proADM. | Both mid-regional pro-atrial natriuretic peptide (MR-proANP) and mid-regional pro-adrenomedullin (MR-proADM) are effective for estimating prognosis in acute decompensated heart failure (ADHF). | [50] |
| 14 | Sjoukje I Lok et al. | 2012 | Original Study | Growth differentiation factor 15. | Growth differentiation factor 15 (GDF) levels are increased in patients with HF and correlate with the extent of myocardial fibrosis, hence they are used as a biomarker for cardiac remodeling | [95] |
| 15 | Natalia Lopez-Andrès et al. | 2012 | RCT | Galectin-3 (Gal-3), N-terminal propeptides of type I and III procollagens (PINP and PIIINP), and matrix metalloproteinase 1 (MMP-1). | Increased galectin-3 (Gal-3) and N-terminal propeptide III procollagen (PIIINP) levels, as well as low (metallic metalloproteinase-1 (MMP-1) levels, have been linked to poor long-term cardiovascular outcomes. | [96] |
| METABOLOMICS | ||||||
| 16 | Olivotto, I. et al. | 2020 | RCT | Selective allosteric inhibitor of cardiac myosin ATPase. | Patients with obstructive hypertrophic cardiomyopathy saw improvements in exercise capacity, LVOT blockage, NYHA functional class, and health status after treatment with mavacamten, a small molecule modulator of β-cardiac myosin | [97] |
| 17 | Hunter WG et al. | 2016 | Original Study | Medium- and long-chain acylcarnitines and ketone bodies. | Patients with HFpEF had higher levels of medium- and long-chain acylcarnitines and ketone bodies than those with HFrEF. | [98] |
| 18 | Du Z et al. | 2014 | Original Study | 3-hydroxybutyrate, acetone, and succinate, were significantly elevated in patients with HFrEF. | Patients with HFrEF had higher levels of 3-hydroxybutyrate, acetone, and succinate, all of which are predictive of HF outcomes. | [99] |
| 19 | Wang Li et al. | 2013 | Original Study | Lactate, alanine, creatinine, proline, isoleucine, and leucine in plasma. | Levels of lactate, alanine, creatine, proline, isoleucine, and leucine in plasma were all greater in patients with ischemic HFrEF compared to healthy controls. | [100] |
| 20 | Desmoulin F et al. | 2013 | Original Study | Plasma lactate and total cholesterol. | Patients with acute decompensated HF who have a high ratio of plasma lactate to total cholesterol have a significantly higher risk of dying within 30 days (ADHF). | [101] |
| MICROBIOMICS | ||||||
| 21 | W H Wilson Tang et al. | 2014 | Original Study | Trimethylamine N-oxide (TMAO). | Increased mortality risk over the long run was associated with elevated TMAO levels. | [102] |
2.3.1. Genomics
2.3.2. Proteomics
2.3.3. Metabolomics
2.3.4. Microbiomics
2.4. Precision Medicine and Aortic Diseases
3. Precision Cardiology and Artificial Intelligence
4. Cardiovascular Pharmacology and Precision Medicine
5. Precision Cardiology and the Omics
6. Evolving Understanding of the Immune Cells and the Future of Precision Cardiology
7. Challenges to PM in Cardiology
8. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vos, T.; Allen, C.; Arora, M.; Barber, R.M.; Bhutta, Z.A.; Brown, A.; Carter, A.; Casey, D.C.; Charlson, F.J.; Chen, A.Z.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 310 Diseases and Injuries, 1990–2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1545–1602. [Google Scholar] [CrossRef]
- Roth, G.A.; Johnson, C.O.; Abajobir, A.A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.Y.; Ahmed, M.B.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef]
- Roth, G.A.; Mensah, G.A.; Johnson, C.O.; Addolorato, G.; Ammirati, E.; Baddour, L.M.; Barengo, N.C.; Beaton, A.Z.; Benjamin, E.J.; Benziger, C.P.; et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019. J. Am. Coll. Cardiol. 2020, 76, 2982–3021. [Google Scholar] [CrossRef]
- WHO. WHO Top10 Causes of Death; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- CDC Health and Economic Costs of Chronic Diseases. Available online: https://www.cdc.gov/chronicdisease/about/costs/index.htm (accessed on 10 February 2023).
- Masaebi, F.; Salehi, M.; Kazemi, M.; Vahabi, N.; Azizmohammad Looha, M.; Zayeri, F. Trend Analysis of Disability Adjusted Life Years Due to Cardiovascular Diseases: Results from the Global Burden of Disease Study 2019. BMC Public Health 2021, 21, 1268. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The Growing Role of Precision and Personalized Medicine for Cancer Treatment. Technology 2018, 06, 79–100. [Google Scholar] [CrossRef] [PubMed]
- Leopold, J.A.; Loscalzo, J. Emerging Role of Precision Medicine in Cardiovascular Disease. Circ. Res. 2018, 122, 1302–1315. [Google Scholar] [CrossRef] [PubMed]
- Antman, E.M.; Loscalzo, J. Precision Medicine in Cardiology. Nat. Rev. Cardiol. 2016, 13, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Currie, G.; Delles, C. Precision Medicine and Personalized Medicine in Cardiovascular Disease. Adv. Exp. Med. Biol. 2018, 1065, 589–605. [Google Scholar] [PubMed]
- Krittanawong, C.; Zhang, H.; Wang, Z.; Aydar, M.; Kitai, T. Artificial Intelligence in Precision Cardiovascular Medicine. J. Am. Coll. Cardiol. 2017, 69, 2657–2664. [Google Scholar] [CrossRef]
- GALLONE, G.; BRUNO, F.; D’ASCENZO, F.; de Ferrari, G.M. What Will We Ask to Artificial Intelligence for Cardiovascular Medicine in the next Decade? Minerva Cardiol. Angiol. 2022, 70, 92–101. [Google Scholar] [CrossRef]
- Simeon, M.; Dangwal, S.; Sachinidis, A.; Doss, M.X. Application of the Pluripotent Stem Cells and Genomics in Cardiovascular Research-What We Have Learnt and Not Learnt until Now. Cells 2021, 10, 3112. [Google Scholar] [CrossRef] [PubMed]
- Pullamsetti, S.S.; Tello, K.; Seeger, W. Utilising Biomarkers to Predict Right Heart Maladaptive Phenotype: A Step toward Precision Medicine. Eur. Respir. J. 2021, 57, 2004506. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, F.; Lupieri, A.; Aikawa, M.; Aikawa, E. Harnessing Single-Cell RNA Sequencing to Better Understand How Diseased Cells Behave the Way They Do in Cardiovascular Disease. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 585. [Google Scholar] [CrossRef] [PubMed]
- Schiano, C.; Benincasa, G.; Franzese, M.; della Mura, N.; Pane, K.; Salvatore, M.; Napoli, C. Epigenetic-Sensitive Pathways in Personalized Therapy of Major Cardiovascular Diseases. Pharmacol. Ther. 2020, 210, 107514. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Zeller, T. Biomarkers in Primary Prevention. Herz 2019, 45, 10–16. [Google Scholar] [CrossRef]
- Gruson, D.; Bernardini, S.; Dabla, P.K.; Gouget, B.; Stankovic, S. Collaborative AI and Laboratory Medicine Integration in Precision Cardiovascular Medicine. Clin. Chim. Acta 2020, 509, 67–71. [Google Scholar] [CrossRef]
- Visco, V.; Ferruzzi, G.J.; Nicastro, F.; Virtuoso, N.; Carrizzo, A.; Galasso, G.; Vecchione, C.; Ciccarelli, M. Artificial Intelligence as a Business Partner in Cardiovascular Precision Medicine: An Emerging Approach for Disease Detection and Treatment Optimization. Curr. Med. Chem. 2021, 28, 6569–6590. [Google Scholar] [CrossRef]
- AMBROSINI, S.; MOHAMMED, S.A.; COSTANTINO, S.; PANENI, F. Disentangling the Epigenetic Landscape in Cardiovascular Patients: A Path toward Personalized Medicine. Minerva Cardiol. Angiol. 2021, 69, 331–345. [Google Scholar] [CrossRef]
- Pan, Z.; Ebert, A.; Liang, P. Human-Induced Pluripotent Stem Cells as Models for Rare Cardiovascular Diseases: From Evidence-Based Medicine to Precision Medicine. Pflugers Arch. 2021, 473, 1151–1165. [Google Scholar] [CrossRef]
- Gualandro, D.M.; Twerenbold, R.; Boeddinghaus, J.; Nestelberger, T.; Puelacher, C.; Müller, C. Biomarkers in Cardiovascular Medicine: Towards Precision Medicine. Swiss Med. Wkly. 2019, 149, w20125. [Google Scholar] [CrossRef]
- Rodríguez-Carrio, J.; López, P.; Suárez, A. EPC Dysfunction and Immune Networks: Translating Opportunities for Clinical Setting in Personalized Medicine. Curr. Med. Chem. 2018, 25, 4497–4506. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.D.; Rayner, B.L. Hypertension in Blacks. Hypertension 2018, 18, 607–622. [Google Scholar] [CrossRef] [PubMed]
- Halushka, P.V.; Goodwin, A.J.; Halushka, M.K. Opportunities for MicroRNAs in the Crowded Field of Cardiovascular Biomarkers. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 211–238. [Google Scholar] [CrossRef] [PubMed]
- Zaiou, M.; el Amri, H. Cardiovascular Pharmacogenetics: A Promise for Genomically-guided Therapy and Personalized Medicine. Clin. Genet 2017, 91, 355–370. [Google Scholar] [CrossRef]
- MacRae, C.A.; Roden, D.M.; Loscalzo, J. The Future of Cardiovascular Therapeutics. Circulation 2016, 133, 2610–2617. [Google Scholar] [CrossRef]
- Nishikimi, T.; Kuwahara, K.; Nakagawa, Y.; Kangawa, K.; Nakao, K. Adrenomedullin in Cardiovascular Disease: A Useful Biomarker, Its Pathological Roles and Therapeutic Application. Curr. Protein. Pept. Sci. 2013, 14, 256–267. [Google Scholar] [CrossRef]
- Pollard, T.J. THE ACUTE MYOCARDIAL INFARCTION. Prim. Care Clin. Off. Pract. 2000, 27, 631–649. [Google Scholar] [CrossRef]
- Mohan, S.; Lynch, S.; Cummings, T.A. Time Equals Myocardium: Are We in Time? West Indian Med. J. 2010, 59, 680–685. [Google Scholar]
- Andreotti, F.; Maggioni, A.P.; Scambia, G. Sex- and Gender-specific Precision Medicine for Chronic Coronary Syndromes: Challenges and Opportunities. Kardiol. Pol. 2021, 79, 373–375. [Google Scholar] [CrossRef]
- Wen, Z.; Zhan, J.; Li, H.; Xu, G.; Ma, S.; Zhang, J.; Li, Z.; Ou, C.; Yang, Z.; Cai, Y.; et al. Dual-Ligand Supramolecular Nanofibers Inspired by the Renin-Angiotensin System for the Targeting and Synergistic Therapy of Myocardial Infarction. Theranostics 2021, 11, 3725–3741. [Google Scholar] [CrossRef]
- Tong, G.; Liang, Y.; Xue, M.; Chen, X.; Wang, J.; An, N.; Wang, N.; Chen, Y.; Wang, Y.; Jin, L.; et al. The Protective Role of BFGF in Myocardial Infarction and Hypoxia Cardiomyocytes by Reducing Oxidative Stress via Nrf2. Biochem. Biophys. Res. Commun. 2020, 527, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.; Marzolla, V.; Weidmann, H.; Caprio, M.; Tregouet, D.-A.; Zeller, T.; Karakas, M. Design and Rationale of the ERA-CVD Consortium PREMED-CAD—Precision Medicine in Coronary Artery Disease. Biomolecules 2020, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Crea, F.; Vergallo, R. Plaque Erosion: Towards Precision Medicine in Acute Coronary Syndromes. Int. J. Cardiol. 2019, 288, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Montone, R.A.; Russo, M.; Niccoli, G. MINOCA: Current Perspectives. Aging 2018, 10, 3044–3045. [Google Scholar] [CrossRef] [PubMed]
- Mangion, K.; Gao, H.; Husmeier, D.; Luo, X.; Berry, C. Advances in Computational Modelling for Personalised Medicine after Myocardial Infarction. Heart 2018, 104, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Pasea, L.; Chung, S.-C.; Pujades-Rodriguez, M.; Moayyeri, A.; Denaxas, S.; Fox, K.A.A.; Wallentin, L.; Pocock, S.J.; Timmis, A.; Banerjee, A.; et al. Personalising the Decision for Prolonged Dual Antiplatelet Therapy: Development, Validation and Potential Impact of Prognostic Models for Cardiovascular Events and Bleeding in Myocardial Infarction Survivors. Eur. Heart J 2017, 38, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Goretti, E.; Wagner, D.R.; Devaux, Y. MiRNAs as Biomarkers of Myocardial Infarction: A Step Forward towards Personalized Medicine? Trends Mol. Med. 2014, 20, 716–725. [Google Scholar] [CrossRef]
- Kaschina, E.; Lauer, D.; Schmerler, P.; Unger, T.; Steckelings, U.M. AT2 Receptors Targeting Cardiac Protection Post-Myocardial Infarction. Curr. Hypertens Rep. 2014, 16, 441. [Google Scholar] [CrossRef]
- Oni-Orisan, A.; Alsaleh, N.; Lee, C.R.; Seubert, J.M. Epoxyeicosatrienoic Acids and Cardioprotection: The Road to Translation. J. Mol. Cell Cardiol. 2014, 74, 199–208. [Google Scholar] [CrossRef]
- Arenja, N.; Mueller, C.; Ehl, N.F.; Brinkert, M.; Roost, K.; Reichlin, T.; Sou, S.M.; Hochgruber, T.; Osswald, S.; Zellweger, M.J. Prevalence, Extent, and Independent Predictors of Silent Myocardial Infarction. Am. J. Med. 2013, 126, 515–522. [Google Scholar] [CrossRef]
- Pereira, N.L.; Farkouh, M.E.; So, D.; Lennon, R.; Geller, N.; Mathew, V.; Bell, M.; Bae, J.-H.; Jeong, M.H.; Chavez, I.; et al. Effect of Genotype-Guided Oral P2Y12 Inhibitor Selection vs Conventional Clopidogrel Therapy on Ischemic Outcomes After Percutaneous Coronary Intervention. JAMA 2020, 324, 761. [Google Scholar] [CrossRef] [PubMed]
- Claassens, D.M.F.; Vos, G.J.A.; Bergmeijer, T.O.; Hermanides, R.S.; van ’t Hof, A.W.J.; van der Harst, P.; Barbato, E.; Morisco, C.; Tjon Joe Gin, R.M.; Asselbergs, F.W.; et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N. Engl. J. Med. 2019, 381, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.A.; Freitag, D.F.; Li, L.; Chu, A.Y.; Surendran, P.; Young, R.; Grarup, N.; Stancáková, A.; Chen, Y.; Varga, T.v.; et al. A Genomic Approach to Therapeutic Target Validation Identifies a Glucose-Lowering GLP1R Variant Protective for Coronary Heart Disease. Sci. Transl. Med. 2016, 8, 341ra76. [Google Scholar] [CrossRef] [PubMed]
- Tanzilli, G.; Arrivi, A.; Placanica, A.; Viceconte, N.; Cammisotto, V.; Nocella, C.; Barillà, F.; Torromeo, C.; Pucci, G.; Acconcia, M.C.; et al. Glutathione Infusion Before and 3 Days After Primary Angioplasty Blunts Ongoing NOX2-Mediated Inflammatory Response. J. Am. Heart Assoc. 2021, 10, e020560. [Google Scholar] [CrossRef] [PubMed]
- Bicciré, F.G.; Pastori, D.; Tanzilli, A.; Pignatelli, P.; Viceconte, N.; Barillà, F.; Versaci, F.; Gaudio, C.; Violi, F.; Tanzilli, G. Low Serum Albumin Levels and In-Hospital Outcomes in Patients with ST Segment Elevation Myocardial Infarction. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 2904–2911. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Granger, C.B.; Craig, D.; Haynes, C.; Bain, J.; Stevens, R.D.; Hauser, E.R.; Newgard, C.B.; Kraus, W.E.; Newby, L.K.; et al. Validation of the Association between a Branched Chain Amino Acid Metabolite Profile and Extremes of Coronary Artery Disease in Patients Referred for Cardiac Catheterization. Atherosclerosis 2014, 232, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Cavalu, S.; Popa, A.; Bratu, I.; Borodi, G.; Maghiar, A. New Evidences of Key Factors Involved in “Silent Stones” Etiopathogenesis and Trace Elements: Microscopic, Spectroscopic, and Biochemical Approach. Biol. Trace Elem. Res 2015, 168, 311–320. [Google Scholar] [CrossRef]
- Shah, R.V.; Truong, Q.A.; Gaggin, H.K.; Pfannkuche, J.; Hartmann, O.; Januzzi, J.L. Mid-Regional pro-Atrial Natriuretic Peptide and pro-Adrenomedullin Testing for the Diagnostic and Prognostic Evaluation of Patients with Acute Dyspnoea. Eur. Heart J. 2012, 33, 2197–2205. [Google Scholar] [CrossRef]
- Cavalu, S.; Damian, G.; Dansoreanu, M. EPR study of non-covalent spin labeled serum albumin and hemoglobin. Biophys. Chem. 2002, 99, 181–188. [Google Scholar] [CrossRef]
- WHO. Global Health Observatory (GHO) Data World Health Organization. Available online: https://www.who.int/data/gho (accessed on 10 February 2023).
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Després, J.-P.; Fullerton, H.J.; et al. Heart Disease and Stroke Statistics—2016 Update. Circulation 2016, 133, e38–e360. [Google Scholar] [CrossRef]
- Research Councils UK. King’s College London. Ancestry and Biological Informative Markers for Stratification of HYpertension: The AIM HY Study. Available online: https://gtr.ukri.org/projects?ref=MR%2FM016560%2F1 (accessed on 10 February 2023).
- Rydberg, D.M.; Mejyr, S.; Loikas, D.; Schenck-Gustafsson, K.; von Euler, M.; Malmström, R.E. Sex Differences in Spontaneous Reports on Adverse Drug Events for Common Antihypertensive Drugs. Eur. J. Clin. Pharmacol. 2018, 74, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Wang, Z.; Beitelshees, A.L.; McDonough, C.W.; Langaee, T.Y.; Hall, K.; Schmidt, S.O.F.; Curry, R.W.; Gums, J.G.; Bailey, K.R.; et al. Pharmacogenomic Genome-Wide Meta-Analysis of Blood Pressure Response to β-Blockers in Hypertensive African Americans. Hypertension 2016, 67, 556–563. [Google Scholar] [CrossRef]
- Zhao, Y.; Peng, J.; Lu, C.; Hsin, M.; Mura, M.; Wu, L.; Chu, L.; Zamel, R.; Machuca, T.; Waddell, T.; et al. Metabolomic Heterogeneity of Pulmonary Arterial Hypertension. PLoS ONE 2014, 9, e88727. [Google Scholar] [CrossRef] [PubMed]
- Brewster, L.M.; Seedat, Y.K. Why Do Hypertensive Patients of African Ancestry Respond Better to Calciumblockers and Diuretics than to ACE Inhibitors and β-Adrenergic Blockers? Asystematic Review. BMC Med. 2013, 11, 141. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Frye, R.F.; Zhu, H.; Gong, Y.; Boyle, S.; Churchill, E.; Cooper-Dehoff, R.M.; Beitelshees, A.L.; Chapman, A.B.; Fiehn, O.; et al. Pharmacometabolomics Reveals Racial Differences in Response to Atenolol Treatment. PLoS ONE 2013, 8, e57639. [Google Scholar] [CrossRef]
- Loganathan, L.; Gopinath, K.; Sankaranarayanan, V.M.; Kukreti, R.; Rajendran, K.; Lee, J.-K.; Muthusamy, K. Computational and Pharmacogenomic Insights on Hypertension Treatment: Rational Drug Design and Optimization Strategies. Curr. Drug Targets 2019, 21, 18–33. [Google Scholar] [CrossRef]
- Kronish, I.M.; Cheung, Y.K.; Shimbo, D.; Julian, J.; Gallagher, B.; Parsons, F.; Davidson, K.W. Increasing the Precision of Hypertension Treatment Through Personalized Trials: A Pilot Study. J. Gen. Intern. Med. 2019, 34, 839–845. [Google Scholar] [CrossRef]
- Hoh, B.-P.; Abdul Rahman, T.; Yusoff, K. Natural Selection and Local Adaptation of Blood Pressure Regulation and Their Perspectives on Precision Medicine in Hypertension. Hereditas 2019, 156, 1. [Google Scholar] [CrossRef] [PubMed]
- Kokubo, Y.; Padmanabhan, S.; Iwashima, Y.; Yamagishi, K.; Goto, A. Gene and Environmental Interactions According to the Components of Lifestyle Modifications in Hypertension Guidelines. Environ. Health Prev. Med. 2019, 24, 19. [Google Scholar] [CrossRef] [PubMed]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic Analysis of over 1 Million People Identifies 535 New Loci Associated with Blood Pressure Traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef]
- Mattson, D.L.; Liang, M. From GWAS to Functional Genomics-Based Precision Medicine. Nat. Rev. Nephrol. 2017, 13, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Surendran, P.; Drenos, F.; Young, R.; Warren, H.; Cook, J.P.; Manning, A.K.; Grarup, N.; Sim, X.; Barnes, D.R.; Witkowska, K.; et al. Trans-Ancestry Meta-Analyses Identify Rare and Common Variants Associated with Blood Pressure and Hypertension. Nat. Genet. 2016, 48, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W. Epigenetics of Epithelial Na + Channel-Dependent Sodium Uptake and Blood Pressure Regulation. World J. Nephrol. 2015, 4, 363. [Google Scholar] [CrossRef] [PubMed]
- Chittani, M.; Zaninello, R.; Lanzani, C.; Frau, F.; Ortu, M.F.; Salvi, E.; Fresu, G.; Citterio, L.; Braga, D.; Piras, D.A.; et al. TET2 and CSMD1 Genes Affect SBP Response to Hydrochlorothiazide in Never-Treated Essential Hypertensives. J. Hypertens 2015, 33, 1301–1309. [Google Scholar] [CrossRef]
- Trudu, M.; Janas, S.; Lanzani, C.; Debaix, H.; Schaeffer, C.; Ikehata, M.; Citterio, L.; Demaretz, S.; Trevisani, F.; Ristagno, G.; et al. Common Noncoding UMOD Gene Variants Induce Salt-Sensitive Hypertension and Kidney Damage by Increasing Uromodulin Expression. Nat. Med. 2013, 19, 1655–1660. [Google Scholar] [CrossRef]
- Muskalla, A.M.; Suter, P.M.; Saur, M.; Nowak, A.; Hersberger, M.; Krayenbuehl, P.-A. G-Protein Receptor Kinase 4 Polymorphism and Response to Antihypertensive Therapy. Clin. Chem. 2014, 60, 1543–1548. [Google Scholar] [CrossRef]
- Choi, H.D.; Suh, J.H.; Lee, J.Y.; Bae, S.K.; Kang, H.E.; Lee, M.G.; Shin, W.G. Effects of ACE and ADD1 Gene Polymorphisms on Blood Pressure Response to Hydrochlorothiazide: A Meta-Analysis. Int. J. Clin. Pharmacol. Ther. 2013, 51, 718–724. [Google Scholar] [CrossRef]
- Duarte, J.D.; Turner, S.T.; Tran, B.; Chapman, A.B.; Bailey, K.R.; Gong, Y.; Gums, J.G.; Langaee, T.Y.; Beitelshees, A.L.; Cooper-Dehoff, R.M.; et al. Association of Chromosome 12 Locus with Antihypertensive Response to Hydrochlorothiazide May Involve Differential YEATS4 Expression. Pharm. J. 2013, 13, 257–263. [Google Scholar] [CrossRef]
- Bazzell, B.G.; Rainey, W.E.; Auchus, R.J.; Zocco, D.; Bruttini, M.; Hummel, S.L.; Byrd, J.B. Human Urinary MRNA as a Biomarker of Cardiovascular Disease. Circ. Genom. Precis. Med. 2018, 11, e002213. [Google Scholar] [CrossRef]
- Arnett, D.K.; Davis, B.R.; Ford, C.E.; Boerwinkle, E.; Leiendecker-Foster, C.; Miller, M.B.; Black, H.; Eckfeldt, J.H. Pharmacogenetic Association of the Angiotensin-Converting Enzyme Insertion/Deletion Polymorphism on Blood Pressure and Cardiovascular Risk in Relation to Antihypertensive Treatment. Circulation 2005, 111, 3374–3383. [Google Scholar] [CrossRef]
- Turner, S.T.; Bailey, K.R.; Fridley, B.L.; Chapman, A.B.; Schwartz, G.L.; Chai, H.S.; Sicotte, H.; Kocher, J.-P.; Rodin, A.S.; Boerwinkle, E. Genomic Association Analysis Suggests Chromosome 12 Locus Influencing Antihypertensive Response to Thiazide Diuretic. Hypertension 2008, 52, 359–365. [Google Scholar] [CrossRef] [PubMed]
- LIU, J.; LIU, Z.; YU, B.; XU, F.; MO, W.; ZHOU, G.; LIU, Y.; LI, Q.; ZHOU, H. B1-Adrenergic Receptor Polymorphisms Influence the Response to Metoprolol Monotherapy in Patients with Essential Hypertension. Clin. Pharmacol. Ther. 2006, 80, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.K.; Tragante, V.; Guo, W.; Guo, Y.; Lanktree, M.B.; Smith, E.N.; Johnson, T.; Castillo, B.A.; Barnard, J.; Baumert, J.; et al. Loci Influencing Blood Pressure Identified Using a Cardiovascular Gene-Centric Array. Hum. Mol. Genet. 2013, 22, 1663–1678. [Google Scholar] [CrossRef] [PubMed]
- Rimpelä, J.M.; Kontula, K.K.; Fyhrquist, F.; Donner, K.M.; Tuiskula, A.M.; Sarin, A.P.; Mohney, R.P.; Stirdivant, S.M.; Hiltunen, T.P. Replicated evidence for aminoacylase 3 and nephrin gene variations to predict antihypertensive drug responses. Pharmacogenomics 2017, 18, 445–458. [Google Scholar] [CrossRef]
- Ferrandi, M.; Molinari, I.; Torielli, L.; Padoani, G.; Salardi, S.; Rastaldi, M.P.; Ferrari, P.; Bianchi, G. Adducin- and Ouabain-Related Gene Variants Predict the Antihypertensive Activity of Rostafuroxin, Part 1: Experimental Studies. Sci. Transl. Med. 2010, 2, e01815. [Google Scholar] [CrossRef]
- Horigan, G.; McNulty, H.; Ward, M.; Strain, J.; Purvis, J.; Scott, J.M. Riboflavin Lowers Blood Pressure in Cardiovascular Disease Patients Homozygous for the 677C→T Polymorphism in MTHFR. J. Hypertens. 2010, 28, 478–486. [Google Scholar] [CrossRef]
- Alderman, M. Plasma Renin Activity Levels in Hypertensive Persons: Their Wide Range and Lack of Suppression in Diabetic and in Most Elderly Patients. Am. J. Hypertens 2004, 17, 1–7. [Google Scholar] [CrossRef]
- Egan, B.M.; Basile, J.N.; Rehman, S.U.; Davis, P.B.; Grob, C.H.; Riehle, J.F.; Walters, C.A.; Lackland, D.T.; Merali, C.; Sealey, J.E.; et al. Plasma Renin Test-Guided Drug Treatment Algorithm for Correcting Patients With Treated but Uncontrolled Hypertension: A Randomized Controlled Trial. Am. J. Hypertens 2009, 22, 792–801. [Google Scholar] [CrossRef]
- Dumeny, L.; Vardeny, O.; Edelmann, F.; Pieske, B.; Duarte, J.D.; Cavallari, L.H. NR3C2 Genotype Is Associated with Response to Spironolactone in Diastolic Heart Failure Patients from the Aldo-DHF Trial. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2021, 41, 978–987. [Google Scholar] [CrossRef]
- Maurer, M.S.; Schwartz, J.H.; Gundapaneni, B.; Elliott, P.M.; Merlini, G.; Waddington-Cruz, M.; Kristen, A.V.; Grogan, M.; Witteles, R.; Damy, T.; et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N. Engl. J. Med. 2018, 379, 1007–1016. [Google Scholar] [CrossRef]
- Dominiczak, A.; Delles, C.; Padmanabhan, S. Genomics and Precision Medicine for Clinicians and Scientists in Hypertension. Hypertension 2017, 69, e08252. [Google Scholar] [CrossRef] [PubMed]
- Núñez, J.; Llàcer, P.; García-Blas, S.; Bonanad, C.; Ventura, S.; Núñez, J.M.; Sánchez, R.; Fácila, L.; de la Espriella, R.; Vaquer, J.M.; et al. CA125-Guided Diuretic Treatment Versus Usual Care in Patients With Acute Heart Failure and Renal Dysfunction. Am. J. Med. 2020, 133, 370–380.e4. [Google Scholar] [CrossRef] [PubMed]
- Pellicori, P.; Ferreira, J.P.; Mariottoni, B.; Brunner-La Rocca, H.; Ahmed, F.Z.; Verdonschot, J.; Collier, T.; Cuthbert, J.J.; Petutschnigg, J.; Mujaj, B.; et al. Effects of Spironolactone on Serum Markers of Fibrosis in People at High Risk of Developing Heart Failure: Rationale, Design and Baseline Characteristics of a Proof-of-concept, Randomised, Precision-medicine, Prevention Trial. The Heart OMics in AGing (HOMAGE) Trial. Eur. J. Heart Fail. 2020, 22, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.-D.; Jiang, Y.; Lin, Z.-H.; Lin, P.-H.; Lin, S.-M.; Liu, Q.-C. Diagnostic Value of Brain Natriuretic Peptide and β-Endorphin Plasma Concentration Changes in Patients with Acute Left Heart Failure and Atrial Fibrillation. Medicine 2017, 96, e7526. [Google Scholar] [CrossRef]
- Drum, C.L.; Tan, W.K.Y.; Chan, S.; Pakkiri, L.S.; Chong, J.P.C.; Liew, O.; Ng, T.; Ling, L.; Sim, D.; Leong, K.G.; et al. Thymosin Beta-4 Is Elevated in Women With Heart Failure With Preserved Ejection Fraction. J. Am. Heart Assoc. 2017, 6, 05586. [Google Scholar] [CrossRef]
- Felker, G.M.; Mentz, R.J.; Teerlink, J.R.; Voors, A.A.; Pang, P.S.; Ponikowski, P.; Greenberg, B.H.; Filippatos, G.; Davison, B.A.; Cotter, G.; et al. Serial High Sensitivity Cardiac Troponin T Measurement in Acute Heart Failure: Insights from the RELAX-AHF Study. Eur. J. Heart Fail. 2015, 17, 1262–1270. [Google Scholar] [CrossRef]
- Pozsonyi, Z.; Förhécz, Z.; Gombos, T.; Karádi, I.; Jánoskuti, L.; Prohászka, Z. Copeptin (C-Terminal pro Arginine-Vasopressin) Is an Independent Long-Term Prognostic Marker in Heart Failure with Reduced Ejection Fraction. Heart Lung Circ. 2015, 24, 359–367. [Google Scholar] [CrossRef]
- Glick, D.; deFilippi, C.R.; Christenson, R.; Gottdiener, J.S.; Seliger, S.L. Long-Term Trajectory of Two Unique Cardiac Biomarkers and Subsequent Left Ventricular Structural Pathology and Risk of Incident Heart Failure in Community-Dwelling Older Adults at Low Baseline Risk. JACC Heart Fail. 2013, 1, 353–360. [Google Scholar] [CrossRef]
- Gaggin, H.K.; Motiwala, S.; Bhardwaj, A.; Parks, K.A.; Januzzi, J.L. Soluble Concentrations of the Interleukin Receptor Family Member ST2 and β-Blocker Therapy in Chronic Heart Failure. Circ. Heart Fail. 2013, 6, 1206–1213. [Google Scholar] [CrossRef]
- Lok, S.I.; Winkens, B.; Goldschmeding, R.; van Geffen, A.J.P.; Nous, F.M.A.; van Kuik, J.; van der Weide, P.; Klöpping, C.; Kirkels, J.H.; Lahpor, J.R.; et al. Circulating Growth Differentiation Factor-15 Correlates with Myocardial Fibrosis in Patients with Non-Ischaemic Dilated Cardiomyopathy and Decreases Rapidly after Left Ventricular Assist Device Support. Eur. J. Heart Fail. 2012, 14, 1249–1256. [Google Scholar] [CrossRef]
- Lopez-Andrès, N.; Rossignol, P.; Iraqi, W.; Fay, R.; Nuée, J.; Ghio, S.; Cleland, J.G.F.; Zannad, F.; Lacolley, P. Association of Galectin-3 and Fibrosis Markers with Long-Term Cardiovascular Outcomes in Patients with Heart Failure, Left Ventricular Dysfunction, and Dyssynchrony: Insights from the CARE-HF (Cardiac Resynchronization in Heart Failure) Trial. Eur. J. Heart Fail. 2012, 14, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Olivotto, I.; Oreziak, A.; Barriales-Villa, R.; Abraham, T.P.; Masri, A.; Garcia-Pavia, P.; Saberi, S.; Lakdawala, N.K.; Wheeler, M.T.; Owens, A.; et al. Mavacamten for Treatment of Symptomatic Obstructive Hypertrophic Cardiomyopathy (EXPLORER-HCM): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2020, 396, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.G.; Kelly, J.P.; McGarrah, R.W.; Khouri, M.G.; Craig, D.; Haynes, C.; Ilkayeva, O.; Stevens, R.D.; Bain, J.R.; Muehlbauer, M.J.; et al. Metabolomic Profiling Identifies Novel Circulating Biomarkers of Mitochondrial Dysfunction Differentially Elevated in Heart Failure With Preserved Versus Reduced Ejection Fraction: Evidence for Shared Metabolic Impairments in Clinical Heart Failure. J. Am. Heart Assoc. 2016, 5, e03190. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Shen, A.; Huang, Y.; Su, L.; Lai, W.; Wang, P.; Xie, Z.; Xie, Z.; Zeng, Q.; Ren, H.; et al. 1H-NMR-Based Metabolic Analysis of Human Serum Reveals Novel Markers of Myocardial Energy Expenditure in Heart Failure Patients. PLoS ONE 2014, 9, e88102. [Google Scholar] [CrossRef]
- Wang, J.; Li, Z.; Chen, J.; Zhao, H.; Luo, L.; Chen, C.; Xu, X.; Zhang, W.; Gao, K.; Li, B.; et al. Metabolomic Identification of Diagnostic Plasma Biomarkers in Humans with Chronic Heart Failure. Mol. Biosyst. 2013, 9, 2618. [Google Scholar] [CrossRef]
- Desmoulin, F.; Galinier, M.; Trouillet, C.; Berry, M.; Delmas, C.; Turkieh, A.; Massabuau, P.; Taegtmeyer, H.; Smih, F.; Rouet, P. Metabonomics Analysis of Plasma Reveals the Lactate to Cholesterol Ratio as an Independent Prognostic Factor of Short-Term Mortality in Acute Heart Failure. PLoS ONE 2013, 8, e60737. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Wang, Z.; Fan, Y.; Levison, B.; Hazen, J.E.; Donahue, L.M.; Wu, Y.; Hazen, S.L. Prognostic Value of Elevated Levels of Intestinal Microbe-Generated Metabolite Trimethylamine-N-Oxide in Patients With Heart Failure. J. Am. Coll. Cardiol. 2014, 64, 1908–1914. [Google Scholar] [CrossRef]
- Domínguez, F.; Cuenca, S.; Bilińska, Z.; Toro, R.; Villard, E.; Barriales-Villa, R.; Ochoa, J.P.; Asselbergs, F.; Sammani, A.; Franaszczyk, M.; et al. Dilated Cardiomyopathy Due to BLC2-Associated Athanogene 3 (BAG3) Mutations. J. Am. Coll. Cardiol. 2018, 72, 2471–2481. [Google Scholar] [CrossRef]
- Shah, S.; Henry, A.; Roselli, C.; Lin, H.; Sveinbjörnsson, G.; Fatemifar, G.; Hedman, Å.K.; Wilk, J.B.; Morley, M.P.; Chaffin, M.D.; et al. Genome-Wide Association and Mendelian Randomisation Analysis Provide Insights into the Pathogenesis of Heart Failure. Nat. Commun. 2020, 11, 163. [Google Scholar] [CrossRef]
- Albert, C.L.; Tang, W.H.W. Metabolic Biomarkers in Heart Failure. Heart Fail. Clin. 2018, 14, 109–118. [Google Scholar] [CrossRef]
- Ahmad, T.; Voora, D.; Becker, R.C. The Pharmacogenetics of Antiplatelet Agents: Towards Personalized Therapy? Nat. Rev. Cardiol. 2011, 8, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Biddinger, A.; Rocklin, M.; Coselli, J.; Milewicz, D.M. Familial Thoracic Aortic Dilatations and Dissections: A Case Control Study. J. Vasc. Surg. 1997, 25, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Brownstein, A.; Kostiuk, V.; Ziganshin, B.; Zafar, M.; Kuivaniemi, H.; Body, S.; Bale, A.; Elefteriades, J. Genes Associated with Thoracic Aortic Aneurysm and Dissection: 2018 Update and Clinical Implications. AORTA 2018, 06, 013–020. [Google Scholar] [CrossRef] [PubMed]
- Renard, M.; Francis, C.; Ghosh, R.; Scott, A.F.; Witmer, P.D.; Adès, L.C.; Andelfinger, G.U.; Arnaud, P.; Boileau, C.; Callewaert, B.L.; et al. Clinical Validity of Genes for Heritable Thoracic Aortic Aneurysm and Dissection. J. Am. Coll. Cardiol. 2018, 72, 605–615. [Google Scholar] [CrossRef]
- Wolford, B.N.; Hornsby, W.E.; Guo, D.; Zhou, W.; Lin, M.; Farhat, L.; McNamara, J.; Driscoll, A.; Wu, X.; Schmidt, E.M.; et al. Clinical Implications of Identifying Pathogenic Variants in Individuals With Thoracic Aortic Dissection. Circ. Genom. Precis. Med. 2019, 12, e02476. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Sethi, Y.; Patel, N.; Kaka, N.; Desai, A.; Kaiwan, O.; Sheth, M.; Sharma, R.; Huang, H.; Chopra, H.; Khandaker, M.U.; et al. Artificial Intelligence in Pediatric Cardiology: A Scoping Review. J. Clin. Med. 2022, 11, 7072. [Google Scholar] [CrossRef]
- de Marvao, A.; Dawes, T.J.; Howard, J.P.; O’Regan, D.P. Artificial Intelligence and the Cardiologist: What You Need to Know for 2020. Heart 2020, 106, 399–400. [Google Scholar] [CrossRef]
- Johnson, K.W.; Torres Soto, J.; Glicksberg, B.S.; Shameer, K.; Miotto, R.; Ali, M.; Ashley, E.; Dudley, J.T. Artificial Intelligence in Cardiology. J. Am. Coll Cardiol. 2018, 71, 2668–2679. [Google Scholar] [CrossRef]
- Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. N. Engl. J. Med. 2009, 360, 753–764. [CrossRef]
- Jorgensen, A.L.; FitzGerald, R.J.; Oyee, J.; Pirmohamed, M.; Williamson, P.R. Influence of CYP2C9 and VKORC1 on Patient Response to Warfarin: A Systematic Review and Meta-Analysis. PLoS ONE 2012, 7, e44064. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M.; Johnson, J.A.; Langaee, T.Y.; Feng, H.; Stanaway, I.B.; Schwarz, U.I.; Ritchie, M.D.; Stein, C.M.; Roden, D.M.; Smith, J.D.; et al. A Genome-Wide Scan for Common Genetic Variants with a Large Influence on Warfarin Maintenance Dose. Blood 2008, 112, 1022–1027. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, F.; McGinnis, R.; Bourgeois, S.; Barnes, C.; Eriksson, N.; Soranzo, N.; Whittaker, P.; Ranganath, V.; Kumanduri, V.; McLaren, W.; et al. A Genome-Wide Association Study Confirms VKORC1, CYP2C9, and CYP4F2 as Principal Genetic Determinants of Warfarin Dose. PLoS Genet. 2009, 5, e1000433. [Google Scholar] [CrossRef] [PubMed]
- Oqueli, E.; Hiscock, M.; Dick, R. Clopidogrel Resistance. Heart Lung Circ. 2007, 16, S17–S28. [Google Scholar] [CrossRef]
- Collet, J.-P.; Hulot, J.-S.; Pena, A.; Villard, E.; Esteve, J.-B.; Silvain, J.; Payot, L.; Brugier, D.; Cayla, G.; Beygui, F.; et al. Cytochrome P450 2C19 Polymorphism in Young Patients Treated with Clopidogrel after Myocardial Infarction: A Cohort Study. Lancet 2009, 373, 309–317. [Google Scholar] [CrossRef]
- Mega, J.L.; Hochholzer, W.; Frelinger, A.L.; Kluk, M.J.; Angiolillo, D.J.; Kereiakes, D.J.; Isserman, S.; Rogers, W.J.; Ruff, C.T.; Contant, C.; et al. Dosing Clopidogrel Based on CYP2C19 Genotype and the Effect on Platelet Reactivity in Patients With Stable Cardiovascular Disease. JAMA 2011, 306, 2221–2228. [Google Scholar] [CrossRef]
- Kimmel, S.E.; French, B.; Kasner, S.E.; Johnson, J.A.; Anderson, J.L.; Gage, B.F.; Rosenberg, Y.D.; Eby, C.S.; Madigan, R.A.; McBane, R.B.; et al. A Pharmacogenetic versus a Clinical Algorithm for Warfarin Dosing. N. Engl. J. Med. 2013, 369, 2283–2293. [Google Scholar] [CrossRef]
- Pirmohamed, M.; Burnside, G.; Eriksson, N.; Jorgensen, A.L.; Toh, C.H.; Nicholson, T.; Kesteven, P.; Christersson, C.; Wahlström, B.; Stafberg, C.; et al. A Randomized Trial of Genotype-Guided Dosing of Warfarin. N. Engl. J. Med. 2013, 369, 2294–2303. [Google Scholar] [CrossRef]
- Garmaroudi, F.S.; Handy, D.E.; Liu, Y.-Y.; Loscalzo, J. Systems Pharmacology and Rational Polypharmacy: Nitric Oxide−Cyclic GMP Signaling Pathway as an Illustrative Example and Derivation of the General Case. PLoS Comput. Biol. 2016, 12, e1004822. [Google Scholar] [CrossRef]
- Lau, E.; Wu, J.C. Omics, Big Data, and Precision Medicine in Cardiovascular Sciences. Circ. Res. 2018, 122, 1165–1168. [Google Scholar] [CrossRef]
- Sohag, M.M.H.; Raqib, S.M.; Akhmad, S.A. OMICS Approaches in Cardiovascular Diseases: A Mini Review. Genom. Inform. 2021, 19, e13. [Google Scholar] [CrossRef] [PubMed]
- Frostegård, J. Immunity, Atherosclerosis and Cardiovascular Disease. BMC Med. 2013, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Rurik, J.G.; Aghajanian, H.; Epstein, J.A. Immune Cells and Immunotherapy for Cardiac Injury and Repair. Circ. Res. 2021, 128, 1766–1779. [Google Scholar] [CrossRef]
- Lu, Y.; Xia, N.; Cheng, X. Regulatory T Cells in Chronic Heart Failure. Front. Immunol. 2021, 12, 732794. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Rosenzweig, R.; Asalla, S.; Nehra, S.; Prabhu, S.D.; Bansal, S.S. TNFR1 Contributes to Activation-Induced Cell Death of Pathological CD4+ T Lymphocytes During Ischemic Heart Failure. JACC Basic Transl. Sci. 2022, 7, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Zhang, H.; Xu, T.; Hao, J.; Chen, X.; Sun, S.; Yang, J.; Sun, J.; Lin, H.; Guo, H. Identification and Verification of Immune-Related Biomarkers and Immune Infiltration in Diabetic Heart Failure. Front. Cardiovasc. Med. 2022, 9, 931066. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Prabhu, S.D.; Bansal, S.S. CD4+ T-Lymphocytes Exhibit Biphasic Kinetics Post-Myocardial Infarction. Front. Cardiovasc. Med. 2022, 9, 992653. [Google Scholar] [CrossRef]
- Hayward, S.L.; Bautista-Lopez, N.; Suzuki, K.; Atrazhev, A.; Dickie, P.; Elliott, J.F. CD4 T Cells Play Major Effector Role and CD8 T Cells Initiating Role in Spontaneous Autoimmune Myocarditis of HLA-DQ8 Transgenic IAb Knockout Nonobese Diabetic Mice. J. Immunol. 2006, 176, 7715–7725. [Google Scholar] [CrossRef]
- Fuster, V. A Second Dilemma in Cardiovascular Medicine. J. Am. Coll Cardiol. 2014, 64, 1292–1293. [Google Scholar] [CrossRef]
- National Human Genome Research Institute. Inter-Society Coordinating Committee for Practitioner Education in Genomics (ISCC). Available online: https://www.genome.gov/For-Health-Professionals/Inter-Society-Coordinating-Committee-for-Practitioner-Education-in-Genomics (accessed on 10 February 2023).
- Korf, B.R.; Berry, A.B.; Limson, M.; Marian, A.J.; Murray, M.F.; O’Rourke, P.P.; Passamani, E.R.; Relling, M.v.; Tooker, J.; Tsongalis, G.J.; et al. Framework for Development of Physician Competencies in Genomic Medicine: Report of the Competencies Working Group of the Inter-Society Coordinating Committee for Physician Education in Genomics. Genet. Med. 2014, 16, 804–809. [Google Scholar] [CrossRef]
- Dickson, D.; Pfeifer, J. Real-World Data in the Molecular Era-Finding the Reality in the Real World. Clin. Pharmacol. Ther. 2016, 99, 186–197. [Google Scholar] [CrossRef] [PubMed]



| S.N. | Authors | Year | Results | Clinical Significance | References |
|---|---|---|---|---|---|
| 1 | Guglielmo Gallone et al. | 2022 | The research focuses on how AI may bridge the gap between data-rich technologies and their deployment. | AI has the potential to use data-rich technologies in patient care, cardiovascular research, and health policy research. | [12] |
| 2 | Michael Simeon et al. | 2021 | It will be possible to use human pluripotent stem cells (hPSCs) in cardiovascular clinical care by developing isogenic hPSC cell lines as a control for hPSCs with disease-specific mutations and a large number of hPSC lines with gene mutations, for the in vitro modeling of human diseases with complex genotypes and phenotypes. | hPSCs with disease-specific mutations have application for use in cardiovascular clinical care. | [13] |
| 3 | Soni Savai Pullamsetti et al. | 2021 | Novel biomarkers can distinguish between left and right ventricular hypertrophy/failure. | Novel biomarkers can distinguish left ventricular hypertrophy/failure from right ventricular hypertrophy/failure, assess right ventricular disease severity, and potentially identify maladaptive changes in RV size, function, and architecture. | [14] |
| 4 | Farwah Iqbal et al. | 2021 | In both healthy and pathological cardiovascular tissues, scRNA-seq has enabled the characterization of heterogeneous cell subpopulations with distinct genetic profiles. | These can shed light on the pathological mechanisms underlying atherosclerosis and suggest new potential treatments for calcific aortic valve disease. | [15] |
| 5 | Concetta Schiano et al. | 2021 | Numerous ncRNAs, including miR-93, miR-340, miR-433, miR-765, CHROME, and large epigenetic changes in DNA methylation have been linked to atherogenesis in endothelial, smooth muscle, and macrophage cells. | In pro-inflammatory macrophages of the human carotid plaque, elevated HDAC9 was related to matrix metalloproteinase 1 (MMP1) and MMP2 production, while decreased HDAC9 was seen to promote resolution of inflammation and reverse cholesterol transfer, which may halt or reverse the disease process. | [16] |
| 6 | Christian Schulte et al. | 2020 | Emerging biomarkers—cardiac myosin binding protein C, SNPs, and non-coding RNAs. | Newer biomarkers, adding more specificity to the diagnosis. | [17] |
| 7 | Damien Gruson et al. | 2020 | In cardiovascular medicine, AI has shown promise as a tool to improve patient care and increase the efficiency of cardiologists. | The integration of AI and laboratory medicine can enable personalized care in cardiovascular medicine. | [18] |
| 8 | Valeria Visco et al. | New technologies in heart failure, atrial fibrillation, and cardiac rehabilitation offer a low-cost, non-invasive solution to long-term monitoring and management. | The availability of current devices with massive datasets offers a valuable tool for predicting the progression and outcome of various cardiovascular illnesses. Due to continuous monitoring, this new guided therapy can provide rapid and tailored treatment while also having a substantial psychological impact on patients. | [19] | |
| 9 | Samuele Ambrosini et al. | 2020 | Building personalized maps of cardiovascular risk and designing individualized diagnostic and treatment approaches can be made possible by the integration of epigenetic and genetic data. | Genomics—customized diagnostic and therapeutic strategies. | [20] |
| 10 | Ziwei Pan et al. | 2020 | Induced pluripotent stem cells (iPSC)—functional evaluation of genes, patient risk stratification, testing of drugs, and personalized medicine. | With the help of this overview, readers should be better able to comprehend the value of iPSC-CM models, their various features, and their potential. | [21] |
| 11 | Danielle Menosi Gualandro et al. | 2019 | 1. The interpretation of Hs-cTnT/I concentrations should always be performed quantitatively, and not in a binary fashion, as with pregnancy tests; 2. New hs-cTnT/I elevations show that “false elevations” from analytical issues are less likely than the “true elevations” due to an implicit or underestimated cardiac disorder such as heart failure causing myocardial injury. | The interpretation of Hs-cTnT/I concentrations should always be carried out within the context of the clinical presentation of the patient, using all other clinical information available and not in isolation. | [22] |
| 12 | Javier Rodríguez-Carrio et al. | 2018 | The CXCR4 pathway was associated with the effect of aging on Endothelial Progenitor Cells, whereas TNFα was found to be associated with hypertension. The response to dyslipidemia and diabetes-related traits was observed to be the activation of Akt/eNOS. The EPC dysfunction at the time of smoking is due to inflammation and oxidative stress. | Feasible biomarkers for risk stratification in personalized medicine schemes are suggested to be inflammatory and immune networks. | [23] |
| 13 | J. David Spence et al. | 2018 | Phenotype-based approach for HTN: 1. Regarding the Liddle phenotype (low renin/low aldosterone due to ENaC overactivity)—the specific therapy is amiloride; 2. In terms of the phenotype in primary aldosteronism—aldosterone antagonists; adrenalectomy; In terms of the renal phenotype—the renin/angiotensin system antagonists; renal revascularization. | Therapeutic approaches for hypertension can be best chosen based on specific phenotypes. The Liddle phenotype is more likely to be present in black hypertensives. They also tend to retain salt and water. | [24] |
| 14 | Perry V. Halushka et al. | 2018 | Evolving role of miRNA biomarkers as a diagnostic tool in diseases such as coronary artery vasculopathy, diabetic cardiomyopathy, aortic stenosis, and atrial fibrillation. | miRNA-based biomarkers can help as tools for specific early diagnosis. | [25] |
| 15 | Gemma Currie et al. | 2018 | It is suggested that the following steps should allow the translation of precision medicine into practical cardiology: (1) Make use of existing data; (2) Improve diagnostic tests; (3) Standardize phenotypes; (4) Techniques for the standardization and simplification of omics to support large-scale epidemiological studies; (5) Change the mindset in cardiovascular medicine toward molecular diagnostics; (6) Conduct stratified clinical trials. | Sex/gender have not yet been completely studied in precision medicine; nonetheless, the prospect of using molecular data to better properly manage men and women with cardiovascular disorders has been recognized. | [10] |
| 16 | M Zaiou et al. | 2018 | Genetic defects may be included in clinical practice since they may affect how common cardiovascular medicines such as aspirin, clopidogrel, warfarin, and statins interact with different individuals and influence how CVDs develop. | Knowledge of genomics can help the process of individualizing therapy. | [26] |
| 17 | Calum A. MacRae et al. | 2016 | The mainstay of modern cardiovascular therapeutics is small molecules such as antibodies against ANGPTL4 or Statins and PCSK9 inhibitors for lipid lowering. Genome-wide association studies (GWAS) have been successful in defining new loci leading to common diseases. | The potential for precision medicine-based therapeutic approach to cardiovascular disease care is immense, but so are the challenges for the same. | [27] |
| 18 | Toshio Nishikimi et al. | 2013 | Adrenomedullin (AM)—a potent vasodilatory peptide. It can also act as an autocrine and/or paracrine factor. Increased adrenomedullin is associated with the defense mechanism against further elevation of peripheral vascular resistance in cases of heart failure. | AM can be used in cardiovascular diseases for diagnosis and treatment. | [28] |
| Clinical Applicability |
|---|
| 1. Precision disease stratification |
| 2. AI-aided diagnostics—probable diagnosis and risk stratification from imaging and investigations |
| 3. Continuous remote monitoring and wearable devices |
| 4. Telemedicine and remote diagnosis |
| 5. Extension of Physician efficiency and efficacy |
| 6. Integration of multi-omic data |
| 7. Therapy selection |
| 8. Database integration and reporting |
| 9. Healthcare research and analysis |
| 10. Patient education and information |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sethi, Y.; Patel, N.; Kaka, N.; Kaiwan, O.; Kar, J.; Moinuddin, A.; Goel, A.; Chopra, H.; Cavalu, S. Precision Medicine and the future of Cardiovascular Diseases: A Clinically Oriented Comprehensive Review. J. Clin. Med. 2023, 12, 1799. https://doi.org/10.3390/jcm12051799
Sethi Y, Patel N, Kaka N, Kaiwan O, Kar J, Moinuddin A, Goel A, Chopra H, Cavalu S. Precision Medicine and the future of Cardiovascular Diseases: A Clinically Oriented Comprehensive Review. Journal of Clinical Medicine. 2023; 12(5):1799. https://doi.org/10.3390/jcm12051799
Chicago/Turabian StyleSethi, Yashendra, Neil Patel, Nirja Kaka, Oroshay Kaiwan, Jill Kar, Arsalan Moinuddin, Ashish Goel, Hitesh Chopra, and Simona Cavalu. 2023. "Precision Medicine and the future of Cardiovascular Diseases: A Clinically Oriented Comprehensive Review" Journal of Clinical Medicine 12, no. 5: 1799. https://doi.org/10.3390/jcm12051799
APA StyleSethi, Y., Patel, N., Kaka, N., Kaiwan, O., Kar, J., Moinuddin, A., Goel, A., Chopra, H., & Cavalu, S. (2023). Precision Medicine and the future of Cardiovascular Diseases: A Clinically Oriented Comprehensive Review. Journal of Clinical Medicine, 12(5), 1799. https://doi.org/10.3390/jcm12051799